Introduction
Bladder cancer is one of the most significant
worldwide health problems, accounting for an estimated 386,300 new
cases it is the ninth most common cause of cancer, and was
responsible for 150,200 cancer-associated deaths in 2008 (1). Histologically, bladder urothelial
carcinoma accounts for >90% of newly diagnosed bladder
malignancies (2,3). However, the cause of bladder cancer,
like the majority of other types of human cancer, remains to be
defined, although tobacco smoking is linked to ~50% of bladder
cases (4). Clinically, early symptoms
of bladder cancer include macroscopic and/or microscopic blood in
the urine, pain during urination and frequent urination, all of
which are non-specific (5). The early
stage of bladder cancer recurs in 50–70% of patients. Seeing as
advanced stages of the disease remain difficult to control,
effective therapeutic control of tumor recurrence is required at an
early stage (5). Thus, further
studies of bladder cancer are required to better understand the
molecular mechanisms underlying urothelial carcinogenesis in order
to provide novel strategies to prevent bladder carcinogenesis,
diagnose bladder cancer early and effectively treat or cure bladder
cancer in clinic.
Cell adhesion molecule 1 (CADM1) is a novel tumor
suppressor gene that is localized at chromosome 11q23.2. CADM1
protein is a transmembrane glycoprotein with 442 amino acids and
belongs to an immunoglobulin cell adhesion superfamily (6,7); thus,
CADM1 protein has structural homology to the neural cell adhesion
molecules and cell signaling transduction (8) and is implicated in calcium ion
independent cell-cell adhesion (9).
CADM1 was reported to be abrogated or significantly reduced in a
number of human cancer tissues and cell lines, including esophageal
cancer (10), cutaneous melanoma
(11), hepatocellular carcinoma
(12), ovarian carcinoma (13), breast cancer (14,15),
pancreatic ductal adenocarcinoma (16), lung cancer (17), laryngeal squamous cell carcinoma
(18), colorectal cancer (19), prostate cancer (20), neuroblastoma (21) and nasopharyngeal carcinoma (22,23). Lost
CADM1 expression was associated with poor prognosis and development
of esophageal cancer (10) and
ovarian cancer (13). CADM1 promoter
hypermethylation may be responsible for lost CADM1 expression
(16,19,20). In
bladder cancer, numerous previous studies analyzed altered
methylation of CADM1 promoter in non-muscle-invasive bladder
cancer (24–26) and bladder cancer with various stages
(27). The results revealed an
increase in methylation of CADM1 promoter in
non-muscle-invasive bladder cancer, with the exception of the study
by Hellwinkel et al (27). To
assess the contradiction, the present study further investigated
the expression and the function of CADM1 in bladder tissues. In
order to assess the associations with clinicopathological data of
patients, we detected the methylation status of GpG islands of
CADM1 promoter and protein expression levels of bladder cancer
tissues. Then we investigated the effects of CADM1 overexpression
on the regulation of bladder cancer cell viability, apoptosis,
invasion and gene expression in vitro. The present study is
expected to provide useful evidence for CADM1 as a biomarker for
early detection of bladder cancer and prediction of disease
progression and it may be a novel target for future control of
bladder cancer in clinical practice.
Materials and methods
Tissue specimens
A total of 84 bladder cancer tissues from patients
with bladder cancer (58 male with mean age of 68.5±9.9 years and 26
female with mean age of 69.4±7.3 years) and 20 normal bladder
mucosae were collected from patients with benign prostate
hyperplasia were collected from the Department of Urology, The
Second Hospital of Tianjin Medical University (Tianjin, China)
between March 2012 and April 2014. All tissue specimens were
collected within 30 min following surgery, snap-frozen and stored
in an −80°C freezer. Bladder cancer was histopathologically
confirmed by Jiwu Chang and Aixiang Wang, pathologists of The
Tianjin Institute of Urology, according to the 2004 WHO
classification of tumor of urinary system (28) and 2009 TNM classification of malignant
tumors (29). The present study also
obtained 2 ml peripheral blood samples from each of the 20
cancer-free individuals. The present study was approved by the
Ethics Committee of The Second Hospital of Tianjin Medical
University and written informed consent was obtained from each
patient prior to enrollment in the present study.
Cell line and culture
The human bladder cancer T24 cell line was obtained
from Tianjin Institute of Urology (Tianjin, China) and cultured in
Dulbecco's modified Eagle's medium (DMEM) supplemented with 10%
fetal bovine serum (Invitrogen; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA), penicillin (100 U/ml; Sigma-Aldrich; Merck KGaA,
Darmstadt, Germany) and streptomycin (100 ·g/ml; Sigma-Aldrich;
Merck KGaA) in a humidified incubator with 5% CO2 at
37°C.
Bisulfite modification and methylation
specific PCR
Genomic DNA was extracted from bladder cancer
tissues, normal bladder mucosae and peripheral blood mononuclear
cells using the E.Z.N.A.™ Tissue DNA kit (Omega Bio-Tek, Norcross,
GA, USA). The purity and concentration of DNA was measured using a
DU800 ultraviolet spectrophotometer (Beckman-Coulter, Inc., Brea,
CA, USA). Following quantification, these DNA samples were
subjected to bisulfite modification using EZ DNA Methylation-Gold
kit TM (Omega Bio-Tek). Briefly, 1 µg genomic DNA sample of each
was incubated with the kit reagents, according to the
manufacturer's protocol, and 4 µl of the treated DNA samples were
amplified by PCR using primers specific for either the unmethylated
or methylated template in a 50 µl PCR mixture. Primers used for
this methylated reaction were as follows: Forward,
5′-TTTTAATTATTATTTTCGAGTTTATCG-3′ and reverse,
5′-TCTTTAAAAACGAAAACTATACG-3′, and primers for the unmethylated
reaction were: Forward, 5′-TTTTTTAATTATTTTTGAGTTTATTG-3′ and
reverse, 5′-AAATCTTTAAAAACAAAAACTATAC-3′. PCR amplification
conditions were set for an initial denaturation at 95°C for 10 min,
followed by 35 cycles of 94°C for 30 sec, 57°C (for methylated
primers) or 55°C (for unmethylated primers) for 30 sec and a final
extension at 72°C for 3 min. Genomic DNA from normal pancreatic
tissues treated with or without SssI methyltransferase was used as
a positive or negative control, respectively. The PCR products were
then separated in 2% agarose gels, stained with ethidium bromide
and visualized under UV illumination.
