Introduction
Somatostatin (SST) receptors (SSTRs) are
G-protein-coupled plasma membrane receptors with two forms of SST
peptides, SS-14 and SS-28, as their natural ligands (1). The two peptides produced by SST cells
act as neurotransmitters or paracrine/autocrine regulators,
respectively, via five different subtypes of human SSTR (SSTR1-5),
encoded by five distinct SSTR genes segregated on chromosomes 14,
16, 17, 20 and 22, respectively (2).
Activation of SSTRs frequently results in inhibition of cell
proliferation and secretion (3). It
is generally accepted that all five SSTR subtypes are involved in
the inhibition of the adenylate cyclase-cyclic adenosine
3′5′-monophosphate pathway and stimulate protein tyrosine
phosphatases (3). However, a number
of effects demonstrated subtype selectivity, and subtype-specific
signaling has also been reported (4,5). For
instance, SSTR1, 2, 4 and 5 frequently interfere with the
mitogen-activate protein kinase pathway to modulate cell
proliferation, whereas SSTR3 was indicated to have an increased
potential to induce apoptosis (6,7).
Additionally, owing to multiple SSTRs being frequently expressed in
the same cell, and the existence of ligand-induced dimerization
proposed for G-protein-coupled receptors, it is hypothesized that
SSTRs are functionally redundant and act in concert (8,9).
Expression levels of SSTRs have been determined in
multiple human tissues as well as in the majority of neuroendocrine
and non-endocrine tumor types, including hepatocellular carcinoma,
pancreatic cancer and breast cancer (10–17).
Activation of SSTRs in SSTR-expressing tumors frequently results in
marked inhibition of tumor cell proliferation via indirect
activities of inhibiting growth hormone secretion and direct
activity through SSTR signaling pathways (18). Therefore, SST and SST analogues (SSAs)
with improved metabolic stabilities have frequently been used in
the treatment of SSTR-positive tumors (19–22).
However, the therapeutic results of SSA treatments varied markedly
due to the loss-of-expression of SSTRs, different SSTR expression
patterns and reasons that are not fully understood (4,23–25).
In the present study, the expression levels of the
five different SSTR subtypes were determined in 160 primary ductal
breast tumor samples using immunohistology. All five SSTR subtypes
were expressed in the tumor tissues. The expression levels of SSTR1
and SSTR4 were detected in 90.0 and 71.3% of tumor tissues,
respectively. The expression levels of SSTR1 and SSTR4 were
determined to be negatively associated with cancer cell
differentiation, but were independent of patient age and the cancer
stage. SSTR1 and SSTR4 were subsequently overexpressed in cultured
MDA-MB-435S cells, which have previously been demonstrated to
exhibit decreased endogenous SSTR expression (26). The potential interaction of SSTR1 and
SSTR4 was analyzed using immunofluorescence and
co-immunoprecipitation. The overexpressed SSTRs were then activated
with the subtype-specific SSA L-803087, which has previously been
identified to exhibit high selective binding affinity with SSTR1
and SSTR4 (27). The influence of
receptor expression and activation on cell proliferation was
investigated further using flow cytometry. The results of the
present study indicated a ligand-induced heterodimerization of
SSTR1 and SSTR4, and the functional significance of the receptor
dimerization in regulating cell proliferation. Future
investigations on receptor dimerization between other SSTR subtypes
and the subsequent effect on cell proliferation will provide
valuable references for selection of breast cancer cases suitable
for SSA treatment.
Materials and methods
Breast tumor samples and the clinical
information
Sections of all breast tumor samples (fixed with 4%
paraformaldehyde in PBS at 4°C overnight and embedded in paraffin)
were obtained from the Department of Pathology of The First
Affiliated Hospital of Jinan University (Guangzhou, China) from
January 2010 to December 2015. A total of 160 primary ductal breast
cancer cases, confirmed by pathology, were selected. The clinical
references including ages of patients, tumor type and steroid
receptor expression levels were provided by the Department of
Pathology.
Reagents
Dulbecco's modified Eagle's medium (DMEM), PBS and
fetal bovine serum (FBS) used in tissue culture were purchased from
Invitrogen; Thermo Fisher Scientific, Inc. (Waltham, MA, USA).
Additionally, 3,3-diaminobenzidine tetrachloride,
1,4-dithiothreitol (DTT), dimethylsulfoxide, mouse monoclonal
anti-hemagglutinin (anti-HA; cat. no. H3663; 1:1,000), protease
inhibitor cocktail, penicillin and streptomycin were purchased from
Sigma-Aldrich; Merck KGaA (Darmstadt, Germany). Polyethyleneimine
(PEI) was purchased from Polyplus-transfection SA (Illkirch,
France). L-803087 and Protein A/G PLUS-Agarose were purchased from
Santa Cruz Biotechnology, Inc. (Dallas, TX, USA). Blue Range™
Prestained Protein Molecular Marker, used for SDS-PAGE, was
purchased from Pierce; Thermo Fisher Scientific, Inc. Rabbit
monoclonal anti-SSTR1 (cat. no. ab137083; 1:500), rabbit monoclonal
anti-SSTR2 (cat. no. ab134152; 1:1,000), rabbit monoclonal
anti-SSTR3 (cat. no. ab137026; 1:10,000), rabbit polyclonal
anti-SSTR4 (cat. no. ab28578; 1:1,000) and rabbit monoclonal
anti-SSTR5 (cat. no. ab109495; 1:1,000) were purchased from Abcam
(Cambridge, MA, USA). The horseradish peroxidase (HRP)-conjugated
goat anti-rabbit immunoglobulin G (cat. no. TA140003; 1:10,000),
HRP-conjugated goat anti-mouse IgG (cat. no. TA130004; 1:20,000)
used for western blot analysis were purchased from OriGene
Technologies, Inc. (Rockville, MD, USA). The
tetramethylrhodamine-conjugated goat anti-rabbit IgG (cat. no.
111-025-003; 1:100) and the fluorescein isothiocyanate-conjugated
mouse anti-rabbit IgG (cat. no. 111-095-003; 1:100) used in
immunofluorescence were purchased from Jacksons ImmunoResearch
Europe, Ltd. (Newmarket, UK). PrimeSTAR DNA Polymerase, EasyTaq DNA
Polymerase, EcoRI, XhoI and T4 DNA ligase were
purchased from Takara Bio, Inc. (Otsu, Japan). Ethanol, xylene and
KCl were purchased from Guangzhou Chemical Reagent Factory
(Guangzhou, China). EDTA, 0.1% Triton X-100, hematoxylin and
glycerol were purchased from Beijing Dingguo Changsheng
Biotechnology Co., Ltd. (Beijing, China). MDA-MB-435S cells were
purchased from American Type Culture Collection (Manassas, VA, USA)
and maintained in our laboratory according to the instruction.
