Introduction
Vascular endothelial cells participate in numerous
physiological processes, such as endocrine modulation and
inflammatory reactions (1).
Atherosclerosis, which can lead to heart failure, is a primary
vascular disease caused by endothelial dysfunction (2). Previous findings have demonstrated that
atherosclerosis has a high morbidity and mortality (3). During the process of atherosclerosis,
platelet aggregation occurs, and lipids invade vascular cells
leading to hypoxia; thereby inducing endothelial cell dysfunction,
and atypical electron leaking from the mitochondria will produce
the reactive oxygen species (ROS) (4,5), which can
result in endothelial cell dysfunction.
MicroRNAs (miRNAs) not only block messenger RNA
(mRNA) duplication and translation, they can also affect cell
proliferation, differentiation, apoptosis and autophagy by
regulating the expression of target genes (6). miR-155 is a multifunctional miRNA, which
is transcribed from chromosome 21 of the B-cell Integration Cluster
(7). Previous research demonstrated
that miR-155 is abnormally expressed in several types of human
cancer, including endometrial cancer and lymphoma (8), with studies indicating that miR-155 has
a potential therapeutic effect on vascular disease by enhancing the
viability of vascular cells (6,9).
Autophagy is an evolutionarily conserved stress
response in eukaryotes (10), and can
improve cell viability under conditions of cellular stress. An
autophagosome is a vacuole structure with a double-membrane that is
located in the cytoplasm, which can digest dysfunctional proteins
and organelles (11,12). Autophagy-related genes can regulate
autophagosome formation to impact the process of autophagy
(13). However, the effects and
mechanism of miR-155 in endothelial cells through autophagy under
the conditions of oxidative stress have not been completely
elucidated.
In the present study, miR-155 was transfected into
H2O2-treated endothelial cells. The CCK8
assay was used to examine the effect of miR-155 on the
proliferation of H2O2-treated cells, and the
expressions of Microtubule Associated Protein 1 Light Chain 3 α
(LC3) and (Nucleoporin 62) P62 were detected to examine the effect
of miR-155 on the autophagy of Human umbilical vein endothelial
cells (HUVECs), and the tandem-fluorescent-adenovirus was used to
observe the intracellular autophagosome formation. The results
demonstrated that the proliferation of endothelial cells under
oxidative stress was promoted and the expression of LC3 and P62 was
increased and decreased respectively. The results also demonstrated
that miR-155 modulated autophagy via promoting autophagy-related
genes (ATG5) expression.
Materials and methods
Cell culture
Human umbilical vein endothelial cells (HUVECs) were
obtained from iCell Bioscience, Inc. (Shanghai, China), The cells
were cultured in a 7.5 cm2 culture flask (Corning
Incorporated, Corning, NY, USA) in (450 mg/l) high-glucose DMEM
(Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA)
supplemented with 10% fetal bovine serum (FBS; Hyclone; GE
Healthcare Life Sciences, Logan, UT, USA), 1% endothelial growth
factor and 1% penicillin and streptomycin (Hyclone; GE Healthcare
Life Sciences). All the cells were cultured in an incubator at 37°C
with 5% CO2. The experiments were conducted in
triplicate at least 3 times.
Treatment of the HUVECs by use of
H2O2
Hydrogen peroxide (H2O2,
Sinopharm Chemical Reagent Co., Ltd., Shanghai, China) at 30% was
used to treat the HUVECs at concentrations of 0, 100, 200, 400 and
800 nmol/l for 24 h at 37°C prior to experimentation. The suit
concentration of H2O2, which produced
cytotoxic ROS was selected to treat the HUVECs in order to
establish the oxidative stress cell model. A final concentration of
5 µM 3-methyladenine (3-MA; Beyotime Institute of Biotechnology,
Haimen, China) was used to treat the cells of the contrast group at
37°C. The experiments were conducted in triplicate at least 3
times.
Transfection of small interfering RNA
(siRNA)
The sequence of miR-155-5p mimics was
5′-UUAAUGCUAAUCGUCAUAGGGGU-3′, the sequence of miR-155-5p inhibitor
was 5′-ACCCCUAUCACGAUUAGCAUUAA-3′ and the siRNA sequence of miR-155
was
5′-GACAAUUACGAUUAGCACUAUCCCCAAAAACGGAGGUUGACUGAGGAUGUAUAAUCGUAAUUGUC-3′,
miR-155-5p mimics, miR-155-5p inhibitor and siRNA of miR-155 was
purchased from Generay Biotech Co., Ltd. (Shanghai, China), The
concentration of siRNA was 5 uM. The HUVECs were cultured in a 6
cm2 petri dish (Corning Life Sciences, Wujiang, China)
at a density of 5×105 cell/cm2, the cells
were transfected with 100 µmol/l siRNA miR-155 as the miR-155
negative control, miR-155 inhibitor and miR-155 mimic groups, by
using a ribo FECT™ CP Transfection kit (Guangzhou
RiboBio Co., Ltd., Guangzhou, China) according to the
manufacturer's protocol, and the cells were subsequently incubated
in fresh High Glucose-DMEM (Gibco; Thermo Fisher Scientific, Inc.)
for 24 h at 37°C prior to treatment with
H2O2. The experiments were conducted in
triplicate at least 3 times.