Construction of lentiviral vector
carrying CADM1 cDNA and short interfering (si)RNAs, production of
viral particles and viral infection
Human CADM1 full-length cDNA (accession number
NM_014333) was amplified using reverse transcription (RT)-PCR with
CADM1-specific primers containing XhoI and NotI
restriction enzyme sites (forward,
5′-AATCTCGAGATGGCGAGTGTAGTGCTGCC-3′ and reverse,
5′-ATAGCGGCCGCCTAGATGAAGTACTCTTTCT-3′). PCR was performed using the
LA Taq™ System (Takara Biotechnology Co., Ltd., Dalian, China) for
30 cycles of 94°C for 40 sec, 55°C for 60 sec and 72°C for 50 sec,
according to the manufacturer's protocol. The PCR products were
ligated into the pHBLV-IRES-ZsGreen-PGK-puro vector (Hanbio
Biotechnology Co., Ltd., Shanghai, China) to obtain the Ad-CADM1
vector. Furthermore, 3 siRNAs were also designed to silence CADM1
expression and the DNA sequences of these siRNAs were: siRNA1,
5′-CAGATGACTTATCCTCTACAA-3′; siRNA2, 5′-CCAGACATAAAGGTACATACT-3′;
and siRNA3, 5′-AGACGCAGACACAGCTATAAT-3′. These DNA sequences were
ligated into the pHBLV-U6-ZsGreen-Puro vector (Hanbio Biotechnology
Co., Ltd.) to obtain sh-CADM1. These lentivirus
pHBLV-IRES-ZsGreen-PGK-puro- CADM1 (Ad-CADM1),
pHBLV-U6-ZsGreen-Puro-sh1/2/3 (Ad-sh1/2/3) or pHBLV-U6-ZsGreen-Puro
(Ad-GFP2), and lentivirus packaging plasmid pSPAX2 and pMD2G or
pHBLV-IRES-ZsGreen-PGK-puro (Ad-GFP1) were then using
Lipofectamine™ 2000 (Life Technologies) according to the
manufacturer's instructions into human embryonic kidney 293T cells
for homologous recombination and production of lentiviral
particles. Each recombinant virus was purified by
ultracentrifugation at 4°C and at 72,000 × g for 120 min. Titers of
these viral particles were assessed for multiplicity of infection
(MOI).
Human bladder cancer T24 cells were plated at a
density 5×105 cell in 6-wells plates and grown overnight
prior to infection with these lentiviruses at an MOI of 20 for 24 h
at 36°C, 48 and 72 h at 36°C. Expression of green fluorescence
protein (GFP) was detected under a fluorescence microscope for
infection efficiency in ≥5 fields of view.
RT-qPCR
Total RNA was isolated from parental T24,
T24-Ad-CADM1, T24-Ad-sh1/2/3 and T24-Ad-GFP1/2 cells using the
TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.),
according to the manufacturer's protocol. qPCR was performed on ABI
7900 96 HT series PCR machine (Applied Biosystems; Thermo Fisher
Scientific, Inc.) using FastStart Universal SYBR Green Master (ROX)
kit (Roche). qPCR was then performed with the specific CADM1
primers (forward, 5′-ATGGCGAGTGTAGTGCTGC-3′ and reverse,
5′-GATCACTGTCACGTCTTTCGT-3′). Relative CADM1 expression level was
then normalized to levels of GAPDH (forward,
5′-GGAGCGAGATCCCTCCAAAAT-3′ and reverse,
5′-GGCTGTTGTCATACTTCTCATGG-3′) as an internal control. PCR
conditions were 50°C for 2 min and 95°C for 10 min followed by 40
cycles of 95°C for 15 sec, and 60°C for 1 min. The expression
levels were normalized against glyceraldehyde
3-phosphatedehydrogenase (GAPDH) (Human GAPDH Endogenous Control,
Applied Biosystems; Thermo Fisher Scientific, Inc.). The results
were summarized from triplicate experiments using the
2−∆∆Cq method (30).
Protein extraction and western
blotting
Total cellular protein was extracted from parental
T24, T24-Ad-CADM1, T24-Ad-sh1/2/3 and T24-Ad-GFP1/2 cells or from
tissue specimens using a lysis buffer containing PBS, Triton X-100,
PMSF, leupeptine and pepstain (Boster Biological Technology,
Pleasanton, CA, USA). Following quantification using a Bio-Rad
Protein Assay kit II (Bio-Rad Laboratories, Inc., Hercules, CA,
USA), these protein samples (15 µg of each tissue or cell lysate)
were subjected to 10–15% SDS-PAGE and transferred onto
polyvinylidene fluoride membranes (EMD Millipore, Billerica, MA,
USA). The Western blotting was performed according to standard
methods using antibodies against CADM1 (1:1,000 dilution; catalog
no. H00023705-M02; Abnova, Taipei, Taiwan) and GAPDH (1:1,000
dilution; catalog no. ab70699), Caspase-3 (1:1,000 dilution;
catalog no. ab2171), Bcl-2 (1:1,000 dilution; catalog no. ab32124),
Bax (1:1,000 dilution; catalog no. ab32503E-cadherin (1:1,000
dilution; catalog no. ab1416), β-catenin (1:1,000 dilution; catalog
no. ab16051), vimentin (1:1,000 dilution; catalog no. ab92547), p27
(1:1,000 dilution; catalog no. ab32034), cyclinD1 (1:1,000
dilution; catalog no. ab134175), cyclinE1 (1:1,000 dilution;
catalog no. ab3927), CDK2 (1:1,000 dilution; catalog no. ab32147)
(all Abcam, Cambridge, UK). Briefly, the membranes were blocked
with 5% skimmed milk solution in PBS for 1 h at the room
temperature and incubated with a rat anti-CADM1, followed by
biotinylated goat anti rat IgG secondary antibody (1:1,000
dilution; catalog no. SAB4600463; Sigma-Aldrich; Merck KGaA) for 2
h at 37°C. The membranes were incubated with enhanced
chemiluminescence solution (PerkinElmer, Inc., Waltham, MA, USA).
The images were acquired using an Image Quant350 digital image
system (GE Healthcare Life Sciences, Chalfont, UK). Expression
levels of proteins were semi-quantified by evaluating the gray
scale using Image-ProPlus version 5.0 software (Media Cybernetics,
Inc., Rockville, MD, USA). Expression levels of GAPDH protein were
used as a loading control. The relative expression levels of
protein were determined as the CADM1/GAPDH ratio.