Expression constructs of hSSTRs
Total mRNA was extracted from cultured HeLa cells
using TRIzol® (Invitrogen; Thermo Fisher Scientific,
Inc.) reagent, according to the manufacturer's protocol. A total of
2 µg total RNA in each reaction was reverse-transcribed into cDNA
with oligo(dT) primers using Moloney Murine Leukemia Virus Reverse
Transcriptase kit (Promega Corporation, Madison, WI, USA). The cDNA
of hSSTR1 and hSSTR4 was amplified with PrimeSTAR using the
polymerase chain reaction (PCR) with the following primers: hSSTR1
(1,176 bp), sense, 5′-CCGGAATTCGCCACCATGTTCCCCAATGGCACCG-3′, and
antisense, 5′-CCGCTCGAGTCAGAGCGTCGTGATCCGG-3′; and hSSTR4 (1,167
bp), sense, 5′-CCGGAATTCGCCACCATGAGCGCCCCCTCGACG-3′, and antisense,
5′-CCGCTCGAGTCAGAAGGTGGTGGTCCTGG-3′. PCR was performed in a Peltier
Thermal Cycler 200 programmed with an initial denaturation at 98°C
for 5 min followed by 35 cycles of denaturation at 98°C for 30 sec,
annealing at 55–58°C for 30 sec and extension at 72°C for 90 sec.
The PCR products were verified by electrophoresis on 1% agarose gel
and visualized by ethidium bromide staining under ultraviolet
light. The purified PCR products were then digested with
EcoRI/XhoI and cloned using T4 ligase into
pcDNA6-Myc/His (Invitrogen; Thermo Fisher Scientific, Inc.) and
pCMV-HA (Clontech Laboratories, Inc., Mountainview, CA, USA). The
constructs were screened using PCR with Taq polymerase and the
sequences of the constructs were verified by DNA sequencing
(RuibioBiotech, Beijing, China).
Cell culture and transfection
Human cancer cell line MDA-MB-435S was maintained in
DMEM, supplemented with 10% FBS, 100 U/ml penicillin and 100 µg/ml
streptomycin in an atmosphere containing 5% CO2 at 37°C.
Cells were transfected with the indicated constructs using PEI
according to the manufacturer's protocol.
Histology and
immunohistochemistry
A total of five 6 µm paraffin sections were prepared
for each sample. The paraffin sections of breast tumor tissues were
baked in an oven at 65°C for 4 h prior to being deparaffinized in
xylene for 20 min twice at room temperature. The samples were
subsequently rehydrated by incubating at room temperature in an
ethanol gradient series for 3 min at each step (100, 95, 80, 70 and
50%, and distilled water). The tissue sections were then washed
with PBS three times prior to being boiled in a microwave in 0.01 M
citric acid (pH 6.0) for 20 min. The samples were then washed once
with PBS.
For immunostaining, the sections were treated with
3% H2O2 for 10 min at room temperature and
blocked with 1% bovine serum albumin (Thermo Fisher Scientific,
Inc.) plus 0.1% Triton X-100 in PBS for 30 min at room temperature.
The sections were incubated with the indicated primary antibodies
(with PBS in blank control) at 4°C overnight and then washed with
3X PBS. The slides were probed with HRP-conjugated secondary
antibodies for 1 h at room temperature and the excess antibody was
washed off with PBS. Subsequently, 250 µl DAB per section was then
added for color reaction. Following an intensive wash with PBS, the
slides were counterstained with hematoxylin for 8 min at room
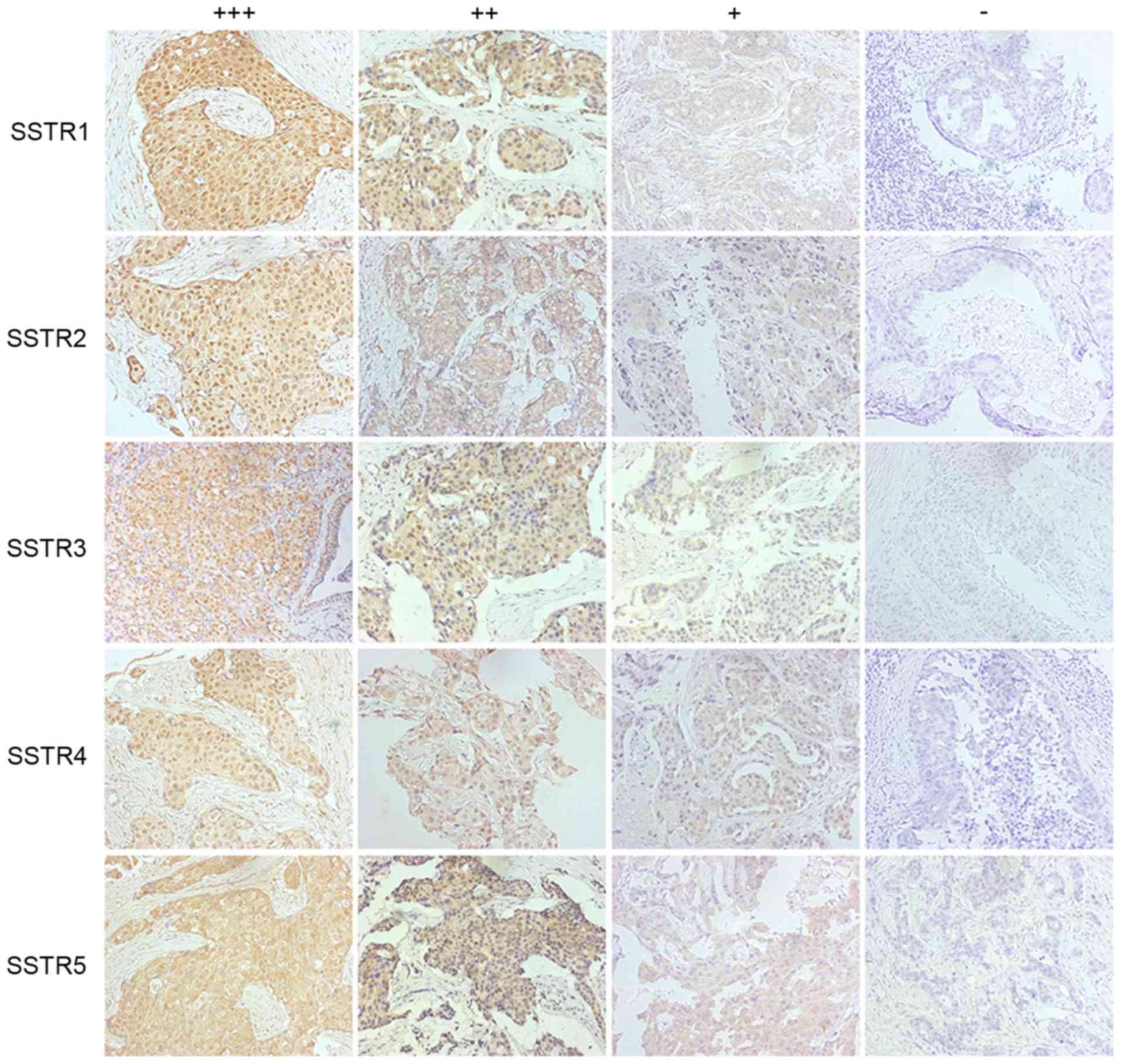
temperature. The expression levels of SSTRs were scored according
to the color and number of positive cells (positive cells <5%,
0; positive cells 5–25%, 1; positive cells >25–50%, 2; positive
cells >50%, 3; light tea color, 1; yellow-brownish, 2; brown,
3). An overall score of 0–1 represented negative, a score of 2–3
points represented +, a score of 4–6 represented ++ and a score of
>6 represented +++. The incidences of SSTR expressions were
presented in percentages. The association between SSTR expression
levels and other clinical indexes were analyzed using a
χ2 test and P<0.05 was used as statistical
significance. A colon cancer tissue sample that was confirmed to
exhibit positive SSTR1-5 expression was stored in our lab and
included in every experiment as a positive control.