Transfection of
fluorescent-mRFP-GFP-LC3 adenovirus
To detect intracellular autophagy, the
fluorescent-mRFP-GFP-LC3 adenovirus (Hanbio Biotechnology Co.,
Ltd., Shanghai, China) was transfected into the HUVECs, miR-155 was
transfected into four groups of cells, comprising miR-155 inhibitor
negative control, miR-155 inhibitor, miR-155 mimic negative control
and miR-155 mimic groups, prior to treatment with
H2O2, then 50 µl adenovirus and 8 µl
polybrane were added. Following co-culture for 48 h at 37°C, the
number of fluorescent puncta in cells were counted using
immunofluorescence laser confocal laser scanning microscopy
(magnification, ×40). Green fluorescent protein (GFP), displayed as
green dots, represent the acidic lysosome. The red fluorescent
proteins (RFP) represent the autophagy lysosome. When the number of
green spots increased, the level of autophagy was reduced. mRFP
merged with GFP, displayed as yellow dots, and represented the
autophagosome. The experiments were repeated 3 times.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
The total RNA of each group (miR-155 inhibitor
negative control, miR-155 inhibitor, miR-155 mimic negative control
and miR-155 mimic groups) was extracted from cultured cells using
TRIzol reagent (Thermo Fisher Scientific, Inc.). The reverse
transcription was performed by use of PrimeScript™ RT
reagent kit (Takara Biotechnology Co., Ltd., Dalian, China) to
synthesize complementary DNA (cDNA), according to the
manufacturer's protocol. The cDNA reverse transcription procedure
was as follows: 42°C for 15 min followed with 85°C for 2 min. The
primer sequences used are listed in Table
I. The qPCR Master Mix (Promega Corporation, Madison, WI, USA)
was used according to the manufacturer's protocol, RT-qPCR was
performed using a LightCycler 480 Real-Time system. The
thermocycling conditions were as follows: 95°C for 5 min, and then
40 cycles of 95°C for 15 sec and 60°C for 1 min, and finally 1
cycle of 95°C for 15 sec, 60°C for 1 min and 95°C for 15 sec. GAPDH
was a housekeeping gene for normalized the relative target mRNA
expression, and the comparative 2−ΔΔCq method was used
to quantify target gene expression (14). The experiments were repeated 3
times.
 | Table I.Primer sequences used in the present
study. |
Table I.
Primer sequences used in the present
study.
| Target gene | Primer sequence |
|---|
| miR-155 |
|
|
Forward |
5′-GCTTCGGTTAATGCTAATCGTG-3′ |
|
Reverse |
5′-CAGAGCAGGGTCCGAGGTA-3′ |
| LC3A |
|
|
Forward |
5′-GGCTTCCGAGTTGCTGACTG-3′ |
|
Reverse |
5′-CTGGTCGCGGATCTGCTGTA-3′ |
| LC3B |
|
|
Forward |
5′-AGTTGGCACAAACGCAGGGTA-3′ |
|
Reverse |
5′-TTAGGAGTCAGGGACCTTCAGCA-3′ |
| P62 |
|
|
Forward |
5′-AGTCTCTGGCGGAGCAGATGA-3′ |
|
Reverse |
5′-TCTGGCATCTGTAGGGACTGGA-3′ |
| ATG5 |
|
|
Forward |
5′-CCATCAATCGGAAACTCATGGA-3′ |
|
Reverse |
5′-ATCTGCAGCCACAGGACGAA-3′ |
| GAPDH |
|
|
Forward |
5′-GAAGGGCTCATGACCACAGTCCAT-3′ |
|
Reverse |
5′-TCATTGTCGTACCAGGAAATGAGCTT-3′ |
Western blot analysis
To extract proteins from HUVECs, the cells were
washed twice with phosphate-buffered saline (PBS) in 25°C for 5 min
and then lysed in ice-cold RIPA buffer containing 0.5 nM PMSF
(Beyotime Institute of Biotechnology). The lysates were centrifuged
at 15,000 × g in 4°C for 15 min. The BCA protein assay kit (Thermo
scientific, Inc.) was then used to detect the protein
concentrations, and 20 µg protein were separated by 12% SDS-PAGE
gels, then transferred to polyvinylidene difluoride membranes,
following blocking in 5% non-fat milk of TBS with 0.05% Tween-20 at
4°C for 2 h. The membranes were then incubated with primary
antibodies [LC3 A/B rabbit anti-mouse polyclonal antibody (1:1,000;
cat. no. ab128025), ATG5 rabbit anti-mouse monoclonal antibody
(1:1,000; cat. no. ab108327) and GAPDH mouse anti-rabbit monoclonal
antibody (1:2,000; cat. no. ab8245; all Abcam Cambridge, MA, USA)]
and P62 rabbit anti-mouse monoclonal antibody (1:1,000; cat. no.
8025; Cell Signaling Technology, Inc.) at 4°C overnight, followed
by incubation with secondary antibodies [goat polyclonal
anti-rabbit IgG (1:5,000; cat. no. ab6721) and goat polyclonal
anti-mouse IgG (1:2,000; cat. no. ab6789; both Abcam)] for 2 h at
37°C. The results were observed by using an enhanced
chemiluminescence detection system (Thermo Fisher Scientific,
Inc.). The experiments were repeated 3 times.
