Introduction
Hepatocellular carcinoma (HCC) is one of the most
common types of cancer to result in mortality, worldwide. It has
been acknowledged that the primary carcinogenic factors of HCC
include hepatitis B virus (HBV) and hepatitis C virus (HCV)
infection, alcohol and non-alcoholic steatohepatitis (1). Of these principal etiologies, HBV and
HCV predominate in Asian cohorts, whereas European cohorts are
associated with heterogeneous etiologies, including alcohol
consumption, and viral hepatitis within cirrhotic and non-cirrhotic
livers (2). The analysis of patients
with chronic HCV infection, in addition to alcohol abuse and
dependence, indicated that elevated proton pump activity increased
the risk of hepatic decompensation and hepatocellular carcinoma
(3). According to the study of
non-alcoholic fatty liver disease (NAFLD) in Indian patients, 11
patients with a history of considerable alcohol consumption had at
least one risk factor for NAFLD, including alcohol-associated HCC
(4). Asian countries have been
reported to have a high incidence of HBV infection. A previous
study of the Indian population demonstrated a lower prevalence of
HBV infection of unknown origin (4);
however, the direct role of alcohol in the carcinogenesis of HCC,
or the mechanism underlying alcohol-induced hepatotoxicity
associated with alcohol metabolites, oxidative stress and iron
metabolism, are yet to be elucidated (2,5). The
administration of alcohol/diethyl nitrosamine/carbon tetrachloride
in mice successfully induced an α-fetoprotein-secreting HCC model
in adult male BALB/c mice. This research may improve current
knowledge of the stages of occurrence and development associated
with HCC (5). The underlying
mechanism of the alcohol-induced development of liver disease
remains unknown. Recently, the epidemiological evidence regarding
the association of alcohol and liver cancer was summarized; this
publication demonstrated that it is crucial to identify all aspects
associated with the effects of alcohol metabolism on alterations in
hepatocyte metabolism, particularly neoplasia-associated
alterations (6).
Microarray data analysis of the expression of
different genes has provided powerful and feasible evidence for the
clinical diagnosis and treatment of HCC. In the present study, the
GSE50579 dataset was downloaded from the Gene Expression Omnibus
database (GEO; http://www.ncbi.nlm.nih.gov/geo), which includes data
regarding alcohol-associated HCC and HCC liver tissue samples; the
differentially expressed genes (DEGs) were screened, and
bioinformatics analysis was conducted for functional and pathway
enrichment, and for the investigation of protein-protein
interaction (PPI) networks. Statistical analysis and functional
annotation revealed that actin γ1 (ACTG1; DFN20/26) and Toll-like
receptor 3 (TLR3) may be biomarkers for alcohol-induced HCC.
Notably, the present study reported that DEG-associated microRNAs
(miRNAs) may be considered as potential targets in the treatment of
alcohol-induced HCC. In addition, these biomarkers were assessed by
reverse-transcription-quantitative polymerase chain reaction
(RT-qPCR) in samples collected from patients with HCC.
Materials and methods
Ethics statement
The present study was conducted with the approval of
the Ethics Committee of the Affiliated Hospital of Qingdao
University (Qingdao, China). Patients provided informed consent for
the use of their tissue specimens. The mean age of 30 patients,
including 18 males and 12 females, was 52.6±16.8 years. A total of
30 clinical tissue samples, including 12 from patients with
alcohol-associated HCC, 12 from patients with
non-alcohol-associated HCC tissues, and six from healthy
individuals were obtained from the Department of Hepatobiliary
Surgery of the Affiliated Hospital of Qingdao University between
March 2017 and July 2017.
Identification of differentially
expressed genes
The microarray data set GSE50579 (7) was downloaded from the GEO, in which 10
alcohol-associated HCC and 16 clinical HCC liver samples were
analyzed. The platform employed for analysis was the Agilent-028004
SurePrint G3 Human GE 8×60K Microarray (Agilent Technologies, Inc.,
Santa Clara, CA, USA). The annotation file was also acquired; the
original CEL format was converted into an expression matrix using a
function to study RNA with an Affy package (8). Probes were mapped to genes according to
the annotation file in R version 3.3.3 (9). Average expression levels were calculated
for the probes corresponding to the same gene. DEGs between
alcohol-associated HCC tissue samples and non-alcohol associated
individuals were removed via the goodness of fit test; genes that
were not differentially expressed were subject to average
distribution. The R limma package (9)
was adopted for differential analysis. |logFC| (fold change)
>1.0 and P<0.05 were set as the criteria to screen out DEGs
between alcohol-associated HCC liver tissue samples and non-alcohol
associated HCC samples.
Bioinformatics analysis of DEGs
Gene Ontology (GO) enrichment and Kyoto Encyclopedia
of Genes and Genomes (KEGG) pathway enrichment analyses (https://www.genome.jp/kegg) were performed for the
DEGs using the Cluego APP in Cytoscape (version 3.5.1; www.cytoscape.org). A previous study (10) demonstrated that certain genes sharing
the same pathway or similar biological functions and gene
expression patterns exhibit the same pathological effects.
Therefore, the construction of a gene co-expression network may
help identify gene sets associated with specific pathways and
biological processes. In the present study, Pearson correlation
analysis of co-expression was performed with a coefficient >0.85
and P<0.05 as the criteria for investigation. In addition, PPIs
were constructed using Cytoscape software. Hub genes were
identified within the PPIs of DEGs, and the edge length between
nodes from these hub genes was determined.
miRNA analysis of the DEGs
The present study investigated numerous genes,
including, centrin 3 (CETN3), TLR3, erbB2 receptor tyrosine kinase
4 (ERBB4), heat shock protein family member 8 (HSPA8), ACTG1 and
α-smooth muscle actin (ACTA2) on the TargetScan Human 7.1 website
(www.targetscan.org/vert_71) to
predict the associated miRNAs of these genes. A diagram of the
miRNAs and their predicted target genes was produced using Fun Rich
software (http://funrich.org; version 3.0). Scores
≥90 points and overlapping miRNA quantity ≥3 were set to obtain the
desired miRNAs and target DEGs.
RNA extraction
Tissues were stored at −80°C until use. Total RNA
was extracted using TRIzol® (Invitrogen; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) according to the manufacturers
instructions. The RNA solutions were stored at −80°C. The RNA
quality was determined by spectrophotometer.