Co-immunoprecipitation and western
blot analysis
Cultured cells were collected and resuspended in
lysis buffer containing 50 mM Tris/HCl (pH 7.4) (Invitrogen; Thermo
Fisher Scientific, Inc.), 100 mM KCl, 10% glycerol, 1 mM EDTA, 1%
Triton X-100 and 1 mM DTT and protease inhibitor cocktail. The
supernatants were collected by centrifugation at 12,000 × g for 15
min at 4°C. Protein A/G PLUS-Agarose was used for
immunoprecipitation, which was conducted according to the Current
Protocols in Cell Biology (28).
Aliquots (15 µl) of protein samples were then separated by SDS-PAGE
(12% gel) and transferred onto a polyvinylidene fluoride membrane
(Whatman; GE Healthcare Life Sciences, Little Chalfont, UK). The
membrane was then blocked in 5% skimmed milk in PBS plus 1% Triton
X-100 for 1 h at room temperature. Specific primary antibodies and
HRP-conjugated secondary antibodies were then used for probing at
room temperature for 1 h. The immunoblots were visualized by
chemiluminescence with an enhanced chemiluminescent kit (GE
Healthcare Life Sciences) and the results were further analyzed
with AlphaEase FC software 4.1.0 (Protein Simple, San Jose, CA,
USA). Densitometry was performed using a FluorChem™ system (Protein
Simple).
Immunofluorescence analysis of
cultured cells
The cultured MDA-MB-435S cells were plated on
coverslips and transfected using 16 µg PEI per 4 µg of
pCMV-HA-SSTR1 and/or pcDNA-MYC-SSTR4. At 24 h post-transfection,
the SSTR1 and SSTR4 subtype-specific SSA L-803087 was added into
the culture medium to a final concentration of 10 nM (DMEM was
added as mock treatment in control cells). The cells were
maintained for an additional 24 h prior to being analyzed. The
cells were rinsed twice with PBS and fixed with 3.5%
paraformaldehyde in PBS for 15 min at room temperature. The cells
were subsequently permeabilized or not with 0.2% Nonidet-P40 in
PBS, followed by staining for 1 h with the appropriate dilutions of
the indicated primary antibodies at room temperature, according to
the manufacturer's protocol. Excess primary antibodies were washed
off with PBS and attached antibodies were then detected with
appropriate dilutions of HRP-conjugated secondary antibodies for 1
h at room temperature, according to the manufacturer's protocol.
The coverslips were inverted and mounted onto glass slides for
visualization under the 505, 595 and 400 nm wavelengths on a Zeiss
LSM710 confocal microscope (Carl Zeiss AG, Oberkochen, Germany).
The images were obtained using a Zeiss LSM 510 EMTA camera (Carl
Zeiss AG) and processed with Zeiss LSM Image Browser 4.0 (Zeiss
GmbH, Jena, Germany). A total of five different views of
co-transfected cells were randomly selected and the dimerization
between SSTRs was calculated with Pearson's correlation using
Volocity 6.11 (PerkinElmer, Inc., Waltham, MA, USA). Results are
presented as the mean ± standard deviation (SD) and the P-values
were determined with Student's t-test.
Flow cytometry and cell cycle
analysis
In total, 1×105 MDA-MB-435S cells were
used to inoculate 1 ml culture medium per well in 24-well Petri
dishes. The cells were transfected with pCMV-HA-SSTR1 and/or
pcDNA-MYC-SSTR4, and at 24 h post-transfection, the SSTR
subtype-specific SSA, L-803087, was added into the culture medium,
whereas PBS was added in the mock treatment. The cells were
maintained in the aforementioned cell culture conditions for an
additional 24 h prior to further analysis. The cells were then
trypsinized and resuspended in fresh medium followed by flow
cytometry analysis. For the cell cycle assay, cells were fixed with
70% ethanol on ice for 30 min. The cells were then suspended in PBS
and treated with RNase A (final concentration 100 µg/ml; Beijing
Solarbio Science & Technology Co., Ltd., Beijing, China) at
37°C for 30 min. Following removal of RNase A by washing once with
PBS, the cells were stained with propidium iodide (PI) at 5 µg/ml
for 15 min at room temperature and the cell cycle phases were
determined with flow cytometry using a FACSAria I instrument (BD
Biosciences, Franklin Lakes, NJ, USA). The data were further
analyzed using Modfit LT 4.1 (Verity Software House, Inc., Topsham,
ME, USA). The results were obtained from three replicas for each
experiment and are presented as the mean ± SD. P-values were
determined with Student's t-test.
Results
Expression levels of SSTRs in human
breast cancer
Paraffin sections of 160 surgically removed human
breast-infiltrating ductal carcinoma tumor tissues were collected
from the Department of Pathology of The First Affiliated Hospital
of Jinan University. The clinical information, including the
patient age, cell differentiation and the expression levels of the
relevant molecular markers, was provided by the Department of
Pathology of The First Affiliated Hospital of Jinan University.
Among these 160 breast cancer tissue samples, the cancer cells were
determined to be poorly differentiated in 83 samples, moderately
differentiated in 54 samples and well-differentiated in 23 samples,
according to the Scarff-Bloom-Richardson system recommended by
World Health Organization (29).
Histological information of hormone receptor [estrogen receptor
(ER), progesterone receptor (PR) and human epidermal growth factor
receptor-2] expression levels was provided by the Department of
Pathology of The First Affiliated Hospital of Jinan University and
was available for 46 samples. The expression levels of the five
SSTRs were determined for each sample using immunohistology and
their associations with the clinical indexed were further analyzed
(data not shown).