Detecting intracellular ROS by use of
superoxide dismutase (SOD) assay kit
The supernatant of adherence cultured cells was
removed and cells were washed with PBS twice in 25°C for 5 min,
then RIPA buffer (Beyotime Institute of Biotechnology) was used to
split cells, and following centrifugation at 12,000 × g for 10 min
in 4°C, the supernatant was collected. The dye conversion of
4-[3-(4-iodophenyl)-2-(4-nitrophenyl)-2H5-tetrazolium]-1,3-benzene
disulfonate to formazan (WST-1 Cell Proliferation and Cytotoxicity
Assay kit; Beyotime institute of Biotechnology) was used to detect
the intracellular ROS. Cells were plated at 7,500 cells/well into
96 well tissue culture plates and then were allowed to attach at
37°C for 12 h. Subsequently, the cells were treated by 5 µM/l 3-MA
at 37°C until autophagy occurs, and then the media was replaced
with ischemic buffer at 37°C for 30 min followed by being replaced
with High-Glucose DMEM supplemented with 10% FBS, 1% endothelial
growth factor and 1% penicillin and streptomycin in the absence or
presence of BHA (0.1 mM; cat. no. 1009005) and NAC (5 mM; cat. no.
20021; both from Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) for
24 h at 37°C, WST-1 reagent was added in 24-h increments up to 72 h
at 37°C, following the manufacturer's protocols, and absorbance was
recorded using a microplate reader (Thermo Fisher Scientific, Inc.)
at 450 nm wavelength. The experiments were conducted in triplicate
at least 3 times (15).
Cell counting kit-8 assay
The CCK-8 assay kit (Beyotime Institute of
Biotechnology) was used to detect the impact of miR-155 on cell