RT-qPCR
RNA (1 µg) was reverse transcribed into cDNA using
FastQuant RT kit (with gDNase) (Tiangen Biotech Co., Ltd., Beijing,
China), according to the manufacturers protocol. cDNA was amplified
using SYBR Green Real Master mix (Tiangen Biotech Co., Ltd.),
according to the manufacturers protocol. The PCR reaction was
conducted at 95°C for 15 min, followed by 40 cycles at 95°C for 10
sec, 58°C for 20 sec and 72°C for 30 sec. RT-qPCR was performed on
the ABI StepOnePlus™ thermocycler (Thermo Fisher
Scientific, Inc.). U6 single nuclear (sn)RNA was employed as the
loading control for micro (mi)RNA expression, and β-actin for the
expression of genes; stem-loop RT primers were used for miRNAs, and
oligo (dinucleotide) primers for genes. Primers are displayed in
Table I. All reactions were repeated
three times and the data were calculated by the comparative
2−ΔΔCq method (11).
 | Table I.Primers for differentially-expressed
genes and their associated miRNAs. |
Table I.
Primers for differentially-expressed
genes and their associated miRNAs.
| Gene/miRNA
name | Forward primer
(5–3) | Reverse primer
(5–3) | Stem-loop RT primer
(5–3) |
|---|
| ACTG1 |
ATGGAAGGAAACACGGCTC |
CACTCTGTTCTTCCGCCG | – |
| TLR3 |
AGTGCACTTGGTGGTGGAG |
AGGAAAGGCTAGCAGTCATCC | – |
| β-actin |
TGACGGGGTCACCCACACTG |
AAGCTGTAGCCGCGCTCGGT | – |
| hsa-miR-6819 |
AAGCCTCTGTCCCCA |
CAGTGCGTGTCGTGGAGT |
GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGAC |
| hsa-miR-6877 |
CAGCCTCTGCCCTTG |
CAGTGCGTGTCGTGGAGT |
GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGAC |
| U6 |
CTCGCTTCGGCAGCACA |
AACGCTTCACGAATTTGCGT |
AACGCTTCACGAATTTGCGT |
Statistical analysis
All statistical analyses were performed using R
software version 3.3 and GraphPad Prism 6 software (GraphPad
Software, Inc. La Jolla, CA, USA). The data are presented as the
mean ± standard deviation. Statistical significance was examined
using Students t-test or one-way analysis of variance followed by
Dunnetts test. P<0.05 was considered to indicate a statistically
significance difference. To identify the prognostic value of the
differentially expressed coding genes, Kaplan-Meier survival
analysis was conducted.
Results
Identification of DEGs
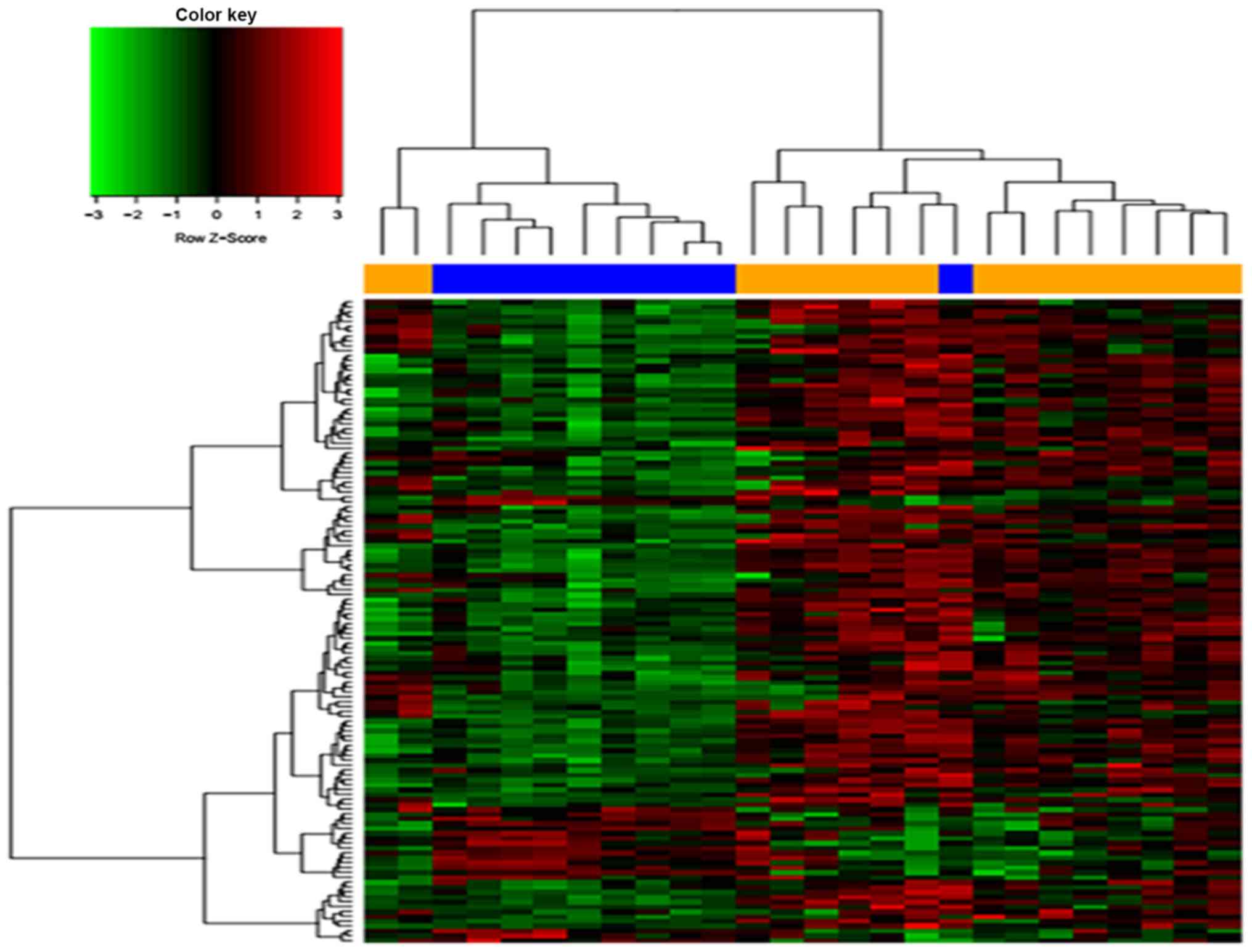
A total of 284 DEGs from the GSE50579 dataset were
obtained, which included 68 upregulated and 216 downregulated DEGs.
The top DEGs were presented as a heat map (Fig. 1).