All five SSTRs were determined to be expressed at
variable levels in breast cancer tissues (Fig. 1), and the expression levels of SSTR1-5
were detected in 90.0, 34.4, 41.9, 71.3 and 44.4% of cancer
tissues, respectively (Table I). The
expression levels of these SSTRs were determined to not be
associated with the ages of patients. Different subtypes of SSTRs
were frequently determined to be co-expressed in tumor tissues.
Only 5 samples were observed to be negative for all five subtypes
of SSTR expression. A total of 19 cases expressed just one subtype
of SSTR, and, among them, 18 were SSTR1-positive and only 1 case
expressed SSTR5 alone. The positive expression instances of all
five SSTRs was negatively associated with cancer tissue
differentiation, and the statistical significance (P<0.05) in
these differences was revealed for the expression levels of SSTR1
and SSTR4 in tumor tissues of different differentiations (Table I). Furthermore, high expression levels
of SSTR2 and SSTR3 were only observed in poorly differentiated
tumor tissues (Fig. 2). No
associations of hormone receptor (estrogen, progesterone and erB-2)
expression levels with SSTR expression levels were identified
(Table II). However, the expression
levels of SSTR2, SSTR3, SSTR4 and SSTR5 exhibited an increased
frequency of observation in ER/PR-negative tumor cases.
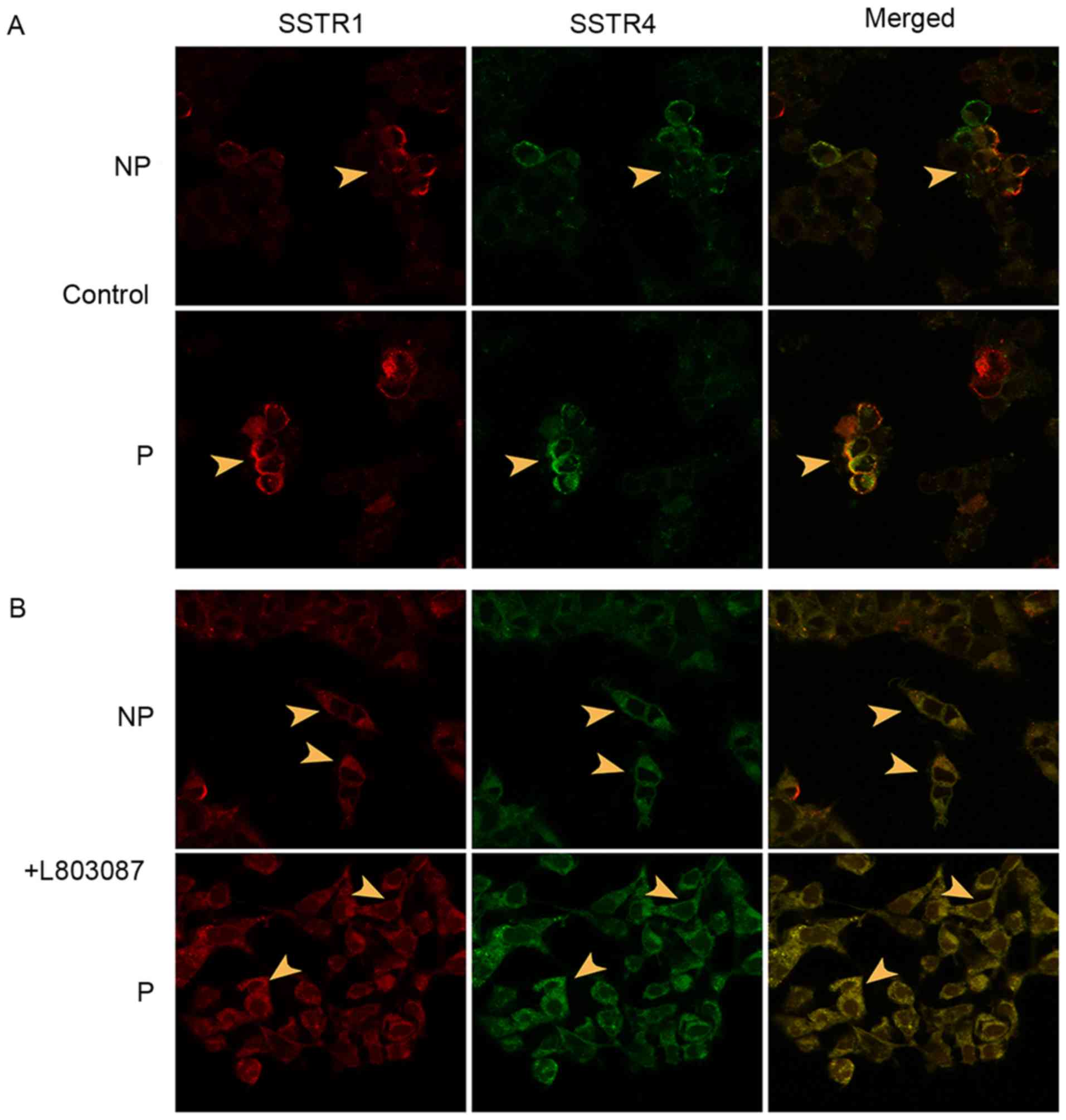 | Figure 2.Association of the cellular
distributions of SSTR1 and SSTR4. HA-SSTR1 and MYC-SSTR4 were
overexpressed in cultured MDA-MB-435S cells. The cellular
distributions of SSTRs were displayed by immunofluorescence using
confocal microscopy and the potential dimerization was calculated
using Pearson's correlation. (A) The cellular distribution of
overexpressed SSTR1 (red) and SSTR4 (green), as indicated with
arrowheads, in control cells that were not activated using the
subtype-specific SSA, L-803087. The cells were non-permeabilized or
permeabilized prior to conducting immunofluorescence. (B) The
cellular distribution of overexpressed SSTR1 (red) and SSTR4
(green), as indicated with arrowheads, in cells that were activated
using the subtype-specific SSA L-803087. The cells were
non-permeabilized or permeabilized prior to conducting
immunofluorescence. Magnification, ×600. SSTR, somatostatin
receptor; NP, unpermeablized; P, permeabilized; SSA, somatostatin
analogue. |
 | Table I.Expression of SSTR subtypes in primary
breast tumor tissues. |
Table I.
Expression of SSTR subtypes in primary
breast tumor tissues.
| Clinical index | No. of cases | SSTR1+ (%) | SSTR2+ (%) | SSTR3+ (%) | SSTR4+ (%) | SSTR5+ (%) |
|---|
| Age, years |
|
|
|
|
|
|
|
<40 | 23 | 21
(91.3) | 8 (34.8) | 9 (39.1) | 18 (78.3) | 12 (52.2) |
|
>40 | 137 | 123 (89.8) | 47 (34.3) | 58 (42.3) | 96 (70.1) | 59 (43.1) |
| Cancer cell
differentiationa |
|
|
|
|
|
|
|
Poor | 83 | 79
(95.2) | 30 (36.1) | 39 (47.0) | 59 (71.1) | 37 (44.6) |
|
Moderate | 54 | 47
(87.0) | 18 (33.3) | 20 (37.0) | 43 (79.6) | 26 (48.1) |
|
Well | 23 | 18
(78.3)b | 7 (30.4) | 8 (34.8) | 12
(52.2)b | 8 (34.8) |
| Total | 160 | 144 (90.0) | 55 (34.4) | 67 (41.9) | 114 (71.3) | 71 (44.4) |
 | Table II.Associations of the expression of
SSTRs with the expression of hormone receptors. |
Table II.