proliferation. HUVECs were plated in 96-well plates at a density of
2×104 cell/well and were cultured for 24 h at 37°C.
Subsequently, 10 µl CCK-8 reagent with 100 µl culture medium was
added to each well for 2 h at 37°C. The cells were measured at a
wavelength of 450 nm using a microplate reader according to the
manufacturer's protocol. The experiments were repeated 3 times.
Statistical analysis
SPSS 16.0 (SPSS, Inc., Chicago, IL, USA) was used to
perform statistical analysis. A Student's paired t-test was used to
compare between two groups by use of an open source software R
language 3.5.0 (https://www.7down.com/soft/288188.html), and one-way
analysis of variance (ANOVA) with Bonferroni post hoc correction
was performed to compare the results between ≤3 groups, The one-way
ANOVA results were conducted by R software written by computer
programmers from Department of Geriatrics in Kunming General
Hospital (Kunming, China). The data are a result of at least three
independent experimental repeats, and are presented as mean ±
standard deviation (SD). P<0.05 was considered to indicate a
statistically significant difference.
Results
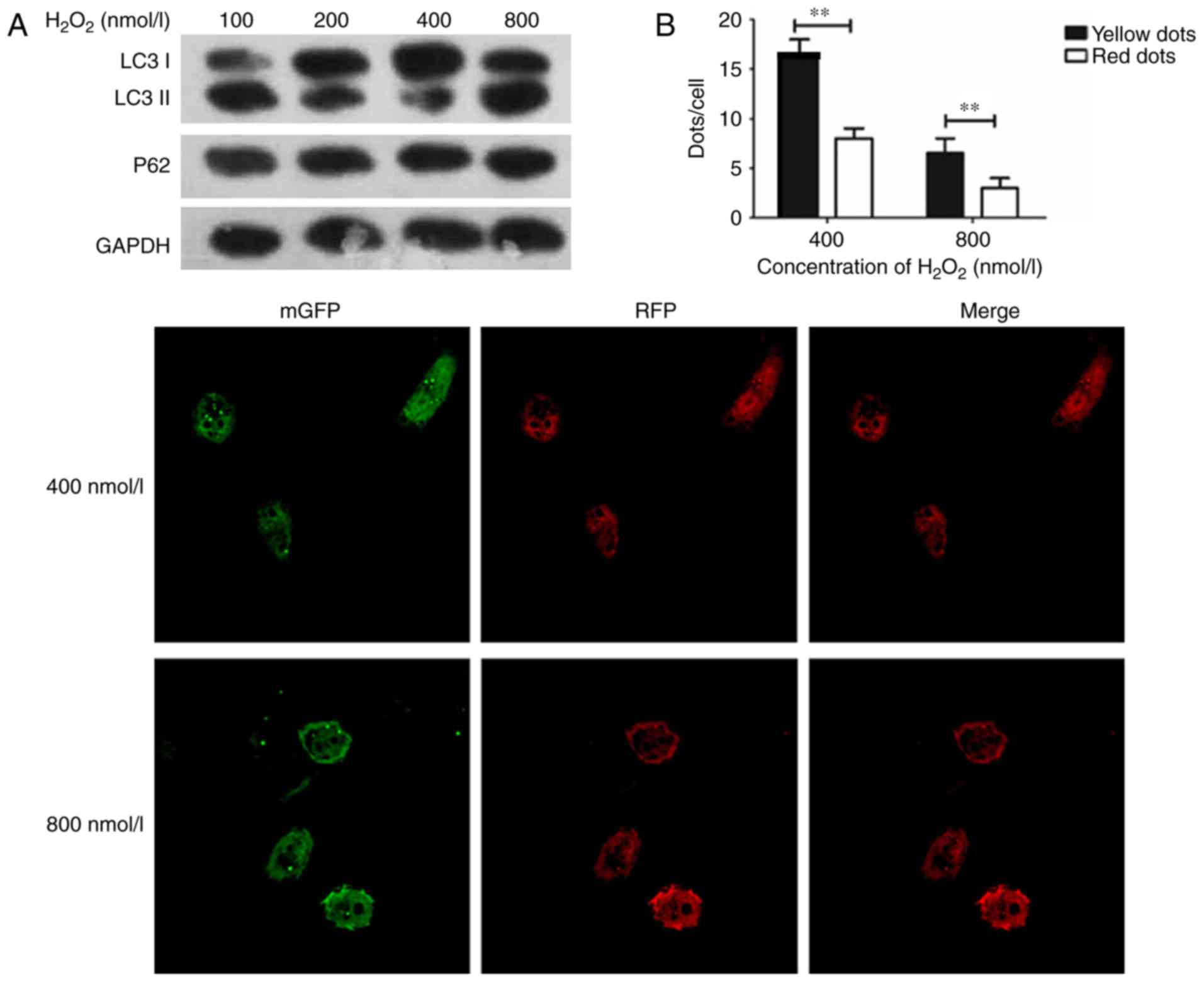
Establishment of oxidative stress
endothelial HUVECs model
A previous study reported that
H2O2-treated cells released ROS, which can
cause damage in cells, and LC3 I will convert into LC3 II when
conjugated with phosphatidylethanolamine (16). The ratio of LC3 II/I was used to
identify the activation of autophagy (17). In the present study,
H2O2 at concentrations of 0, 100, 200, 400,
and 800 nmol/l were used to treat the cultured HUVECs for 24 h, and