Bioinformatics analysis of the
DEGs
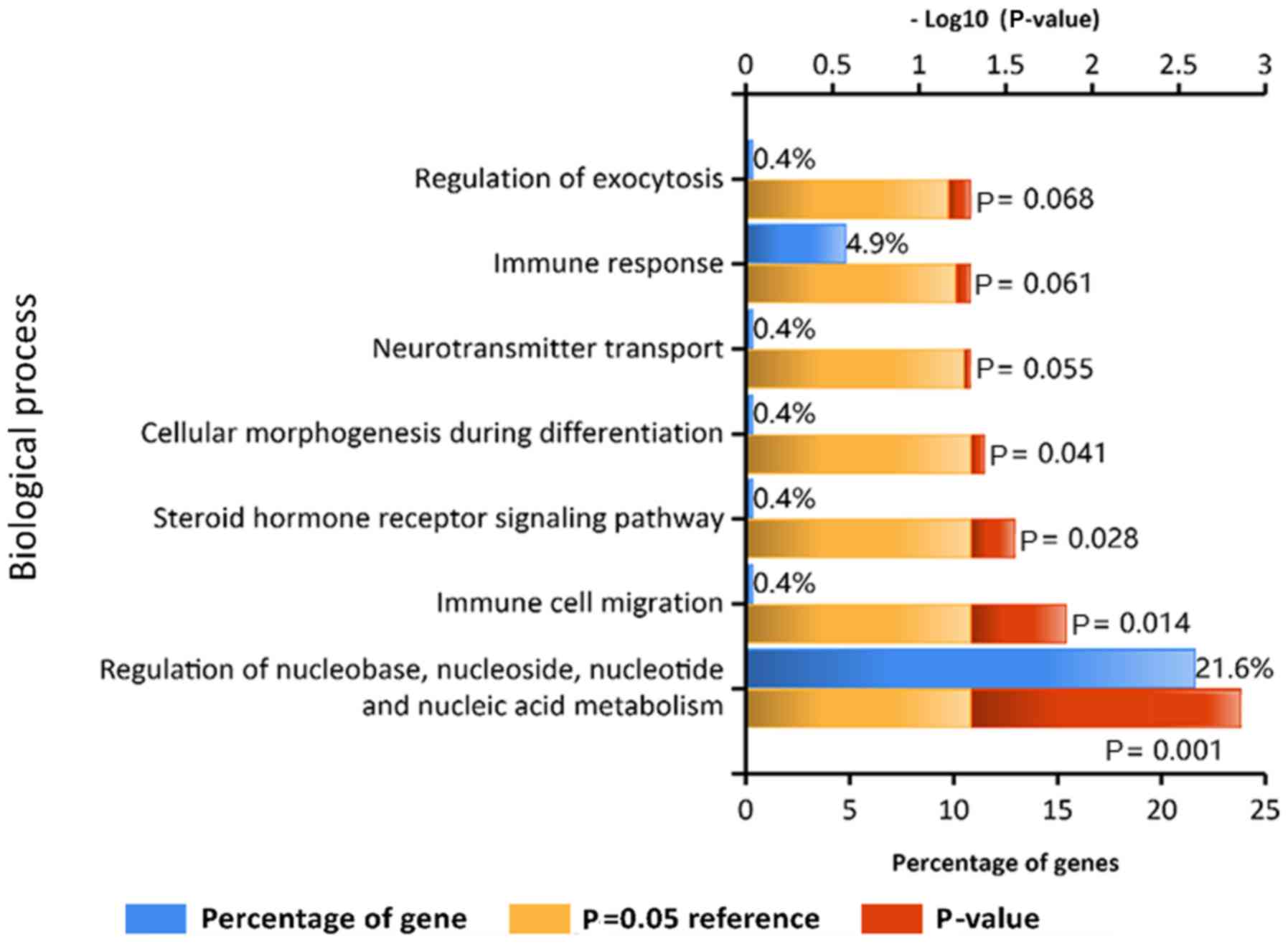
To further understand the functions of the DEGs, GO
and KEGG pathway analyses were conducted. The GO analysis results
indicated that the DEGs were involved in ‘immune response’ and
‘regulation of nucleobase, nucleoside, nucleotide and nucleic acid
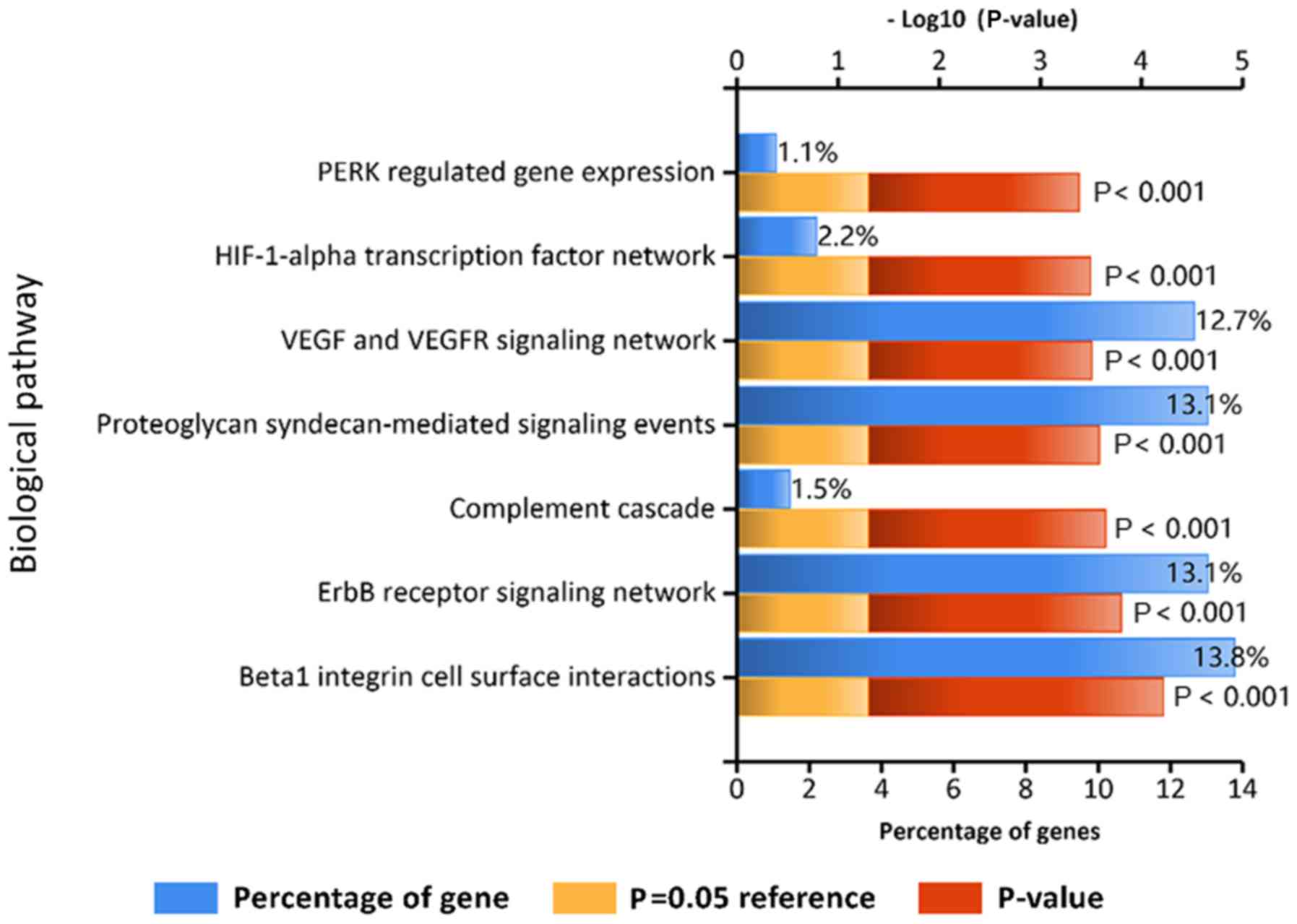
metabolism’ (Fig. 2). In addition,
KEGG pathway analysis revealed that the DEGs were closely
associated with ‘VEGF and VEGFR signaling network’, ‘proteoglycan
syndecan-mediated signaling events’, ‘erbB receptor signaling
network’ and ‘β1 integrin cell surface interactions’ (Fig. 3).