Cell viability assay
Cellular viability was assessed using an MTT assay,
according to the manufacturers protocol. Briefly, parental T24,
T24-Ad-CADM1, T24-Ad-sh1 and T24-Ad-GFP1/2 cells were cultured for
24, 48, 72 and 96 h at 37°C. The cells were further incubated with
0.5 mg/ml MTT for 4 h at 37°C and the culture medium was replaced
with 200 µl dimethyl sulfoxide to dissolve the purple precipitates
for 20 min at room temperature. The absorbance rate was evaluated
at 490 nm using a multi-well plate reader. The experiments were
performed in triplicate and repeated ≥3 times.
Tumor cell invasion assay
Cell invasion capacity was assayed using 24-well
Transwell chambers (8 mm pore size; BD Biosciences, Franklin Lakes,
NJ, USA), according to the manufacturers protocol. In brief, 150 µl
2×105/ml parental T24, T24-Ad-CADM1, T24-Ad-sh1 and
T24-Ad-GFP1/2 cells in serum-free medium were seeded into the upper
chambers, and 800 µl DMEM supplemented with 10% fetal bovine serum
was added to the lower chambers and cultured for 24 h at 37°C.
Cells that remained in the upper chambers were removed with a
cotton swab and the cells that invaded through the Matrigel and
attached to the lower surface of the membrane were fixed and
stained with 1% crystal violet solution. Subsequently, the membrane
of each chamber was removed, mounted on microslides and was fixed
using permount™ mounting medium (Electron Microscopy Sciences,
Hatfield, PA, USA). In total, 6 random fields were used to capture
images under a light microscope and counted. Each cell line
preparation was seeded into 4 chambers and the cell density in each
chamber was 2×105 cells/ml.
Flow cytometric cell cycle assay
Parental T24, T24-Ad- CADM1, T24-Ad-sh1 and
T24-Ad-GFP1/2 cells were harvested and single-cell suspensions were
prepared by passing the cells through a nylon mesh. Subsequently,
cells were fixed in 70% ethanol at −20°C for 2 h, washed 3 times
with ice-cold PBS and suspended in propidium iodide (PI) buffer
(Invitrogen; Thermo Fisher Scientific, Inc.) with cells adjusted to
a density of 4×106/ml. The cells were then incubated
with 5 µg/ml propidium iodide (Sigma-Aldrich; Merck KGaA) at room
temperature for 30 min in the dark. The single cells were then
subjected to FACScan analysis (Becton-Dickinson, Heidelberg,
Germany). Cell cycle distribution was calculated using ModFIT cell
cycle analysis software (version 2.01.2; Becton Dickinson). Each
experiment was repeated at least three times on different days for
validation.
Statistical analysis
All statistical analyses were performed using SPSS
version 17.0 (SPSS Inc., Chicago, IL, USA). The data are expressed
as the mean ± standard deviation, except when otherwise stated. The
Mann-Whitney-U test was performed to compare protein concentrations
in tissue specimens. χ2 test was used to compared the
values of the test and control samples statistically. P<0.05 was
considered to indicate a statistically significant difference.
Results
Association of CADM1 promoter
methylation with bladder cancer progression ex vivo
In the present study, methylation of status CADM1
promoter was assessed in 84 bladder cancer tissues vs. 20 normal
bladder mucosae and it was revealed that CADM1 promoter was
methylated in 43/84 (51.2%) bladder urothelium carcinoma tissues
vs. none of the 20 normal tissues (Table
I). The methylation status of the CADM1 promoter was
significantly different between normal bladder mucosae and bladder
cancer tissues (P<0.01; Table I).
The methylation of CADM1 promoter in bladder cancer tissues was
significantly associated with tumor size, recurrence, pathology
classification and clinical stage (P<0.05; Table I), but not with patients age, gender
and tumor number (P>0.05).
 | Table I.Association of CADM1 promoter
methylation with clinicopathological features from bladder cancer
patients. |
Table I.
Association of CADM1 promoter
methylation with clinicopathological features from bladder cancer
patients.
| Clinicopathological
features | Number | Methylated, n
(%) | Unmethylated, n
(%) | χ2
value | P-value |
|---|
| Gender |
|
|
|
|
|
|
Male | 58 | 34 (58.6) | 24 (41.4) | 1.12 | 0.346 |
|
Female | 26 | 14 (53.8) | 12 (46.2) |
|
|
| Age, years |
|
|
|
|
|
|
≤65 | 33 | 18 (54.5) | 15 (45.5) | 0.044 | 1 |
|
>65 | 51 | 29 (56.9) | 22 (43.1) |
|
|
| Tumor number |
|
|
|
|
|
|
Single | 34 | 20 (58.8) | 14 (41.2) | 0.23 | 0.654 |
|
Multiple | 50 | 32 (64.0) | 18 (36.0) |
|
|
| Tumor
recurrent |
|
|
|
|
|
|
Yes | 49 | 33 (67.3) | 16 (32.7) | 7.51 | 0.008 |
| No | 35 | 13 (37.1) | 22 (62.9) |
|
|
| Tumor size
(cm) |
|
|
|
|
|
| ≤3 | 49 | 21 (42.9) | 28 (57.1) | 5.42 | 0.027 |
|
>3 | 35 | 24 (68.6) | 11 (31.4) |
|
|
| Tumor grade |
|
|
|
|
|
|
Low | 57 | 25 (43.9) | 32 (56.1) | 6.72 | 0.011 |
|
High | 27 | 20 (74.1) | 7 (25.9) |
|
|
| TNM stage |
|
|
|
|
|
|
Ta-T1 | 50 | 22 (44.0) | 28 (56.0) | 8.71 | 0.004 |
|
T2-T4 | 34 | 26 (76.5) | 8 (23.5) |
|
|
Association of CADM1 promoter
methylation with expression level of CADM1 protein in bladder
tissues
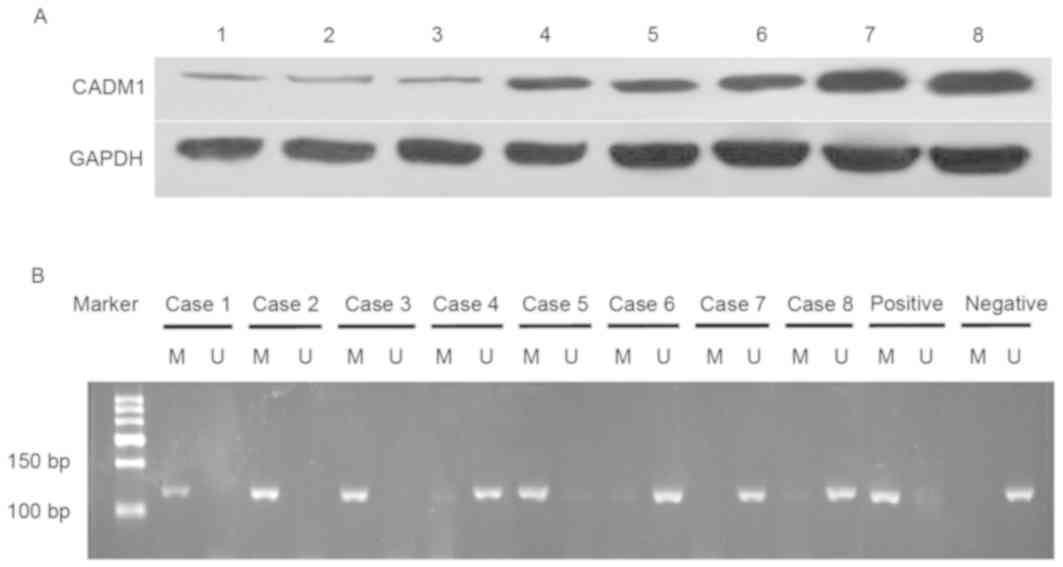
The present study analyzed levels of CADM1 protein
in fresh bladder tissues and demonstrated that CADM1 protein
expression level was lower in bladder cancer tissues compared with
in normal tissues (0.261±0.141 vs. 0.696±0.092; P<0.01; Fig. 1).