the expression of LC3 was detected to study the change of
autophagy. The expression of P62 was also detected as a marker for
autophagy. Furthermore, tandem-fluorescent-adenovirus was used to
observe the intracellular autophagosome information. The SOD assay
was then performed to examine ROS damage in endothelial cells.
The expression of LC3 I/II was its highest in HUVECs
with an H2O2 concentration of 400 nmol/l, and
the expression of LC3 I/II was decreased in HUVECs when
H2O2 was administered at 800 nmol/l, as cell
damage could not be repaired, thus leading to cell apoptosis
(Fig. 1A). The results (Fig. 1B) confirmed the findings of Fig. 1A. Consequently, 400 nmol/l of
H2O2 was used in subsequent experimentations
to induce autophagy in HUVECs.
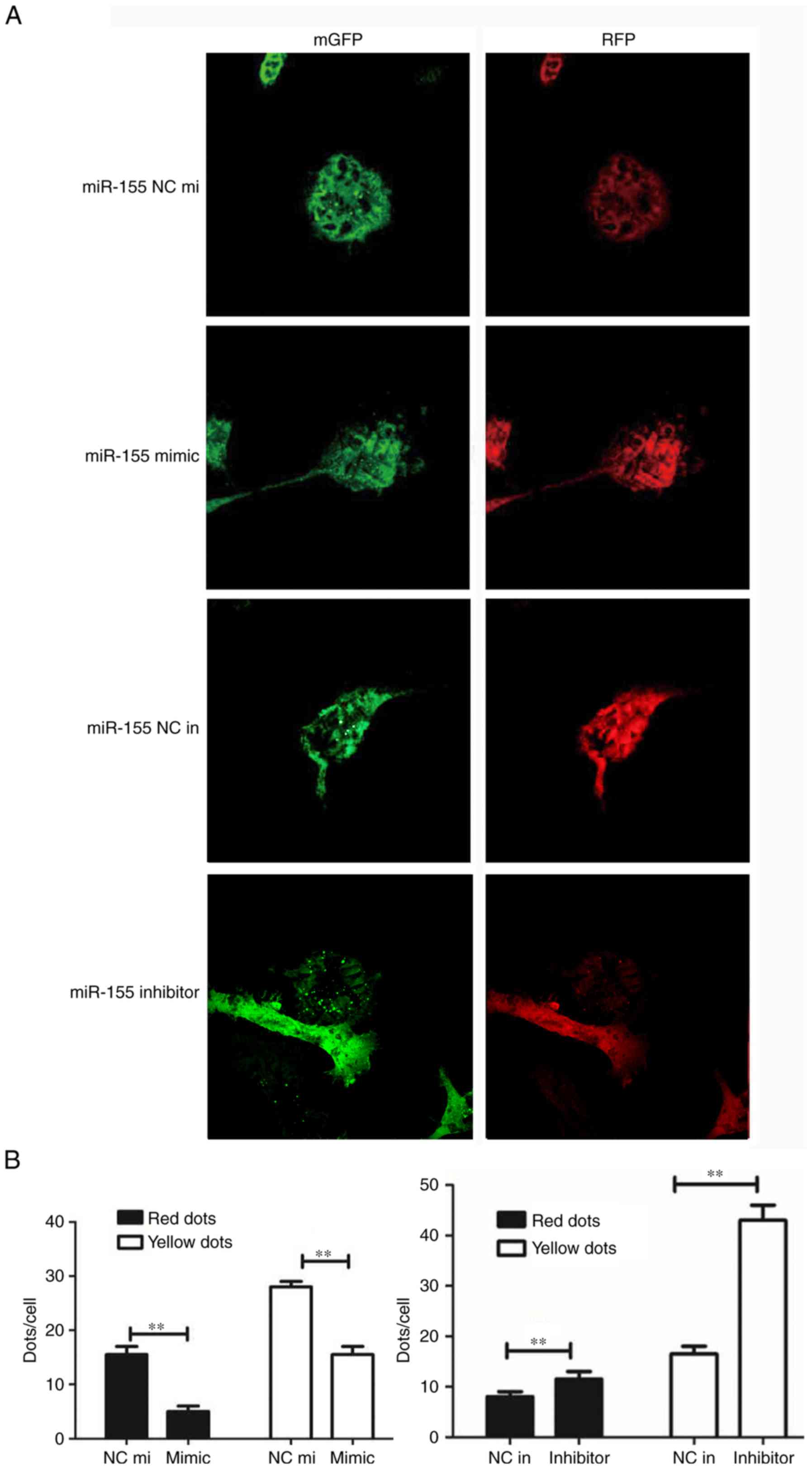
Downregulation of miR-155 can promote
HUVEC proliferation
miR-155, the mimic miR-155 and the negative control
miR-155 were transfected into H2O2-treated
HUVECs. The CCK-8 assay was performed to detect HUVEC
proliferation; the results demonstrated that cell proliferation in
the miR-155 inhibitor group was higher compared with the miR-155
inhibitor negative control group (Fig.
2A). The SOD assay data confirmed that the suppression of
miR-155 can protect HUVECs from oxidative stress (Fig. 2B). Western blotting was used to
determine whether autophagy was involved in miR-155-mediated HUVEC
proliferation and increase the activation of ROS. The result
demonstrated that LC3 was overexpressed and the P62 had low protein
expression levels in the miR-155 inhibitor group (Fig. 2C). The results of RT-qPCR corroborated
these findings (Fig. 2D). The