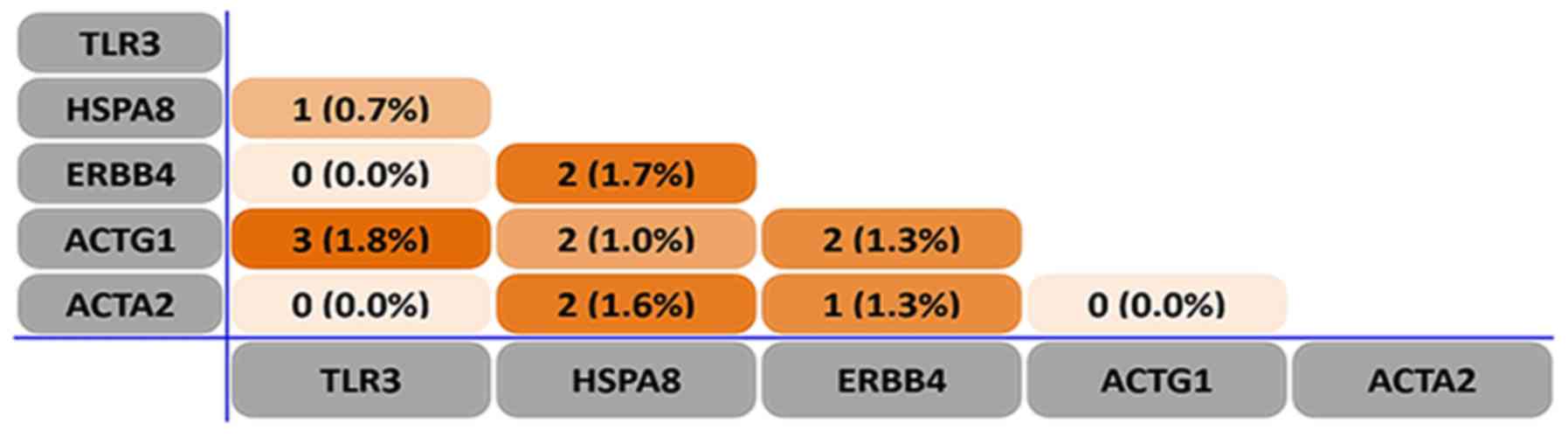
According to the PPI network in Fig. 4, including the upregulated (Fig. 4A) and downregulated PPI network
analyses (Fig. 4B), seven hub genes
were identified, including CETN3, TLR3, ERBB4, HSPA8, ACTG1, ACTA2
and interleukin-8. For consistency with the heat map, CETN3, TLR3,
ERBB4, HSPA8, ACTG1 and ACTA2 were selected for further
investigation.
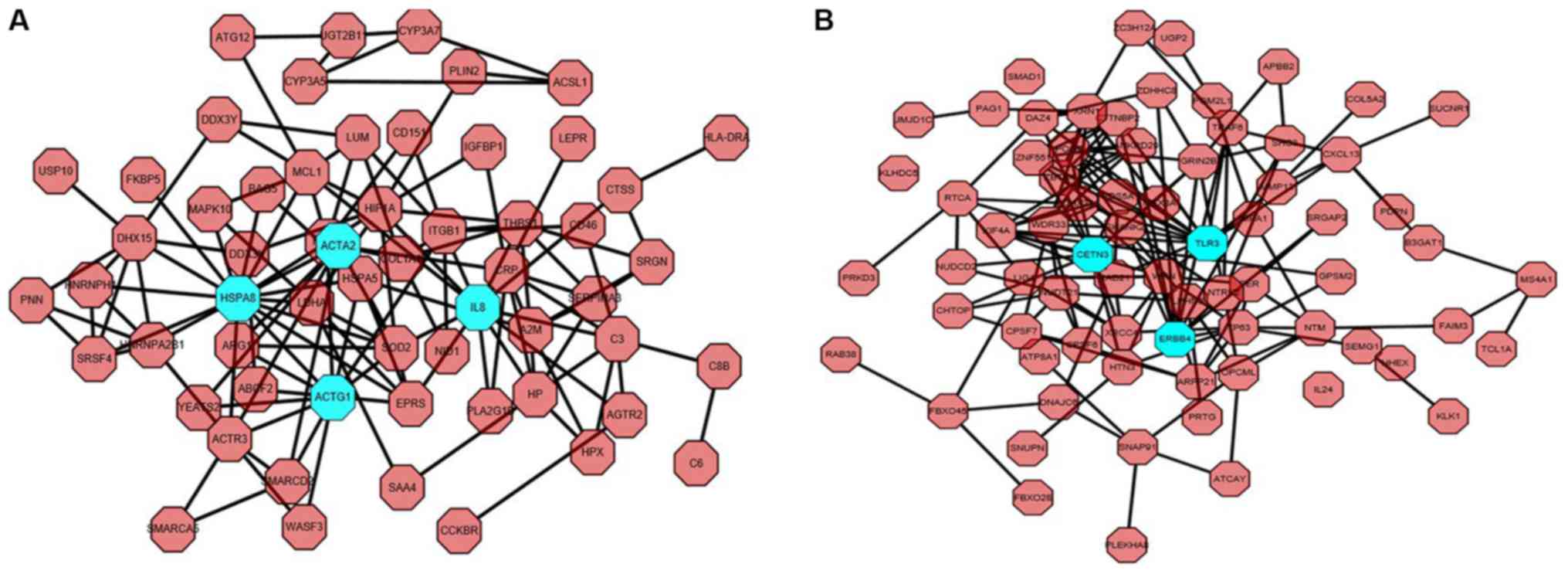 | Figure 4.Protein-protein interaction network
of huh genes obtained using Cytoscape software. The hub genes are
ACTA2, HSPA8, IL-8, ACTG1, CETN3, TLR3 and ERBB4. (A) Network of
upregulated protein-protein interactions. (B) Network of
downregulated protein-protein interactions. Blue nodes represent
hub genes. ACTA2, actin α2; HSPA8, heat shock protein family A
(Hsp70) member 8; IL-8, interleukin 8; ACTG1, actin γ1; CETN3,
centrin 3; TLR3, Toll-like receptor 3; ERBB4, erb-b2 receptor
tyrosine kinase 4. |
A diagram was generated according to the results of
TargetScan analysis of DEGs and their associated miRNAs (Fig. 5). In accordance with the criteria
previously set, miRNA (miR)-6819-3P and miR-6877-3P, and their
common target genes ACTG1 and TLR3, were selected for further
analysis via RT-qPCR.