Effects of CADM1 overexpression and
knockdown on the regulation of bladder cancer cell viability,
apoptosis and cell cycle distribution
The biological effects of CADM1 on bladder cancer
were investigated using infected T24 bladder cancer cells with
lentivirus carrying CADM1 cDNA or siRNAs to established CADM1
stable overexpression cell line T24-Ad-CADM1 and or knockdown cell
line T24-Ad-sh1/2/3, compared with the empty vector-transfected
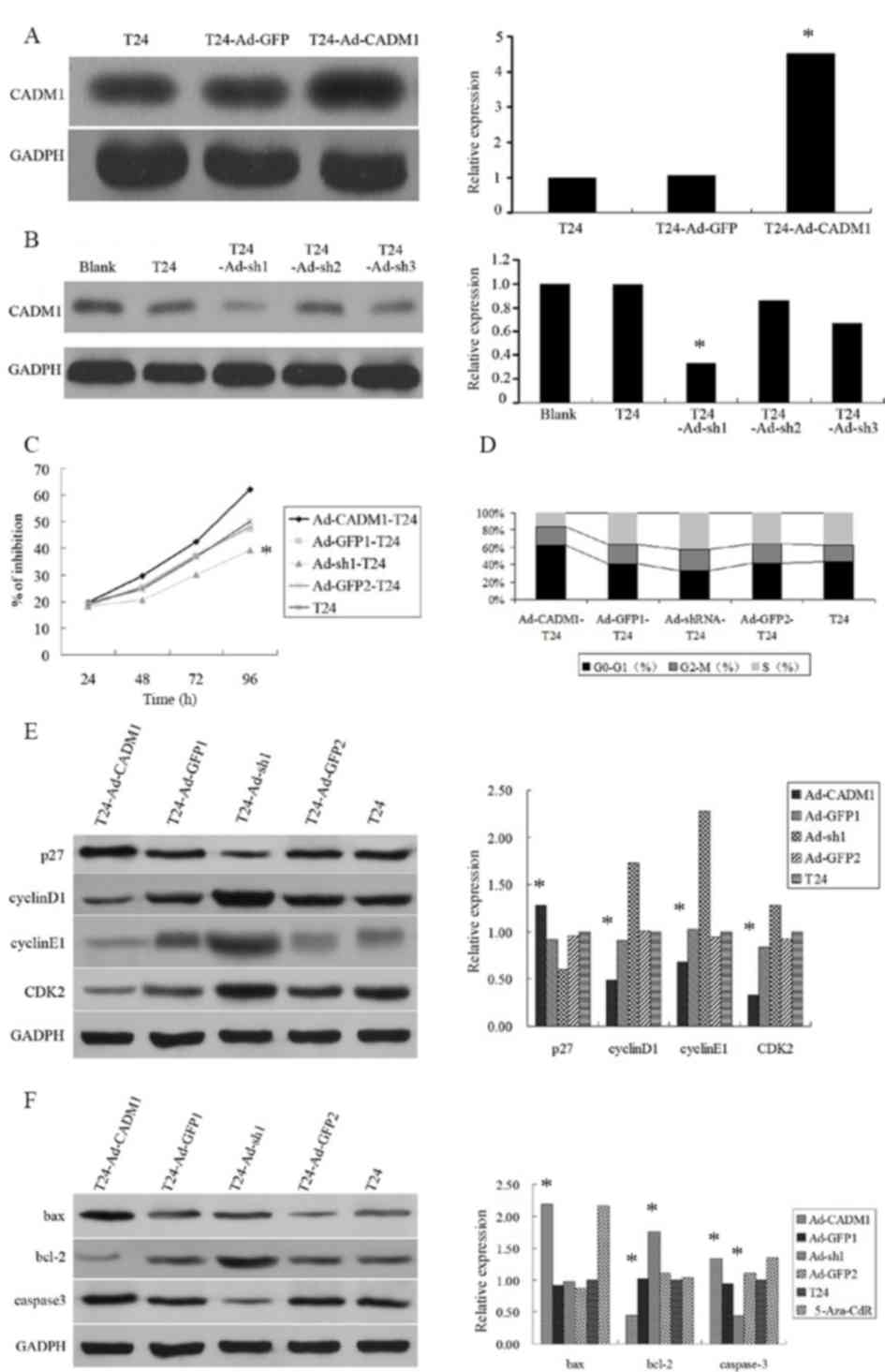
control cell line T24-Ad-GFP1/2. The results revealed that
lentivirus carrying CADM1 cDNA overexpressed CADM1 protein, whereas
lentivirus carrying CADM1 siRNA reduced CADM1 expression level in
T24 bladder cancer cells (Fig. 2A and
B).
The present study then assessed the altered
phenotypes of the T24 bladder cancer cells using MTT and cell cycle
flow cytometric assays and demonstrated that overexpression of
CADM1 reduced cell viability and arrested cells in the
G1/G0 phases of the cell cycle (Fig. 2C and D). However, knockdown of CADM1
expression level promoted cell growth and cell cycle progression to
the S phase (Fig. 2D).