expression of LC3 and number of yellow dots were increased when
miR-155 was downregulated (Fig. 3A and
B). The results indicate that suppression of miR-155 promotes
the proliferation of endothelial cells and decreases intracellular
ROS damage via modulating autophagy.
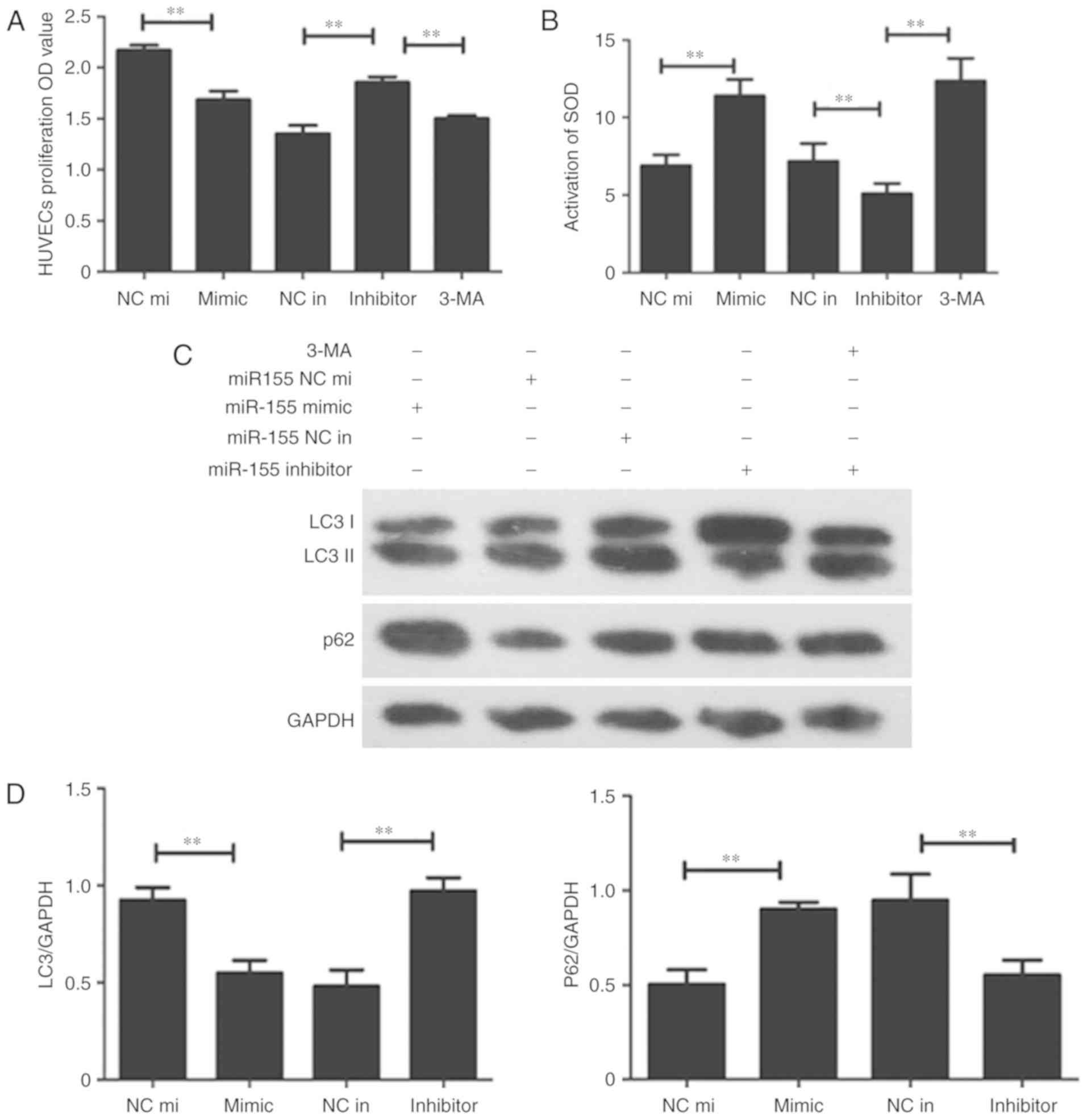 | Figure 2.Downregulation of miR-155 promotes
HUVEC proliferation. (A) Statistical data of the CCK-8 assay. The
OD value at 450 nm was increased in the group transfected with the
miR-155 inhibitor (**P<0.01, n=3). (B) The Superoxide Dismutase
assay was used to detect the activation of intracellular ROS
following transfection with miR-155, the activation of ROS was
decreased in the miR-155 inhibitor group. (C) Western blot
presenting the level of autophagy related proteins expressed in
each transfection group (**P<0.01, n=3). (D) The RT-qPCR data
demonstrated that LC3 I/II was overexpressed while P62 had a low
expression level in the miR-155 inhibitor group (**P<0.01, n=3).
CCK-8, cell counting kit-8; OD, optical density; SOD, Superoxide
Dismutase; HUVEC, Human umbilical vein endothelial cell; LC3,
Microtubule Associated Protein 1 Light Chain 3 α; P62, Nucleoporin
62; 3-MA, 3-methyladenine; miR-155 NC mi, miR-155 mimic negative
control; miR-155 NC in, miR-155 inhibitor, negative control. |
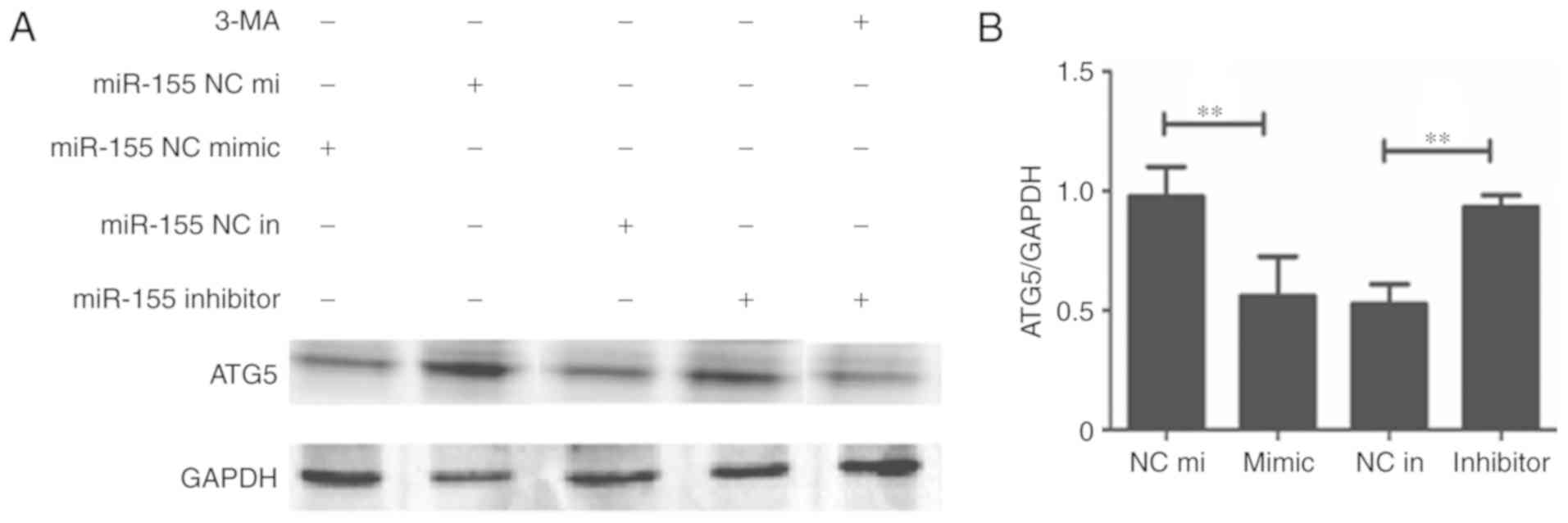
miR-155 regulates autophagy via
modulation of ATG5 in HUVECs
To explore the mechanism of how miR-155 regulates
autophagy in HUVECs, the expression of ATG5 was detected. The
results demonstrated that the expression of ATG5 was decreased when
miR-155 was overexpressed (Fig. 4A).