Associations of the expression of
SSTRs with the expression of hormone receptors.
| Hormone receptor
expression | No. of cases | SSTR1+ (%) | SSTR2+ (%) | SSTR3+ (%) | SSTR4+ (%) | SSTR5+ (%) |
|---|
| ER |
|
|
|
|
|
|
|
Positive | 31 | 27 (87.1) | 7 (22.6) | 4 (12.9) | 16 (51.6) | 7 (22.6) |
|
Negative | 15 | 11 (73.3) | 4 (26.7) | 3 (20.0) | 8
(53.3) | 4 (26.7) |
| PR |
|
|
|
|
|
|
|
Positive | 28 | 24 (85.7) | 5 (17.9) | 4 (12.9) | 14 (45.2) | 6 (19.4) |
|
Negative | 18 | 14 (77.8) | 6 (40.0) | 3 (20.0) | 10 (66.7) | 4 (26.7) |
| Her-2 |
|
|
|
|
|
|
|
Positive | 34 | 28 (82.4) | 8 (23.5) | 4 (11.8) | 17 (50.0) | 7 (20.6) |
|
Negative | 12 | 10 (83.3) | 3 (20.0) | 3 (20.0) | 7
(58.3) | 3 (20.0) |
Heterodimerization of SSTRs
Owing to the common features of ligand-induced
dimerization for G-protein-coupled receptors, the potential
dimerization of the two most commonly expressed SSTRs, SSTR1 and
SSTR4, were investigated further. The full-length coding sequences
of SSTR1 and SSTR4 were cloned into pCMV-HA and pcDNA6-Myc/His,
respectively. Exogenous HA-SSTR1 and MYC-SSTR4 were then
co-expressed in cultured MDA-MB-435S cells. No endogenous
expression of SSTRs was observed, which was verified using
immunoblotting with specific SSTR antibodies (data not shown). The
cells were permeabilized or not permeabilized prior to the
immunofluorescence assay to primarily display the cytoplasmic or
plasma membrane-bound proteins. The target proteins were visualized
using confocal microscopy and the potential protein dimerizations
were calculated with Pearson's correlation. The heterodimerizations
of SSTR1/SSTR4 natively existed on the plasma membrane as well as
in the cytoplasm of cells co-expressing HA-SSTR1 and MYC-SSTR4. The
Pearson's correlation value of SSTR1 and SSTR4 was significantly
increased in permeabilized cells (from 0.74±0.07 to 0.89±0.02) as
well as in non-permeabilized cells (from 0.63±0.04 to 0.83±0.03)
upon subtype-specific SSA L-803087 activation, compared with
non-activated cells. Overexpressed HA-SSTR1 and MYC-SSTR4 were
predominantly observed on cytoplasmic membranes prior to activation
whereas translocation of activated receptors to the cytoplasm was
observed upon ligand induction (Fig.
2).
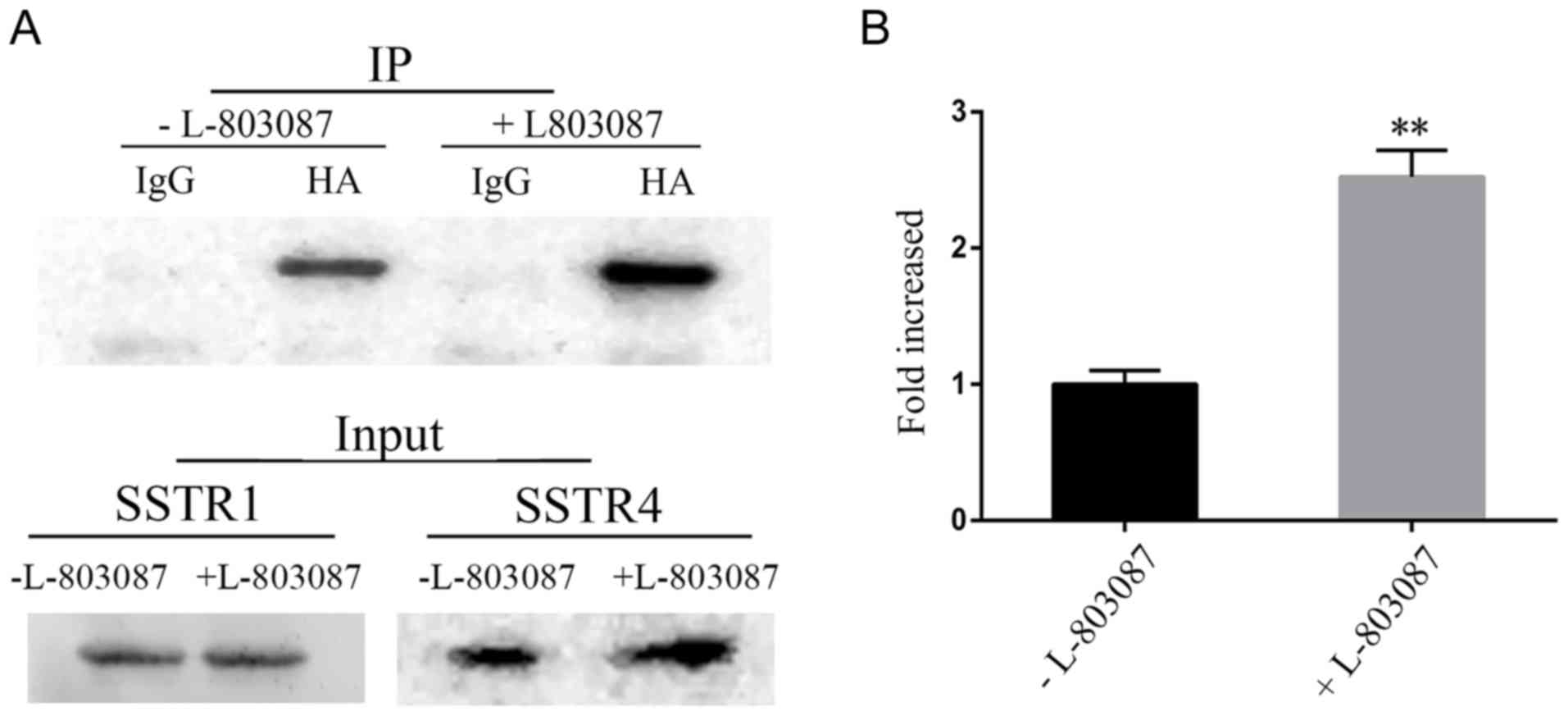
The dimerization of SSTR1 and SSTR4 was also
observed using co-immunoprecipitation. The cell lysates of
MDA-MB-435S cells overexpressing HA-SSTR1 and MYC-SSTR4 were
immunoprecipitated with an anti-HA antibody. Western blot analysis
of the precipitated proteins using an anti-MYC antibody revealed
the association between SSTR1 and SSTR4 (Fig. 3A). The heterodimerization of SSTR1 and
SSTR4 was significantly increased upon receptor activation using
L-803087 (Fig. 3B).