Validation of DEGs and miRNAs via
RT-qPCR
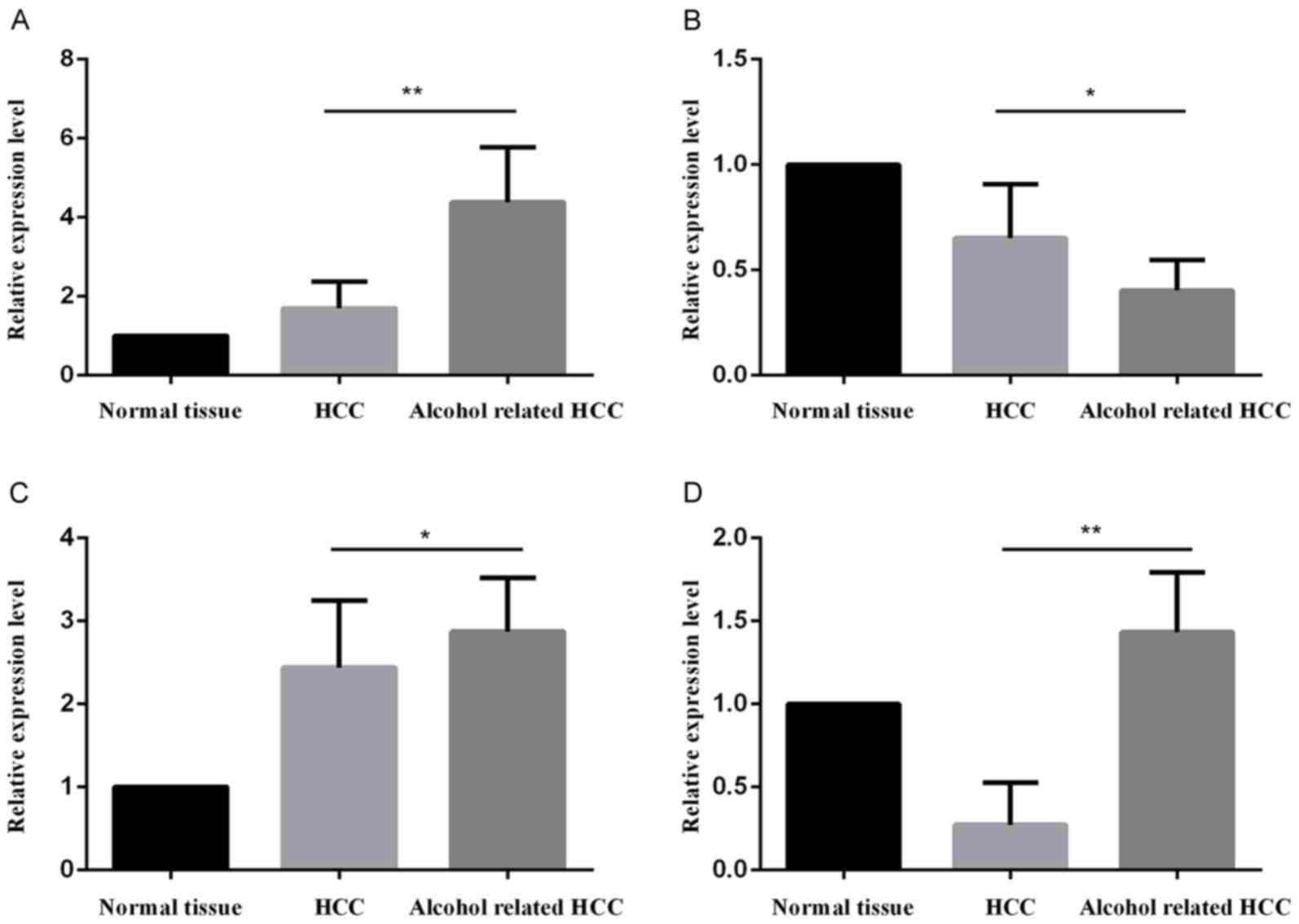
The relative mRNA expression levels of the DEGs and
selected miRNAs are presented in Fig.
6. The significantly upregulated mRNA expression levels of
ACTG1 (Fig. 6A) were consistent with
the bioinformatics analysis. Similarly, the results for TLR3
(Fig. 6B) were in line with the
bioinformatics analysis, in which the gene was downregulated in
alcohol-associated HCC tissues. It was also indicated that miR-6819
(Fig. 6C) and miR-6877 (Fig. 6D) were upregulated in
alcohol-associated HCC compared with non-alcohol-associated HCC.
These results indicated that miR-6819 and miR-6877 may positively
influence the ACTG1 gene, and exert a negative regulatory effect on
TLR3.
Discussion
The role of chronic alcohol consumption in the
induction and development of cancer is well known; heavy alcohol
consumption has been reported to exhibit negative effects on human
health, and there is increasing evidence to suggest that alcohol
increases the risk of carcinogenesis (12).
The global HCC BRIDGE study reported that the most
common risk factor in North America, Europe and Japan is HCV, and
HBV in China, South Korea and Taiwan; together with estimates of
alcohol consumption per capita (World Health Organization), alcohol
has been reported as a strong risk factor for HCC. Additionally,
the reported rate of alcohol abuse in China, Taiwan and Japan was
estimated to be 24, 18 and 2%, respectively (1). Studies of the alcohol-induced liver
injury model revealed that hypothalamic β-endorphin neuron
transplants may reduce liver weight and triglyceride accumulation;
fewer pathological alterations occurred, including infiltration of
inflammatory cells and steatosis of hepatocytes (13). In addition, investigation into the
association between alcohol consumption and the risk of gastric
cancer suggested that aldehyde dehydrogenase [ALDH2; National
Center for Biotechnology Information (NCBI) accession no. rs671],
rather than alcohol dehydrogenase 1B (ADH1B; NCBI accession no.
rs1229984) and 1C (ADH1C; NCBI accession no. rs698), which modify
acetaldehyde, may serve an important role in gastric cancer
(14). Xu et al (15) reported that alcohol may promote the
metastasis of colorectal cancer cells by modulating the glycogen
synthase kinase 3β/β-catenin/monocyte chemoattractant protein-1
signaling pathway. Furthermore, it has been demonstrated that an
alcohol intake >30 g/day may be associated with an increased
risk of colon cancer (12).