Furthermore, when 3-MA was used to inhibit autophagy, ATG5 protein
and mRNA expressions were also decreased (Figs. 4). These results indicate that miR-155
regulates autophagy in HUVECs via decreasing the expression of
ATG5.
Discussion
Oxidative stress can cause endothelial cells to
produce ROS, which can lead to protein dysfunction and DNA
fragmentation, as well as morphological changes in endothelial
cells (3). In the present study,
oxidative stress cell models were established using different
concentrations of H2O2. The expression of LC3
and P62 were detected to explore the suitable concentration, which
can either induce autophagy or protect against death from
apoptosis, culminating in the selection of 400 nmol/l
H2O2 to establish cell models for oxidative
stress.
Several studies have demonstrated that miR-155
serves an essential role in various physiological and pathological
processes, for example, miR-155 is an inhibitor of autophagy in
chondrocytes and contributes to the pathogenesis of OA (9), and miR-155 is downregulated in familial
adenomatous polyposis and modulates WNT signaling by targeting
AXIN1 and transcription factor 4 (18). Previous reports have demonstrated that
miR-155 can inhibit activated caspase-3, and induce apoptosis in
cancer (16,19). Previous research has demonstrated that
there were complex interactions between autophagy and apoptosis.
These studies suggest that miR-155 may also modulate autophagy in
heart disease (20–22).
Autophagy is a highly conserved process that
maintains cellular homeostasis (21).
Under conditions of oxidative stress, autophagy can degrade
proteins to provide essential amino acids and energy (23). A previous report demonstrated that
unregulated autophagy can aid in endothelial cell survival from
intracellular peroxidation damage (24).
In the present study, the effects of miR-155 on
endothelial cells under conditions of oxidative stress were
investigated. Firstly, attenuated miR-155 expression can reduce ROS
overproduction in H2O2-treated HUVECs, which
means modulating miR-155 can prevent HUVECs from intracellular
peroxidative damage. In addition, the activation of ROS was
decreased and cell proliferation was increased when the expression
of miR-155 was inhibited. The results indicated that inhibition of
miR-155 may increase the level of autophagy and promote the
proliferation of HUVECs. Summarily, the data demonstrated that
inhibiting miRNA-155 prevents endothelial cells from oxidative
damage via regulating the level of autophagy.
A previous study reported that, the overexpression
of ATG5 significantly promoted angiogenesis in endothelial
progenitor cells (12). ATG5 can
combine with the proteins autophagy related 12 and autophagy
related 16 to generate a macromolecular complex that promotes the
formation of an autophagosome (23).
It has been reported that endothelial cells will lose the ability
of tubule formation and migration following ATG5 knockdown
(11). Emphasizing that ATG5 can
modulate autophagy and the function of endothelial cells. In the
present study, the mechanism of miR-155 in modulating autophagy was
investigated via detecting the expression of ATG5. The results
demonstrated that inhibited miR-155 increases the expression of
ATG5, which indicated that miR-155 modulates autophagy through
decreasing the expression of ATG5.
The repair of endothelial cell dysfunction is the
foundation for atherosclerosis treatment at the molecular
level.
The present study attempted to verify that
inhibiting the expression of miR-155 may protect endothelial cells
from intracellular peroxidative damage and promote cell
proliferation via modulating autophagy. This provides a novel
insight into miRNAs as potential therapeutics for the treatment of
heart disease.
Acknowledgements
Not applicable.
Funding
The present study was supported by grants from the
National Natural Science Foundation of China (grant no.
81270224).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
HFC and HW conceived and designed the study,
completed the majority of the experiments and wrote the manuscript.
MYLG, LZ, FLH, YKS and XHP provided advice and materials, and
participated in the majority of experiments, data analysis and
writing of the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Liu J, Bi X, Chen T, Zhang Q, Wang SX,
Chiu JJ, Liu GS, Zhang Y, Bu P and Jiang F: Shear stress regulates
endothelial cell autophagy via redox regulation and Sirt1
expression. Cell Death Dis. 6:e18272015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wang R, Yang Q, Wang X, Wang W, Li J, Zhu
J, Liu X, Liu J and Du J: FoxO3α-mediated autophagy contributes to
apoptosis in cardiac microvascular endothelial cells under hypoxia.
Microvasc Res. 104:23–31. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhang L, Wei J, Ren L, Zhang J, Wang J,
Jing L, Yang M, Yu Y, Sun Z and Zhou X: Endosulfan induces
autophagy and endothelial dysfunction via the AMPK/mTOR signaling
pathway triggered by oxidative stress. Environ Pollut. 220:843–852.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
He X, Zhao M, Bi XY, Yu XJ and Zang WJ:
Delayed preconditioning prevents ischemia/reperfusion-induced
endothelial injury in rats: Role of ROS and eNOS. Lab Invest.
93:168–180. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhao H, Han Z, Ji X and Luo Y: Epigenetic
regulation of oxidative stress in ischemic stroke. Aging Dis.