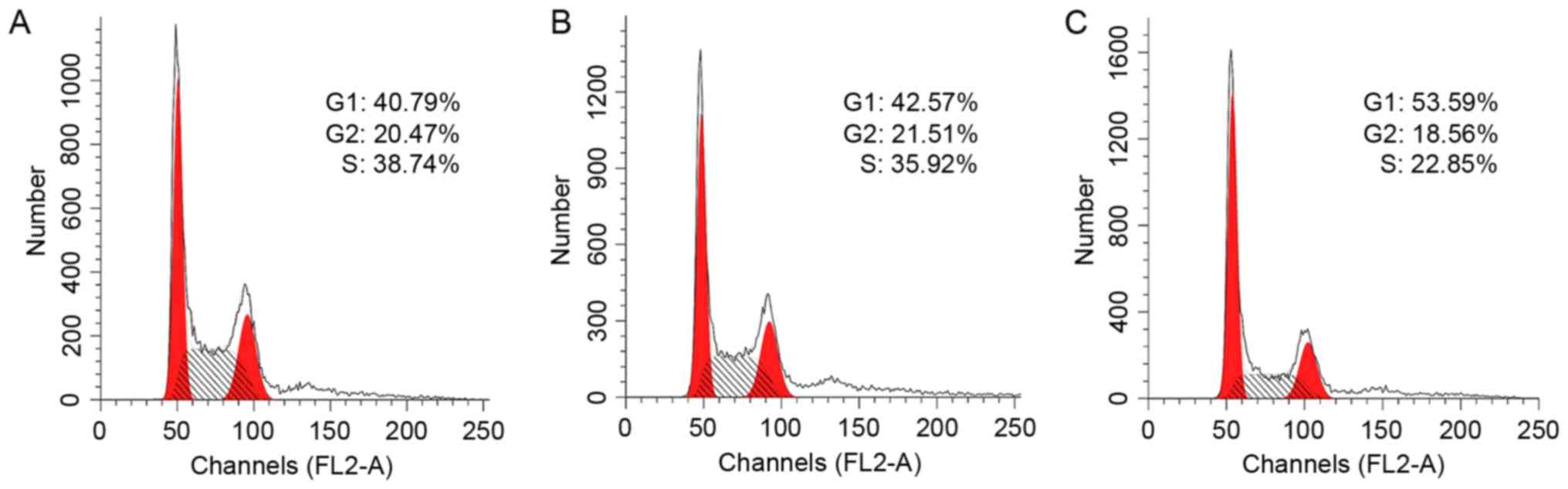
Overexpression and activation of
SSTR1/SSTR4 decreases cancer cell proliferation
The influence of SSTR expression and receptor
activation on cell proliferation was further investigated in
cultured MDA-MB-435S cells using flow cytometry. The cells were
transfected with pCMV-HA-SSTR1 and/or pcDNA-MYC-SSTR4, followed by
receptor activation using 10 nM L-803087. The control cells were
transfected with empty vectors and the mock cells were treated with
buffer only. The cell cycles of these cells were subsequently
analyzed using flow cytometry. Compared with the control
(39.45±0.63% cells in S-phase), the proportion of cells
overexpressing SSTR1 or SSTR4 was slightly decreased in S-phase
(38.36±2.71%; P>0.05; Fig. 4).
However, the proportion of S-phase cells overexpressing SSTR1 and
SSTR4 was significantly decreased when treated with 10 nM L-803087
(26.06±2.79%; P<0.05). The expression levels of the relevant
SSTRs in these cells were verified using western blot analysis
(data not shown). The results indicated that the inhibition of cell
proliferation was mediated via receptor activation and was
subtype-specific.
Discussion
SST is a natural inhibitory peptide and exerts its
anti-secretory/anti-proliferative actions via SSTRs, which have
been determined to be ubiquitously expressed in normal and cancer
tissues. Owing to the anti-proliferative effects of SSTR signaling,
analogs structurally similar to SST that have an increased
half-life and are receptor-subtype-selective have been developed
and frequently used as a complementary treatment in post-surgical
medication for a number of cancer types (1). However, numerous clinical trials
reported insensitivity to the treatment with SSA and the lack of
benefits was considered to be a consequence of the loss of
expression of SSTRs (30).
Furthermore, limited information was available on the detailed
expression patterns of SSTRs in large clinical trials.
Additionally, the underlying molecular mechanisms of receptor
activation, and the interplay between SSTRs and other signaling
pathways have, to the best of our knowledge, rarely been
investigated.
In the present study, the expression pattern of the
five SSTR subtypes in 160 primary ductal breast cancer tissues was
investigated. The results demonstrated that all five subtypes of
SSTR were expressed at variable levels in breast cancer tissues.
The positive expression instance of SSTR subtypes was highest for
SSTR1, followed by SSTR4, SSTR5, SSTR3 and then SSTR2. In contrast
with a previous study (31)
indicating that SSTR2 and SSTR3 were the most frequently expressed
subtypes, the expression levels of SSTR2 and SSTR3 proteins were
detected in only 34.4 and 41.9%, respectively, of breast tumor
tissues in the present study. Additionally, the expression levels
of SSTRs was negatively associated with tumor differentiation and
were independent of patient age. The positive expression levels of
all five SSTRs were decreased in well-differentiated tumor tissues.
Except for SSTR1, the results also indicated that the instances of
positive expression of SSTR2-4 increased in ER- or PR-negative
cells. This result may not reflect any association between SSTR and
ER/PR expression levels, as it may be a coincidence of decreased
expression levels of both in poorly differentiated cancer cells.
However, it could be a true regulation of SSTR expression by
hormone receptors since SST inhibited hormone secretion and
tamoxifen/estradiol-differentially regulated SSTR1/2 expression, as
reported previously (32).
Furthermore, previous studies demonstrated the association of the
expression of SSTR subtypes and ER in breast cancer and
non-functioning pituitary adenomas (33,34). The
detailed regulation of SSTR expression and the potential interplay
between SSTR and hormone receptors require further
investigation.