Similarly, research into breast cancer suggested that the
accumulation of alcohol at a sufficient level may induce cytochrome
P450 2E1, which causes mutagenic DNA adducts (16). In the present study, DEGs were
compared between alcohol-associated HCC (n=10) and HCC (n=16)
tissues in the GSE50579 dataset; ACTG1 and TLR3 were selected among
284 DEGs as potential therapeutic biomarkers in alcohol-associated
HCC, as determined by bioinformatics analysis.
The ACTG1 gene encodes actin γ1, which belongs to
the actin family of six highly conserved proteins. These actin
filaments are critical for the structure of the cytoskeleton and
the shape of the cell, and as such have been reported to regulate
cell motility, contraction and growth (17). In a family with DFN20/26-associated
hearing loss, it was reported that the cause of the condition may
be due to a missense mutation in the ACTG1 gene. The Thr278lle
mutation in ACTG1 has been predicted to affect the structure of the
protein and induce germline mutations in cytoplasmic actin isoforms
(18). Notably, researchers observed
that within this Norwegian family, ACTG1 mutations were not
frequently associated with hereditary hearing impairment. In
addition, the p.V370A mutation may impair actin function, which was
demonstrated by a yeast growth assay (17). K118N and E241K in ACTG1 have been
considered as two novel mutations that cause an aberrant
multi-vacuolar pattern in yeast and mammalian cells (19). According to whole exome sequencing
data and massive parallel sequencing, p.M305T and p.E316k have been
selected as nonsyndromic hearing loss variants (20,21).
Moreover, ACTG1 is positively implicated in the growth and survival
of mammals, as ACTG−/− animals were observed to be
smaller in size and exhibited a delay in the development of the
cardiac outflow tract (22).
Utilizing targeted next generation sequencing analysis, another
study reported that in Japanese families, ACTG1, nonsyndromic
hearing impairment 5, POU class 4 homeobox 3, solute carrier family
26 member 5, SIX homeobox 1, myosin VIIA, cadherin related 23,
protocadherin related 15 and Usher syndrome 2A may be candidate
genes for early-childhood hearing loss (23).
ACTG1 has also been associated with other types of
cancer. Studies have demonstrated that miR-888 may reduce the mRNA
expression levels of all four adherens junction pathway-associated
genes, including E-cadherin, ACTG1, receptor-type tyrosine-protein
phosphatase T and cell division cycle 42 in MCF-7 cells (24). Additionally, ACTG1 has been reported
to exhibit high expression levels in skin cancer tissue, where it
may regulate A431 cell proliferation and migration via the
Rho-associated protein kinase signaling pathway (25). ACTG1 has also been predicted to be a
target gene of miR-145-5P in non-small cell lung cancer (26). Furthermore, ACTG1 and matrix
metalloproteinase 14 (MMP14) were validated as targets of miR-10a
in colorectal cancer cells; ectopic expression of ACTG1 and MMP14
may delay decreases in cell adhesion and anoikis resistance
activity (27). It has also been
reported that in HBx-induced HCC, ATCG1 was upregulated in human
liver tissues (28).
TLR3 belongs to the Toll-like receptor family and is
a receptor for double-stranded (ds)RNA (29); as such, TLR3 is reported to exhibit
protective immunity during viral infection (30). TLR3 has also been reported in a number
of cancer types and the TLR3 signaling pathway may serve an
important role in cell homeostasis. BM-06, a 25-nucleotide dsRNA,
may activate TLR3 to inhibit the proliferation and promote the
apoptosis of HepG2.2.15 cells (29).
TLR3-TIR-domain-containing adapter-inducing interferon-β (TRIF)
signaling may contribute to myocardial inflammation and infarction
when ischemia-reperfusion-associated extracellular RNA is released
(31). In addition, TLR3 is
positively associated with HBV antigen (HBsAg) in HepG2.2.15 cells,
and may promote interstitial immunoreactive cell infiltration and
HCC cell apoptosis (32). According
to RNA-sequencing analysis, TLR3 was proposed as an immune gene,
particularly within BV-2 microglial cells (33). High expression levels of TLR3 are a
distinctive feature of CD8+ dendritic cells, where
activation of the receptor may induce an interferon (IFN)-dependent
antiviral response in dendritic cell subtypes (34). It has been reported that in Chinese
newborns with severe hepatitis, TLR3, TLR2, TLR4 and TLR9 may serve
as potential therapeutic targets for treatment (35). Of note, TLR3 exhibits lower levels of
expression in HCC when compared with adjacent tissues, and is
positively associated with TRIF, nuclear factor-κB and IFN
regulatory factor 3, which may inhibit HCC proliferation and
promote HCC cell apoptosis (36). The
expression levels of TLR3, TRIF and mitochondrial
antiviral-signaling proteins were consistently decreased in chronic
HCV-infected liver tissue, compared to that of than non-diseased
liver tissue (37). Additionally,
endogenous miR-155 can negatively regulate TLR3 expression and
inhibit IFN-β production (38).
Another study has reported that a TLR3 agonist may enhance the
clinical efficacy of sorafenib in treating HCC (29,39). A
TLR3 agonist may also inhibit angiogenesis and induce the apoptosis
of human HCC cells (40). In
addition, the cytoplasmic expression of TLR3 was positively
associated with HBV infection (32,36).
Compared with that of adjacent tissues, the expression of TLR3 is
lower in HCC tissues and is correlated with longer durations of
survival (36). The evidence that
dsRNA induces the activation of TLR3 further strengthens the
hypothesis that TLR3, as a cytotoxic agent, may contribute to
developments in HCC therapy (41).
miRNAs engage in numerous cellular activities and
affect important physiological functions. In the present study,
ACTG1 and TLR3 were proposed to be targets of miR-6819-3P and
miR-6877-3P. In addition, the expression levels of miR-6819-3P and
miR-6877-3P were observed to be higher in alcohol-associated HCC
compared with non alcohol-associated HCC tissues; the upregulation
of miR-6877-3P appeared to be higher compared with the upregulation
of miR-6819-3P. Additionally, ACTG1 and TLR3 may regulate the VEGF
and VEGFR signaling, proteoglycan syndecan-mediated signaling, erbB
receptor signaling, and β1 integrin cell surface interactions.