Effects of CADM1 overexpression and
knockdown on the regulation of gene expression in T24 bladder
cancer cells
To further explore the molecular mechanisms
underlying CADM1 overexpression inhibition of T24 cell
proliferation, expression levels of cell growth and cell
cycle-associated proteins, including p27, cyclinD1, CDK2 and
cyclinE1, were determined. It was revealed that the expression
level of p27 protein in T24-Ad-CADM1 cells was significantly higher
compared with in parental T24 cells and T24-Ad-GFP1 cells
(P<0.05). Whereas, the expression levels of cyclinD1, CDK2, and
cyclinE1 protein in T24-Ad-CADM1 cells was markedly downregulated
compared with in parental T24 cells and T24-Ad-GFP1 cells
(P<0.05; Fig. 2E). Conversely,
CADM1 knockdown had a contrary effect (Fig. 2E). Furthermore, the present study also
assessed the altered apoptosis-associated gene expression level and
the data demonstrated that the expression levels of caspase-3 and
Bax proteins were increased and bcl-2 protein was markedly
decreased in T24-Ad-CADM1 cells compared with in parental T24 cells
and T24-Ad-GFP1 (P<0.05; Fig. 2F).
Conversely, the expression level of caspase-3 protein was markedly
decreased and bcl-2 protein was increased in T24-Ad-sh1 cells
compared with in parental T24 cells and T24-Ad-GFP2 (P<0.05;
Fig. 2F).
Effects of CADM1 overexpression on
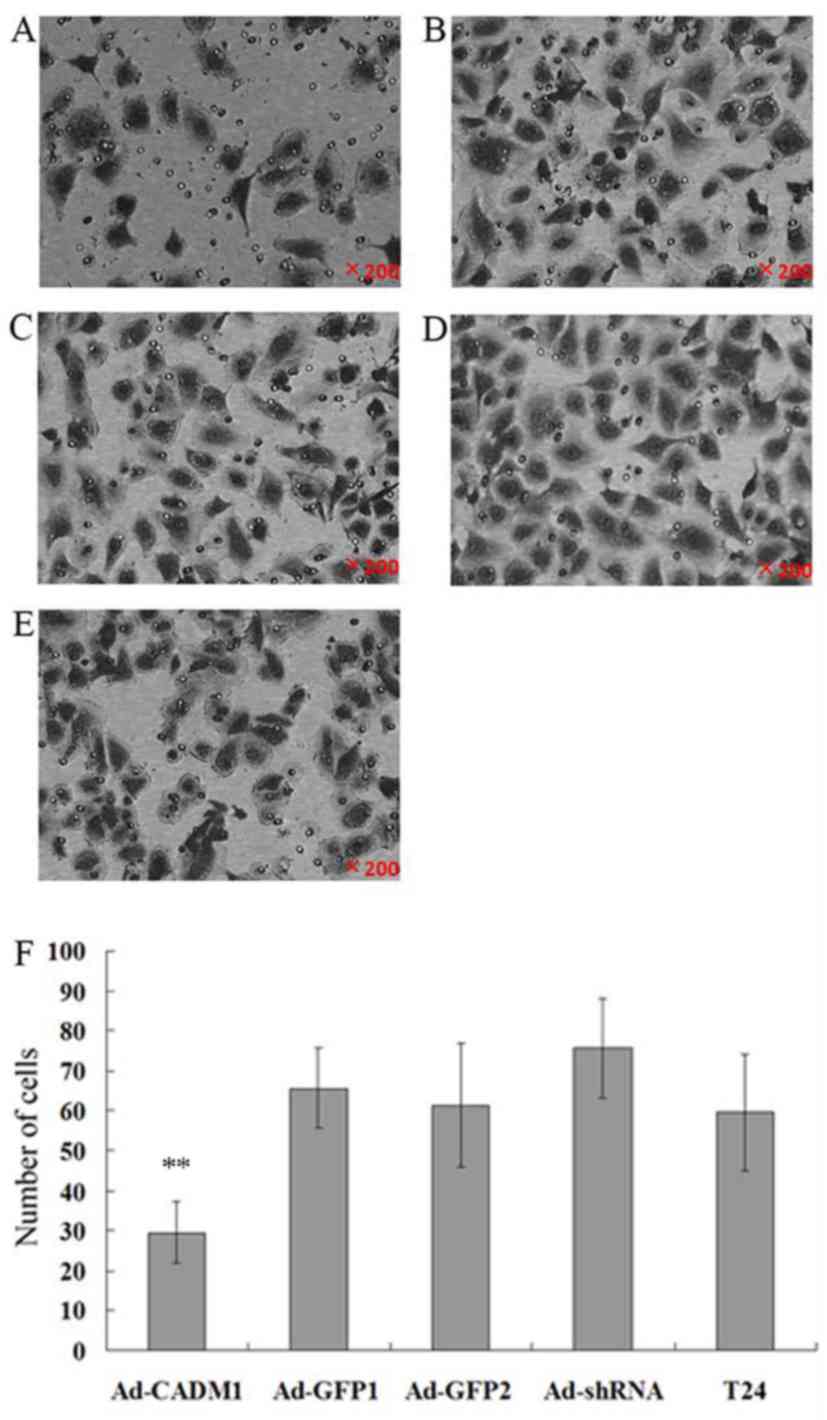
regulation of tumor cell invasion
Since CADM1 functions to regulate cell adhesion and
mobility, the present study assessed CADM1 overexpression or
knockdown in the regulation of tumor cell invasion and gene
expression. The tumor cell invasion results revealed that the
numbers of cells invaded into the lower chamber in T24-Ad-CADM1,
T24-Ad-GFP1, T24-Ad-sh1, T24-Ad-GFP2 and parental T24 cells were
29.54±7.66, 65.57±10.02, 75.61±12.53, 61.32±15.43 and 59.54±14.58,
respectively. Thus, compared with in parental T24 and T24-Ad-GFP1
cells, the overexpression of CADM1 significantly suppressed tumor
cell invasion capacity (P<0.01; Fig.
3). However, CADM1 knockdown significantly increased tumor cell
invasion (P<0.01; Fig. 3).