Owing to the expression pattern of SSTRs in breast
tumor tissues, the potential receptor dimerization between the most
frequently expressed SSTR subtypes was further investigated using
immunofluorescence. MDA-MB-435S cells were originally characterized
as a breast cancer cell line and were later identified to be
cross-contaminated with M14 melanoma cells (35). However, owing to their negative
endogenous expression of SSTR, MDA-MB-435S cells were selected for
the analysis of the potential interaction between SSTR1 and SSTR4
in vitro, to eliminate the interference of other subtypes of
endogenous SSTR. Immunofluorescence using confocal microscopy
revealed an association of the cellular distribution of
overexpressed SSTR1 and SSTR4. Additionally, the associated
distribution of SSTR1 and SSTR4 was significantly increased upon
receptor activation, indicating receptor dimerization as a
mechanism of receptor activation. The direct interaction between
SSTR1 and SSTR4 was further confirmed using co-immunoprecipitation,
which also indicated that the receptor dimerization was notably
induced by the subtype-specific SSA L-803087. Furthermore, instead
of the predominant plasma membrane-associated distribution,
L-803087 induced cytoplasmic translocation of SSTR1/SSTR4,
indicating the functional significance of the receptor
dimerization. The potential influence of SSA-induced SSTR1/SSTR4
dimerization on cell proliferation was investigated further. Using
flow cytometry, the results of the present study indicated that
overexpressing SSTR1 and SSTR4 had limited influence on cell
proliferation, compared with the control cells transfected with the
vector only. However, the proliferation of cells overexpressing
SSTR1 and SSTR4 was significantly inhibited upon
receptor-subtype-specific SSA, L-803087, activation, indicating
that the inhibition of cell proliferation was mediated by receptor
dimerization/activation.
Receptor dimerization has been identified to be
common for G-protein-coupled receptors (36). Dimerizations have been demonstrated
between SSTR subtypes, such as SSTR5 dimerized with SSTR1 and
SSTR2, but not SSTR4, as well as between SSTR and other
G-protein-coupled receptors, including epidermal growth factor
receptor and β1-adrenergic receptor (8,20,37–39). Owing
to the results of the present study and the previous data, it was
proposed that the receptor dimerization between SSTR subtypes could
be a general mechanism for SSTR signaling (8,20,37–39).
Additionally, the cross-talk between SSTR and other associated
G-protein-coupled receptors may be critical for SSTRs to confer
their cell-type-specific effects. Further investigation of the
interaction between other SSTR subtypes and their function will be
beneficial for understanding the subtype-specific cellular effects
and the mechanisms of SSTR signaling pathways, which will be
valuable for optimizing SSA treatment and selection of the patients
suitable for SSA treatment.
Acknowledgements
Not applicable.
Funding
The present study was sponsored by the National
Natural Science Foundation of China (grant no. 81571041) and the
Construction Project for the Key Laboratory of Virology of
Guangzhou (grant no. 201705030003).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
YZou conceived and designed the study, analyzed and
interpreted the data, drafted and revise the manuscript, and
approved the final manuscript. HT acquired the data, analyzed and
interpreted the data, and drafted the manuscript. YZha and YZho
acquired, analyzed and interpreted the data. LC conceived and
designed the study, analyzed and interpreted the data and approved
the final manuscript. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
Ethics approval was obtained from The Committee of
Medical Ethics of the First Affiliated Hospital of Jinan
University. Patients provided written informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Günther T, Tulipano G, Dournaud P,
Bousquet C, Csaba Z, Kreienkamp HJ, Lupp A, Korbonits M, Castaño
JP, Wester HJ, et al: International union of basic and clinical
pharmacology. CV. Somatostatin receptors: Structure, function,
ligands, and new nomenclature. Pharmacol Rev. 70:763–835. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Day R, Dong W, Panetta R, Kraicer J,
Greenwood MT and Patel YC: Expression of mRNA for somatostatin
receptor (sstr) types 2 and 5 in individual rat pituitary cells. A
double labeling in situ hybridization analysis. Endocrinology.
136:5232–5235. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Florio T: Somatostatin/somatostatin
receptor signalling: Phosphotyrosine phosphatases. Mol Cell
Endocrinol. 286:40–48. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zatelli MC, Piccin D, Tagliati F, Bottoni
A, Luchin A, Vignali C, Margutti A, Bondanelli M, Pansini GC and
Pelizzo MR: Selective activation of somatostatin receptor subtypes
differentially modulates secretion and viability in human medullary
thyroid carcinoma primary cultures: Potential clinical
perspectives. J Clin Endocrinol Metab. 91:2218–2224. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ruscica M, Magni P, Steffani L, Gatto F,
Albertelli M, Rametta R, Valenti L, Ameri P, Magnaghi V and Culler
MD: Characterization and sub-cellular localization of SS1R, SS2R,
and SS5R in human late-stage prostate cancer cells: Effect of mono-
and bi-specific somatostatin analogs on cell growth. Mol Cell
Endocrinol. 382:860–870. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Theodoropoulou M and Stalla GK:
Somatostatin receptors: From signaling to clinical practice. Front
Neuroendocrinol. 34:228–252. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
War SA and Kumar U: Coexpression of human
somatostatin receptor-2 (SSTR2) and SSTR3 modulates
antiproliferative signaling and apoptosis. J Mol Signal. 7:1–15.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Rocheville M, Lange DC, Kumar U, Sasi R,
Patel RC and Patel YC: Subtypes of the somatostatin receptor
assemble as functional homo- and heterodimers. J Biol Chem.
275:7862–7869. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Missiaglia E, Dalai I, Barbi S, Beghelli
S, Falconi M, Della PM, Piemonti L, Capurso G, Di FA and Delle FG:
Pancreatic endocrine tumors: Expression profiling evidences a role
for AKT-mTOR pathway. J Clin Oncol. 28:245–255. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Vikić-Topić S, Raisch KP, Kvols LK and
Vuk-Pavlović S: Expression of somatostatin receptor subtypes in
breast carcinoma, carcinoid tumor, and renal cell carcinoma. J Clin
Endocrinol Metab. 80:2974–2979. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Skanberg J, Ahlman H, Benjegard SA,
Fjalling M, Forssell Aronsson EB, Hashemi SH, Nilsson O, Suurkula M
and Jansson S: Indium-111-octreotide scintigraphy, intraoperative
gamma-detector localisation and somatostatin receptor expression in
primary human breast cancer. Breast Cancer Res Treat. 74:101–111.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cakir M, Dworakowska D and Grossman A:
Somatostatin receptor biology in neuroendocrine and pituitary
tumours: Part 1-molecular pathways. J Cell Mol Med. 14:2585–2591.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Callison JC Jr, Walker RC and Massion PP:
Somatostatin receptors in lung cancer: From function to molecular
imaging and therapeutics. J Lung Cancer. 10:69–76. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Watt HL, Kharmate G and Kumar U: Biology
of somatostatin in breast cancer. Mol Cell Endocrinol. 286:251–261.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hicks RJ: Use of molecular targeted agents
for the diagnosis, staging and therapy of neuroendocrine
malignancy. Cancer Imaging 10 Spec no A. S83–S91. 2010. View Article : Google Scholar
|
|
16
|
Reubi JC, Schaer JC, Waser B and Mengod G:
Expression and localization of somatostatin receptor SSTR1, SSTR2,
and SSTR3 messenger RNAs in primary human tumors using in Situ
Hybridization. Cancer Res. 54:3455–3459. 1994.PubMed/NCBI
|
|
17
|
Schmid HA, Lambertini C, Van Vugt HH,
Barzaghi-Rinaudo P, Schäfer J, Hillenbrand R, Sailer AW, Kaufmann M
and Nuciforo P: Monoclonal antibodies against the human
somatostatin receptor subtypes 1-5: Development and
immunohistochemical application in neuroendocrine tumors.