ACTG1 and TLR3 may also function in the immune response and nucleic
acid metabolism. The results of the present study indicated that
miR-6819-3P and miR-6877-3P enhanced the expression of ACTG1 and
inhibited that of TLR3. Further investigation is required to
determine the association between ACTG1, TLR3, miR-6819-3P and
miR-6877-3P in alcohol-associated HCC tissues.
In conclusion, comprehensive bioinformatics analysis
was performed to screen candidate genes for the treatment of
alcohol-associated HCC; the physiological functions and regulatory
mechanisms of TLR3 and ACTG1 require further study. To further
investigate the effects of alcohol on HCC, molecular biological
analysis in established HCC animal models may be advantageous;
however, the findings of the present study may provide novel
insight into the alcohol-associated etiopathogenesis of HCC and
subsequently improve the clinical diagnosis of the disease.
Acknowledgements
Not applicable.
Funding
The present study was supported by funds from the
National Natural Sciences Foundation of China (grant no. 81502065),
the China Postdoctoral Science Foundation Funded Project (grant
nos. 2016T90613 and 2015M580574), the Natural Sciences Foundation
of Shandong Province (grant nos. ZR2014HQ009 and ZR2014HM004), the
Postdoctoral Innovation Project of Shandong Province (grant no.
201602037), the Qingdao Postdoctoral Research Project (grant no.
2015167) and the Qingdao Innovation Project (grant no.
16-5-1-56-JCH).
Availability of data and materials
The datasets used and analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors contributions
JL and LM designed the experiments. BG, SL and ZT
conducted the experiments. SL and ZT collected the clinical data
and were responsible for ethics approval and consent of patients to
participate in the study. BG and SL performed the bioinformatics
and statistical analysis; JL and LM were responsible for the
overall design and funding of the project. JL, LM and BG prepared
the figures and wrote the manuscript. All authors have and approved
the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of the Affiliated Hospital of Qingdao University. Written
informed consent was obtained from patients.
Patient consent for publication
Written informed consent was obtained from all
participating patients.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Park JW, Chen M, Colombo M, Roberts LR,
Schwartz M, Chen PJ, Kudo M, Johnson P, Wagner S, Orsini LS and
Sherman M: Global patterns of hepatocellular carcinoma management
from diagnosis to death: The BRIDGE Study. Liver Int. 35:2155–2166.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Nahon P and Nault JC: Constitutional and
functional genetics of human alcohol-related hepatocellular
carcinoma. Liver Int. 37:1591–1601. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Li DK, Yan P, Abou-Samra AB, Chung RT and
Butt AA: Proton pump inhibitors are associated with accelerated
development of cirrhosis, hepatic decompensation and hepatocellular
carcinoma in noncirrhotic patients with chronic hepatitis C
infection: Results from ERCHIVES. Aliment pharmacol Ther.
47:246–258. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
David D, Raghavendran A, Goel A Bharath,
Kumar C, Kodiatte TA, Burad D, Abraham P, Ramakrishna B, Joseph P,
Ramachandran J and Eapen CE: Risk factors for non-alcoholic fatty
liver disease are common in patients with non-B non-C
hepatocellular carcinoma in India. Indian J Gastroenterol.
36:373–379. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Xin B, Cui Y, Wang Y, Wang L, Yin J, Zhang
L, Pang H, Zhang H and Wang RA: Combined use of alcohol in
conventional chemical-induced mouse liver cancer model improves the
simulation of clinical characteristics of human hepatocellular
carcinoma. Oncol Lett. 14:4722–4728. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ramadori P, Cubero FJ, Liedtke C,
Trautwein C and Nevzorova YA: Alcohol and hepatocellular carcinoma:
Adding fuel to the flame. Cancers (Basel). 9:E1302017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Neumann O, Kesselmeier M, Geffers R,
Pellegrino R, Radlwimmer B, Hoffmann K, Ehemann V, Schemmer P,
Schirmacher P, Lorenzo Bermejo J and Longerich T: Methylome
analysis and integrative profiling of human HCCs identify novel
protumorigenic factors. Hepatology. 56:1817–1827. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Harbig J, Sprinkle R and Enkemann SA: A
sequence-based identification of the genes detected by probesets on
the Affymetrix U133 plus 2.0 array. Nucleic Acids Res. 33:e312005.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wettenhall JM, Simpson KM, Satterley K and
Smyth GK: affylmGUI: A graphical user interface for linear modeling
of single channel microarray data. Bioinformatics. 22:897–899.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Han L, Suzek TO, Wang Y and Bryant SH: The
text-mining based pubchem bioassay neighboring analysis. BMC
Bioinformatics. 11:5492010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cho E, Lee JE, Rimm EB, Fuchs CS and
Giovannucci EL: Alcohol consumption and the risk of colon cancer by
family history of colorectal cancer. Am J Clin Nut. 95:413–419.
2012. View Article : Google Scholar
|
|
13
|
Murugan S, Boyadjieva N and Sarkar DK:
Protective effects of hypothalamic beta-endorphin neurons against
alcohol-induced liver injuries and liver cancers in rat animal
models. Alcohol Clin Exp Res. 38:2988–2997. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hidaka A, Sasazuki S, Matsuo K, Ito H,
Sawada N, Shimazu T, Yamaji T, Iwasaki M, Inoue M and Tsugane S:
JPHC Study Group: Genetic polymorphisms of ADH1B, ADH1C and ALDH2,
alcohol consumption, and the risk of gastric cancer: The Japan
Public Health Center-based prospective study. Carcinogenesis.
36:223–231. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Xu M, Wang S, Qi Y, Chen L, Frank JA, Yang
XH, Zhang Z, Shi X and Luo J: Role of MCP-1 in alcohol-induced
aggressiveness of colorectal cancer cells. Mol Carcinog.