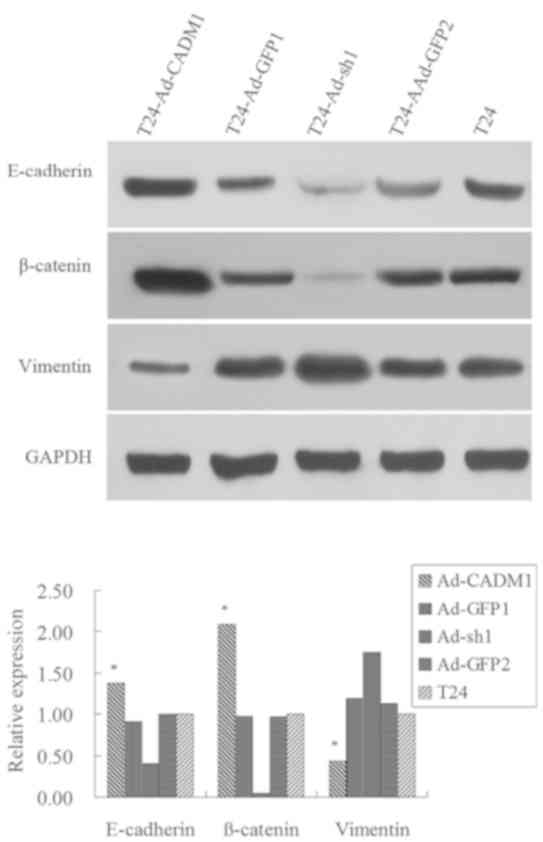
Furthermore, the present study assessed the
expression levels of key factors or markers of epithelial to
mesenchymal transition (EMT), including E-cadherin, β-catenin and
Vimentin. The results revealed that the expression levels of
E-cadherin and β-catenin proteins were markedly increased in
T24-Ad-CADM1 cells compared with those in parental T24 and
T24-Ad-GFP1 cells, whereas the expression level of Vimentin protein
was decreased in T24-Ad-CADM1 cells compared with in parental T24
and T24-Ad-GFP1 cells (P<0.05; Fig.
4). Conversely, CADM1 knockdown had the opposite effects on the
expression level of these proteins (Fig.
4). These results illustrated that the upregulation of CADM1
expression level was able to inhibit EMT in T24 cells.
Discussion
The present study first assessed methylation of
CADM1 promoter and CADM1 protein expression level in bladder
tissues for associations with clinicopathological parameters from
bladder cancer patients. Subsequently, the effects of CADM1
overexpression and knockdown on the regulation of bladder cancer
cell viability, cell cycle distribution, apoptosis, invasion
capacity in vitro and the underlying molecular events, were
investigated. The results demonstrated that the CADM1 promoter was
highly methylated in bladder cancer tissues compared with in normal
mucosae tissues, which induced a reduced CADM1 protein expression
level in bladder cancer tissues. Methylated CADM1 promoter was
significantly associated with tumor size, recurrence, pathology
classification and advanced clinical stages. Furthermore,
overexpression of CADM1 protein inhibited tumor cell proliferation,
reduced cell invasion and induced cell apoptosis, whereas knockdown
of CADM1 expression promoted tumor cell growth, invasion capacity
and cell cycle progression. At the gene level, overexpression of
CADM1 protein increased the expression levels of caspase-3, Bax and
p27 proteins, but decreased the expression levels of bcl-2,
cyclinD1, cyclinE1 and CDK2 proteins. CADM1 knockdown had an
opposite effect on the expression level of these proteins.
Overexpression of CADM1 protein regulated the expression level of
EMT markers, including increased expression levels of E-cadherin
and β-catenin, whereas it decreased the expression levels of
Vimentin. Thus, the present study demonstrated that reduced CADM1
expression levels may contribute to bladder cancer development and
progression and targeting CADM1 expression may be used as a novel
therapeutic strategy in the future for controlling bladder
cancer.
The present study determined methylation status of
the CADM1 promoter for association with clinicopathological
parameters. The results demonstrated that CADM1 promoter
methylation induced lost CADM1 expression in bladder cancer
tissues, which was also associated with tumor progression ex
vivo. Previous studies revealed that methylation of CADM1
promoter occurred in non-muscle-invasive bladder cancer (24–26) and
the present study confirmed that methylation of CADM1 promoter also
increased in Ta and T1 stage bladder cancer; however, these results
were contrary to those by Hellwinkel et al (27) demonstrating that methylation of CADM1
promoter was not significant in bladder cancer with various stages:
Their sample size was small but there were more advanced stage
bladder cancers. Of note, increasing evidence has revealed that
CADM1 is a critical regulator of cell proliferation, invasion and
apoptosis (12,17–19,31,32).
For example, Mao et al (31)
revealed that overexpression of CADM1 inhibited lung cancer cell
proliferation, induced tumor cell apoptosis and increased caspase-3
activity. It was further demonstrated that compared with the
control group, administration of CADM1 suppressed tumor growth in
nude mice to the extent of 70–80% (31). Lu et al (18) reveled that expression levels of CADM1
mRNA and protein was significantly reduced in laryngeal squamous
cell carcinoma tissues, which was significantly associated with
advanced tumor-node-metastasis staging and lymph node metastases,
but not with age, gender and tumor differentiation. They also
demonstrated that elevation of CADM1 expression inhibited tumor
cell proliferation, reduced tumor cell invasion and induced cell
apoptosis (18). Further studies are
required to investigate the underlying mechanism by which CADM1
promoter is methylated and whether it is a common event in various
types of human cancer.
Furthermore, to illustrate the functions of CADM1
protein in T24 bladder cancer cells, the present study performed
cell viability MTT assay following manipulated CADM1 expression
using lentivirus carrying CADM1 cDNA or siRNA vs. negative control
vectors. It was revealed that tumor cell proliferation was markedly
inhibited in T24-Ad-CADM1 compared with parental T24 and
T24-Ad-GFP1 cells. Conversely, knockdown of CADM1 expression level
induced tumor cell proliferation compared with that in parental T24
and T24-Ad-GFP2 cells. The present study demonstrated that CADM1
manipulation altered gene expressions, which further confirmed the
effects of CADM1 on the regulation of tumor cell viability. These
results suggested that CADM1 serves a critical role in regulating
cell proliferation, invasion, apoptosis and cell cycle of T24
bladder cancer cells.