7:295–306. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Donners MM, Wolfs IM, Stöger LJ, van der
Vorst EP, Pöttgens CC, Heymans S, Schroen B, Gijbels MJ and de
Winther MP: Hematopoietic miR155 deficiency enhances
atherosclerosis and decreases plaque stability in hyperlipidemic
mice. PLoS One. 7:e358772012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ma X, Ma C and Zheng X: MicroRNA-155 in
the pathogenesis of atherosclerosis: A conflicting role? Heart Lung
Circ. 22:811–818. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hulsmans M, De Keyzer D and Holvoet P:
MicroRNAs regulating oxidative stress and inflammation in relation
to obesity and atherosclerosis. FASEB J. 25:2515–2527. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
D'Adamo S, Alvarez-Garcia O, Muramatsu Y,
Flamigni F and Lotz MK: MicroRNA-155 suppresses autophagy in
chondrocytes by modulating expression of autophagy proteins.
Osteoarthritis Cartilage. 24:1082–1091. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhang L, Yu Y, Xia X, Ma Y, Chen XW, Ni ZH
and Wang H: Transcription factor E2-2 inhibits the proliferation of
endothelial progenitor cells by suppressing autophagy. Int J Mol
Med. 37:1254–1262. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Du J, Teng RJ, Guan T, Eis A, Kaul S,
Konduri GG and Shi Y: Role of autophagy in angiogenesis in aortic
endothelial cells. Am J Physiol Cell Physiol. 302:C383–C391. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hu N, Kong LS, Chen H, Li WD, Qian AM,
Wang XY, Du XL, Li CL, Yu XB and Li XQ: Autophagy protein 5
enhances the function of rat EPCs and promotes EPCs homing and
thrombus recanalization via activating AKT. Thromb Res.
136:642–651. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang J, Deng H, Liu L, Liu X, Zuo X, Xu
Q, Wu Z, Peng X and Ji A: α-lipoic acid protects against
hypoxia/reoxygenation-induced injury in human umbilical vein
endothelial cells through suppression of apoptosis and autophagy.
Mol Med Rep. 12:180–186. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zeng M, Wei X, Wu Z, Li W, Li B, Fei Y, He
Y, Chen J, Wang P and Liu X: Reactive oxygen species contribute to
simulated ischemia/reperfusion-induced autophagic cell death in
human umbilical vein endothelial cells. Med Sci Monit.
20:1017–1023. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Weber M, Kim S, Patterson N, Rooney K and
Searles CD: MiRNA-155 targets myosin light chain kinase and
modulates actin cytoskeleton organization in endothelial cells. Am
J Physiol Heart Circ Physiol. 306:H1192–H1203. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chen G, Zhang W, Li YP, Ren JG, Xu N, Liu
H, Wang FQ, Sun ZJ, Jia J and Zhao YF: Hypoxia-induced autophagy in
endothelial cells: A double-edged sword in the progression of
infantile haemangioma? Cardiovasc Res. 98:437–448. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Prossomariti A, Piazzi G, D'Angelo L,
Miccoli S, Turchetti D, Alquati C, Montagna C, Bazzoli F and
Ricciardiello L: miR-155 is downregulated in familial adenomatous
polyposis and modulates WNT signaling by targeting AXIN1 and TCF4.
Mol Cancer Res. Aug 2–2018.(Epub ahead of print). View Article : Google Scholar : PubMed/NCBI
|
|
19
|
De Santis R, Liepelt A, Mossanen JC, Dueck
A, Simons N, Mohs A, Trautwein C, Meister G, Marx G,
Ostareck-Lederer A and Ostareck DH: MiR-155 targets Caspase-3 mRNA
in activated macrophages. RNA Biol. 13:43–58. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Jia QW, Chen ZH, Ding XQ, Liu JY, Ge PC,
An FH, Li LH, Wang LS, Ma WZ, Yang ZJ and Jia EZ: Predictive
effects of circulating miR-221, miR-130a and miR-155 for coronary
heart disease: A multi-ethnic study in China. Cell Physiol Biochem.
42:808–823. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Galluzzi L, Pietrocola F, Levine B and
Kroemer G: Metabolic control of autophagy. Cell. 159:1263–1276.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Tang Y, Li J, Li F, Hu CA, Liao P, Tan K,
Tan B, Xiong X, Liu G, Li T and Yin Y: Autophagy protects
intestinal epithelial cells against deoxynivalenol toxicity by
alleviating oxidative stress via IKK signaling pathway. Free Radic
Biol Med. 89:944–951. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang W, Li C, Li W, Kong L, Qian A, Hu N,
Meng Q and Li X: MiR-150 enhances the motility of EPCs in vitro and
promotes EPCs homing and thrombus resolving in vivo. Thromb Res.
133:590–598. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Higdon AN, Benavides GA, Chacko BK, Ouyang
X, Johnson MS, Landar A, Zhang J and Darley-Usmar VM: Hemin causes
mitochondrial dysfunction in endothelial cells through promoting
lipid peroxidation: The protective role of autophagy. Am J Physiol
Heart Circ Physiol. 302:H1394–H1409. 2012. View Article : Google Scholar : PubMed/NCBI
|