55:1002–1011. 2016. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Brooks PJ and Zakhari S: Moderate alcohol
consumption and breast cancer in women: From epidemiology to
mechanisms and interventions. Alcohol Clin Exp Res. 37:23–30. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Rendtorff ND, Zhu M, Fagerheim T, Antal
TL, Jones M, Teslovich TM, Gillanders EM, Barmada M, Teig E, Trent
JM, et al: A novel missense mutation in ACTG1 causes dominant
deafness in a Norwegian DFNA20/26 family, but ACTG1 mutations are
not frequent among families with hereditary hearing impairment. Eur
J Hum Genet. 14:1097–1105. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
van Wijk E, Krieger E, Kemperman MH, De
Leenheer EM, Huygen PL, Cremers CW, Cremers FP and Kremer H: A
mutation in the gamma actin 1 (ACTG1) gene causes autosomal
dominant hearing loss (DFNA20/26). J Med Genet. 40:879–884. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Morín M, Bryan KE, Mayo-Merino F, Goodyear
R, Mencía A, Modamio-Høybjør S, del Castillo I, Cabalka JM,
Richardson G, Moreno F, et al: In vivo and in vitro effects of two
novel gamma-actin (ACTG1) mutations that cause DFNA20/26 hearing
impairment. Hum Mol Genet. 18:3075–3089. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Park G, Gim J, Kim AR, Han KH, Kim HS, Oh
SH, Park T, Park WY and Choi BY: Multiphasic analysis of whole
exome sequencing data identifies a novel mutation of ACTG1 in a
nonsyndromic hearing loss family. BMC Genomics. 14:1912013.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wei Q, Zhu H, Qian X, Chen Z, Yao J, Lu Y,
Cao X and Xing G: Targeted genomic capture and massively parallel
sequencing to identify novel variants causing Chinese hereditary
hearing loss. J Transl Med. 12:3112014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bunnell TM and Ervasti JM: Delayed
embryonic development and impaired cell growth and survival in
Actg1 null mice. Cytoskeleton (Hoboken). 67:564–572. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Mutai H, Suzuki N, Shimizu A, Torii C,
Namba K, Morimoto N, Kudoh J, Kaga K, Kosaki K and Matsunaga T:
Diverse spectrum of rare deafness genes underlies early-childhood
hearing loss in Japanese patients: A cross-sectional, multi-center
next-generation sequencing study. Orphanet J Rare Dis. 8:1722013.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Huang S, Cai M, Zheng Y, Zhou L, Wang Q
and Chen L: miR-888 in MCF-7 side population sphere cells directly
targets E-cadherin. J Genet Genomics. 41:35–42. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Dong X, Han Y, Sun Z and Xu J: Actin Gamma
1, a new skin cancer pathogenic gene, identified by the biological
feature-based classification. J Cell Biochem. 119:1406–1419. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Gan TQ, Xie ZC, Tang RX, Zhang TT, Li DY,
Li ZY and Chen G: Clinical value of miR-145-5p in NSCLC and
potential molecular mechanism exploration: A retrospective study
based on GEO, qRT-PCR, and TCGA data. Tumour Biol.
39:10104283176916832017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Liu Y, Zhang Y, Wu H, Li Y, Zhang Y, Liu
M, Li X and Tang H: miR-10a suppresses colorectal cancer metastasis
by modulating the epithelial-to-mesenchymal transition and anoikis.
Cell Death Dis. 8:e27392017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Sun Q, Wang Y, Zhang Y, Liu F, Cheng X,
Hou N, Zhao X and Yang X: Expression profiling reveals
dysregulation of cellular cytoskeletal genes in HBx–induced
hepatocarcinogenesis. Cancer Biol Ther. 6:668–674. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Xu YY, Chen L, Wang GL, Zhou JM, Zhang YX,
Wei YZ, Zhu YY and Qin J: A synthetic dsRNA, as a TLR3
pathwaysynergist, combined with sorafenib suppresses HCC in vitro
and in vivo. BMC Cancer. 13:5272013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhang SY, Herman M, Ciancanelli MJ, Pérez
de Diego R, Sancho-Shimizu V, Abel L and Casanova JL: TLR3 immunity
to infection in mice and humans. Curr Opin Immunol. 25:19–33. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Chen C, Feng Y, Zou L, Wang L, Chen HH,
Cai JY, Xu JM, Sosnovik DE and Chao W: Role of extracellular RNA
and TLR3-Trif signaling in myocardial ischemia-reperfusion injury.
J Am Heart Assoc. 3:e0006832014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Chen XL, Xu YY, Chen L, Wang GL and Shen
Y: TLR3 plays significant roles against HBV-associated HCC.
Gastroenterol Res Prac. 2015:5721712015.
|
|
33
|
Das A, Chai JC, Kim SH, Lee YS, Park KS,
Jung KH and Chai YG: Transcriptome sequencing of microglial cells
stimulated with TLR3 and TLR4 ligands. BMC Genomics. 16:5172015.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Szeles L, Meissner F, Dunand-Sauthier I,
Thelemann C, Hersch M, Singovski S, Haller S, Gobet F, Fuertes
Marraco SA, Mann M, et al: TLR3-mediated CD8+ dendritic cell
activation is coupled with establishment of a cell-intrinsic
antiviral state. J Immunol. 195:1025–1033. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Qiu X, Dong Y, Cao Y and Luo Y:
Correlation between TLR2, TLR3, TLR4, and TLR9 polymorphisms and
susceptibility to and prognosis of severe hepatitis among the
newborns. J Clin Lab Anal. 32:2018. View Article : Google Scholar
|
|
36
|
Yuan MM, Xu YY, Chen L, Li XY, Qin J and
Shen Y: TLR3 expression correlates with apoptosis, proliferation
and angiogenesis in hepatocellular carcinoma and predicts
prognosis. BMC Cancer. 15:2452015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Kar P, Kumar D, Gumma PK, Chowdhury SJ and
Karra VK: Down regulation of TRIF TLR3, and MAVS in HCV infected
liver correlates with the outcome of infection. J Med Virol.
89:2165–2172. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Hu X, Ye J, Qin A, Zou H, Shao H and Qian
K: Both microRNA-155 and virus-encoded MiR-155 ortholog regulate
TLR3 expression. PLoS One. 10:e01260122015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Ho V, Lim TS, Lee J, Steinberg J, Szmyd R,
Tham M, Yaligar J, Kaldis P, Abastado JP and Chew V: TLR3 agonist
and Sorafenib combinatorial therapy promotes immune activation and
controls hepatocellular carcinoma progression. Oncotarget.
6:27252–27266. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Guo Z, Chen L, Zhu Y, Zhang Y, He S, Qin
J, Tang X, Zhou J and Wei Y: Double-stranded RNA-induced TLR3
activation inhibits angiogenesis and triggers apoptosis of human
hepatocellular carcinoma cells. Oncol Rep. 27:396–402.
2012.PubMed/NCBI
|
|
41
|
Yoneda K, Sugimoto K, Shiraki K, Tanaka J,
Beppu T, Fuke H, Yamamoto N, Masuya M, Horie R, Uchida K and Takei
Y: Dual topology of functional Toll-like receptor 3 expression in
human hepatocellular carcinoma: Differential signaling mechanisms
of TLR3-induced NF-kappaB activation and apoptosis. Int J Oncol.
33:929–936. 2008.PubMed/NCBI
|