In addition, it is well known that EMT serves a
central role in tumor invasion and metastasis. Previous studies
have demonstrated that bladder cancer progression is regulated by
various genes that affect tumor EMT (33–37). The
present study revealed that CADM1 overexpression upregulated the
expression levels of E-cadherin and β-catenin proteins in
T24-Ad-CADM1 cells compared with in parental T24 and T24-Ad-GFP1
cells. Whereas, expression of vimentin protein was decreased in
T24-Ad-CADM1 cells compared with in parental T24 and T24-Ad-GFP1
cells. However, CADM1 knockdown induced an opposite effect on the
expression level of these proteins. The results of the presents
study demonstrated that overexpression of CADM1 inhibited bladder
cancer cell EMT, whereas knockdown of CADM1 expression promoted
EMT.
In conclusion, the present study revealed that CADM1
expression was reduced in bladder cancer tissues and methylation of
CADM1 promoter induced, and was associated with, bladder cancer
progression. Furthermore, overexpression of CADM1 protein inhibited
tumor cell proliferation and decreased tumor cell invasion, but
induced tumor cell apoptosis and cell cycle arrest at the
G1/G0 phase. CADM1 expression also modulated
cell growth, apoptosis and cell cycle-associated proteins as well
as EMT markers. The results of the present study suggested that
CADM1 may be a novel gene target for clinical control of bladder
cancer progression. Further studies may focus on the
CADM1-regulated signaling pathway in bladder cancer.
Acknowledgements
The abstract was presented at the conference of The
2016 Huaxia Medical Forum-Genitourinary Tumor & The 2016
Greatwall International Translational Andrology and Urology Forum
(GUT-HMF 2016 & GITAU 2016). The title of this content as
follows: ‘Lost expression of cell adhesion molecule 1 (CADM1)
associated with bladder cancer progression and recurrence and its
overexpression inhibited tumor cell malignant behaviors’ and was
published as abstract no. AB060 in Transl Androl Urol. 2016 Apr; 5
(Suppl 1): AB060.’
Funding
The present study was supported in by Tianjin
Municipal Science and Technology Commission, Tianjin, China (grant
no. 12JCZDJC23700).
Availability of data and materials
The datasets generated and/or analyzed during this
study are available from the corresponding author on reasonable
request.
Authors' contributions
XQL designed the study. YGC, LL, ZJG and YJY
performed the experiments. YW and YJY were responsible for clinical
diagnosis and sample collection. YGC and YJY contributed to data
collection and statistical analysis. YGC and LL wrote the
manuscript. All the authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The present study was approved and monitored by the
Ethics Committee of Tianjin Medical University (Tianjin, China).
All procedures performed in this study were followed in accordance
with the Helsinki Declaration of 1975, as revised in 2000 (38). Written informed consent was obtained
from all patients and healthy individuals prior to the study.
Patient consent for publication
Written informed consent was obtained from all
individual participants included in the present study.
Competing interests
All authors declare that they have no competing
interest.
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2012. CA Cancer J Clin. 62:10–29. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Pashos CL, Botteman MF, Laskin BL and
Redaelli A: Bladder cancer: Epidemiology, diagnosis, and
management. Cancer Pract. 10:311–322. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zeegers MP, Tan FE, Dorant E and van Den
Brandt PA: The impact of characteristics of cigarette smoking on
urinary tract cancer risk: A meta-analysis ofepidemiologic studies.
Cancer. 89:630–639. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Khadjavi A, Mannu F, Destefanis P,
Sacerdote C, Battaglia A, Allasia M, Fontana D, Frea B, Polidoro S,
Fiorito G, et al: Early diagnosis of bladder cancer through the
detection of urinary tyrosine-phosphorylated proteins. Br J Cancer.
113:469–475. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Murakami Y, Nobukuni T, Tamura K, Maruyama
T, Sekiya T, Arai Y, Gomyou H, Tanigami A, Ohki M, Cabin D, et al:
Localization of tumor suppressor activity important in nonsmall
cell lung carcinoma on chromosome 11q. Proc Natl Acad Sci USA.
95:8153–8158. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kuramochi M, Fukuhara H, Nobukuni T, Kanbe
T, Maruyama T, Ghosh HP, Pletcher M, Isomura M, Onizuka M, Kitamura
T, et al: TSLC1 is a tumor-suppressor gene in human non-small-cell
lung cancer. Nat Genet. 27:427–430. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Shingai T, Ikeda W, Kakunaga S, Morimoto
K, Takekuni K, Itoh S, Satoh K, Takeuchi M, Imai T, Monden M and
Takai Y: Implications of nectin-like molecule-
2/IGSF4/RA175/SgIGSF/TSLC1/SynCAM1 in cell-cell adhesion and
transmembrane protein localization in epithelial cells. J Biol
Chem. 278:35421–35427. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yamada D, Yoshida M, Williams YN, Fukami
T, Kikuchi S, Masuda M, Maruyama T, Ohta T, Nakae D, Maekawa A, et
al: Disruption of spermatogenic cell adhesion and male infertility
in mice lacking TSLC1/IGSF4, animmunoglobulin superfamily cell
adhesion molecule. Mol Cell Biol. 26:3610–3624. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ito T, Shimada Y, Hashimoto Y, Kaganoi J,
Kan T, Watanabe G, Murakami Y and Imamura M: Involvement of TSLC1
in progression of esophageal squamous cell carcinoma. Cancer Res.
63:6320–6326. 2003.PubMed/NCBI
|
|
11
|
You Y, Zhang J, Li Y, Li Y, Shi G, Ma L
and Wei H: CADM1/TSLC1 inhibits melanoma cell line A375 invasion
through the suppression of matrix metalloproteinases. Mol Med Rep.
10:2621–2626. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
He G, Lei W, Wang S, Xiao R, Guo K, Xia Y,
Zhou X, Zhang K, Liu X and Wang Y: Overexpression of tumor
suppressor TSLC1 by a survivin-regulated oncolytic adenovirus
significantly inhibits hepatocellular carcinoma growth. J Cancer
Res Clin Oncol. 138:657–670. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yang G, He W, Cai M, Luo F, Kung H, Guan
X, Zeng Y and Xie D: Loss/down-regulation of tumor suppressor in
lung cancer 1 expression is associated with tumor progression and
is a biomarker of poor prognosis in ovarian carcinoma. Int J
Gynecol Cancer. 21:486–493. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Heller G, Geradts J, Ziegler B, Newsham I,
Filipits M, Markis-Ritzinger EM, Kandioler D, Berger W, Stiglbauer
W, Depisch D, et al: Downregulation of TSLC1 and DAL-1 expression
occurs frequently in breast cancer. Breast Cancer Res Treat.