Neuroendocrinology. 95:232–247. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hofland LJ and Lamberts SW: Somatostatin
receptors and disease: Role of receptor subtypes. Baillières Clin
Endocrinol Metab. 10:163–176. 1996. View Article : Google Scholar
|
|
19
|
Wang S, Bao Z, Liang QM, Long JW, Xiao ZS,
Jiang ZJ, Liu B, Yang J and Long ZX: Octreotide stimulates
somatostatin receptor-induced apoptosis of SW480 colon cancer cells
by activation of glycogen synthase kinase-3beta, A Wnt/beta-catenin
pathway modulator. Hepatogastroenterology. 60:1639–1646.
2013.PubMed/NCBI
|
|
20
|
Grant M, Alturaihi H, Jaquet P, Collier B
and Kumar U: Cell growth inhibition and functioning of human
somatostatin receptor type 2 are modulated by receptor
heterodimerization. Mol Endocrinol. 22:2278–2292. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hasskarl J, Kaufmann M and Schmid HA:
Somatostatin receptors in non-neuroendocrine malignancies: The
potential role of somatostatin analogs in solid tumors. Future
Oncol. 7:895–913. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Raderer M, Hejna MH, Muller C, Kornek GV,
Kurtaran A, Virgolini I, Fiebieger W, Hamilton G and Scheithauer W:
Treatment of hepatocellular cancer with the long acting
somatostatin analog lanreotide in vitro in vivo. Int J
Oncol. 16:1197–1201. 2000.PubMed/NCBI
|
|
23
|
Li M, Zhang R, Li F, Wang H, Kim HJ,
Becnel L, Yao Q, Chen C and Fisher WE: Transfection of SSTR-1 and
SSTR-2 Inhibits Panc-1 Cell proliferation and renders Panc-1 cells
responsive to somatostatin analogue. J Am Coll Surg. 201:571–578.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Arena S, Barbieri F, Thellung S, Pirani P,
Corsaro A, Villa V, Dadati P, Dorcaratto A, Lapertosa G, Ravetti
JL, et al: Expression of somatostatin receptor mRNA in human
meningiomas and their implication in in vitro antiproliferative
activity. J Neurooncol. 66:155–166. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Barbieri F, Pattarozzi A, Gatti M, Aiello
C, Quintero A, Lunardi G, Bajetto A, Ferrari A, Culler MD and
Florio T: Differential efficacy of SSTR1, −2, and −5 agonists in
the inhibition of C6 glioma growth in nude mice. Am J Physiol
Endocrinol Metab. 297:1078–1088. 2009. View Article : Google Scholar
|
|
26
|
Xu Y, Song J, Berelowitz M and Bruno JF:
Estrogen regulates somatostatin receptor subtype 2 messenger
ribonucleic acid expression in human breast cancer cells.
Endocrinology. 137:5634–5640. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Patel YC: Somatostatin and its receptor
family. Front Neuroendocrinol. 20:157–198. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Bonifacino JS, Gershlick DC and
Dell'Angelica EC: Immunoprecipitation. Curr Protoc Cell Biol.
1:712016.
|
|
29
|
Bansal C, Singh US, Misra S, Sharma KL,
Tiwari V and Srivastava AN: Comparative evaluation of the modified
Scarff-Bloom-Richardson grading system on breast carcinoma
aspirates and histopathology. Cytojournal. 9:42012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Mohamed A, Romano D, Saveanu A, Roche C,
Albertelli M, Barbieri F, Brue T, Niccoli P, Delpero JR, Garcia S,
et al: Anti-proliferative and anti-secretory effects of everolimus
on human pancreatic neuroendocrine tumors primary cultures: Is
there any benefit from combination with somatostatin analogs?
Oncotarget. 8:41044–41063. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kumar U, Grigorakis SI, Watt HL, Sasi R,
Snell L, Watson P and Chaudhari S: Somatostatin receptors in
primary human breast cancer: Quantitative analysis of mRNA for
subtypes 1-5 and correlation with receptor protein expression and
tumor pathology. Breast Cancer Res Treat. 92:175–186. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Rivera JA, Alturaihi H and Kumar U:
Differential regulation of somatostatin receptors 1 and 2 mRNA and
protein expression by tamoxifen and estradiol in breast cancer
cells. J Carcinog. 4:102005. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Nishioka H, Tamura K, Iida H, Kutsukake M,
Endo A, Ikeda Y and Haraoka J: Co-expression of somatostatin
receptor subtypes and estrogen receptor-α mRNAs by non-functioning
pituitary adenomas in young patients. Mol Cell Endocrinol.
331:73–78. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Frati A, Rouzier R, Lesieur B, Werkoff G,
Antoine M, Rodenas A, Darai E and Chereau E: Expression of
somatostatin type-2 and −4 receptor and correlation with
histological type in breast cancer. Anticancer Res. 34:3997–4003.
2014.PubMed/NCBI
|
|
35
|
Rae JM, Creighton CJ, Meck JM, Haddad BR
and Johnson MD: MDA-MB-435 cells are derived from M14 melanoma
cells-a loss for breast cancer, but a boon for melanoma research.
Breast Cancer Res Treat. 104:13–19. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wang W, Qiao Y and Li Z: New insights into
modes of GPCR activation. Trends in Pharmacol Sci. 39:367–386.
2018. View Article : Google Scholar
|
|
37
|
Somvanshi RK, War SA, Chaudhari N, Qiu X
and Kumar U: Receptor specific crosstalk and modulation of
signaling upon heterodimerization between β1-adrenergic receptor
and somatostatin receptor-5. Cell Signal. 23:794–811. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Watt HL, Kharmate GD and Kumar U:
Somatostatin receptors 1 and 5 heterodimerize with epidermal growth
factor receptor: Agonist-dependent modulation of the downstream
MAPK signalling pathway in breast cancer cells. Cell Signal.
21:428–439. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Watt HL and Kumar U: Colocalization of
somatostatin receptors and epidermal growth factor receptors in
breast cancer cells. Cancer Cell Int. 6:1–19. 2006. View Article : Google Scholar : PubMed/NCBI
|