103:283–291. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Jansen M, Fukushima N, Rosty C, Walter K,
Altink R, Heek TV, Hruban R, Offerhaus JG and Goggins M: Aberrant
methylation of the 5′ CpG island of TSLC1 is common in pancreatic
ductal adenocarcinoma and is first manifest in high-grade PanlNs.
Cancer Biol Ther. 1:293–296. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Takahashi Y, Iwai M, Kawai T, Arakawa A,
Ito T, Sakurai-Yageta M, Ito A, Goto A, Saito M, Kasumi F and
Murakami Y: Aberrant expression of tumor suppressors CADM1 and 4.1B
in invasive lesions of primary breast cancer. Breast Cancer.
19:242–252. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lei W, Liu HB, Wang SB, Zhou XM, Zheng SD,
Guo KN, Ma BY, Xia YL, Tan WS, Liu XY and Wang YG: Tumor suppressor
in lung cancer-1 (TSLC1) mediated by dual-regulated oncolytic
adenovirus exerts specific antitumor actions in a mouse model. Acta
Pharmacol Sin. 34:531–540. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lu B, Di W, Wang H, Ma H, Li J and Zhang
Q: Tumor suppressor TSLC1 is implicated in cell proliferation,
invasion and apoptosis in laryngeal squamous cell carcinoma by
regulating Akt signaling pathway. Tumour Biol. 33:2007–2017. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chen K, Wang G, Peng L, Liu S, Fu X, Zhou
Y, Yu H, Li A, Li J, Zhang S, et al: CADM1/TSLC1 inactivation by
promoter hypermethylation is a frequent event in colorectal
carcinogenesis and correlates with late stages of the disease. Int
J Cancer. 128:266–273. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Fukuhara H, Kuramochi M, Fukami T,
Kasahara K, Furuhata M, Nobukuni T, Maruyama T, Isogai K, Sekiya T,
Shuin T, et al: Promoter methylation of TSLC1 and tumor suppression
by its gene product in human prostate cancer. Jpn J Cancer Res.
93:605–609. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ochiai H, Takenobu H, Nakagawa A,
Yamaguchi Y, Kimura M, Ohira M, Okimoto Y, Fujimura Y, Koseki H,
Kohno Y, et al: Bmi1 is a MYCN target gene that regulates
tumorigenesis through repression of KIF1Bbeta and TSLC1 in
neuroblastoma. Oncogene. 29:2681–2690. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lung HL, Cheung AK, Xie D, Cheng Y, Kwong
FM, Murakami Y, Guan XY, Sham JS, Chua D, Protopopov AI, et al:
TSLC1 is a tumor suppressor gene associated with metastasis in
nasopharyngeal carcinoma. Cancer Res. 66:9385–9392. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hui AB, Lo KW, Kwong J, Lam EC, Chan SY,
Chow LS, Chan AS, Teo PM and Huang DP: Epigenetic inactivation of
TSLC1 gene in nasopharyngeal carcinoma. Mol Carcinog. 38:170–178.
2003. View
Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sacristan R, Gonzalez C, Fernández-Gómez
JM, Fresno F, Escaf S and Sánchez-Carbayo M: Molecular
classification of non-muscle-invasive bladder cancer (pTa
low-grade, pT1 low-grade, and pT1 high-grade subgroups) using
methylation of tumor-suppressor genes. J Mol Diagn. 16:564–572.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Agundez M, Grau L, Palou J, Algaba F,
Villavicencio H and Sanchez-Carbayo M: Evaluation of the
methylation status of tumour suppressor genes for predicting
bacillus Calmette-Guérin response in patients with T1G3 high-risk
bladder tumours. Eur Urol. 60:131–140. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Casadio V, Molinari C, Calistri D, Tebaldi
M, Gunelli R, Serra L, Falcini F, Zingaretti C, Silvestrini R,
Amadori D and Zoli W: DNA Methylation profiles as predictors of
recurrence in non muscle invasive bladder cancer: An MS-MLPA
approach. J Exp Clin Cancer Res. 32:942013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hellwinkel OJ, Kedia M, Isbarn H, Budäus L
and Friedrich MG: Methylation of the TPEF- and PAX6-promoters is
increased in early bladder cancer and in normal mucosa adjacent to
pTa tumours. BJU Int. 101:753–757. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Eble JS, Epstein JI, Sesterhenn IA and
Sauter G: Pathology and genetics of tumours of the urinary system
and male genital organs. World Health Organization Classification
of Tumours Lyon: IARC Press; 2004
|
|
29
|
Sobin LH, Gospodarowicz MK and Wittekind
C: TNM classification of malignant tumours. (7th). (Weinheim).
Wiley. 2009.
|
|
30
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Mao X, Seidlitz E, Truant R, Hitt M and
Ghosh HP: Re-expression of TSLC1 in a non-small-cell lung cancer
cell line induces apoptosis and inhibits tumor growth. Oncogene.
23:5632–5642. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Qin L, Zhu W, Xu T, Hao Y, Zhang Z, Tian Y
and Yang D: Effect of TSLC1 gene on proliferation, invasion and
apoptosis of human hepatocellular carcinoma cell line HepG2. J
Huazhong Univ Sci Technolog Med Sci. 27:535–537. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Cardif RD: Epithelial to mesenchymal
transition tumors: Fallacious or snail's pace? Clin Cancer Res.
11:8534–8537. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Liang W, Hao Z, Han JL, Zhu DJ, Jin ZF and
Xie WL: CAV-1 contributes to bladder cancer progression by inducing
epithelial-to-mesenchymal transition. Urol Oncol. 32:855–863. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wan F, Cheng C, Wang Z, Xiao X, Zeng H,
Xing S, Chen X, Wang J, Li S, Zhang Y, et al: SATB1 overexpression
regulates the development and progression in bladder cancer through
EMT. PLoS One. 10:e01175182015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhao J, Dong D and Sun L, Zhang G and Sun
L: Prognostic significance of the epithelial-to-mesenchymal
transition markers e-cadherin, vimentin and twist in bladder
cancer. Int Braz J Urol. 40:179–189. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Geng J, Fan J, Ouyang Q, Zhang X, Zhang X,
Yu J, Xu Z, Li Q, Yao X, Liu X and Zheng J: Loss of PPM1A
expression enhances invasion and the epithelial-to-mesenchymal
transition in bladder cancer by activating the TGF-β/Smad signaling
pathway. Oncotarget. 5:5700–5711. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Chhabra R: Cervical cancer stem cells:
Opportunities and challenges. J Cancer Res Clin Oncol.
141:1889–1897. 2015. View Article : Google Scholar : PubMed/NCBI
|