Introduction
Lung cancer has a high mortality rate of ~27% and is
becoming more prevalent in younger populations (1). Despite progress in the diagnosis and
treatment of lung cancer, the 5-year survival rate is only 16%
(2). Individualized therapy is a
promising treatment strategy for non-small cell lung cancer
(3). Mutations in epidermal growth
factor receptor (EGFR) drive the development of lung
adenocarcinoma and have altered the traditional treatment
approaches. Next-generation sequencing revealed that patients with
wild-type EGFR or ALK could present concurrent
oncogenic mutations in KRAS proto-oncogene GTPase
(KRAS) (4),
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit α
(PIK3CA) (5) and tumor
protein p53 (TP53) (6). These
mutations may result in differential clinical features, treatment
outcomes and survival prognoses. The association between KRAS,
PIK3CA and TP53 mutations, clinical features, and the
prognosis of patients with NSCLC is unclear. The present study
retrospectively analyzed 89 cases of NSCLC patients with KRAS,
PIK3CA and TP53 mutations to elucidate the association
between gene mutation, clinical characteristics and survival
prognosis as a basis for individualized treatment.
Patients and methods
Patient selection
A total of 122 patients accepted next-generation
sequencing for advanced NSCLC at Shanghai Changhai Hospital
(Shanghai, China) and were enrolled between January 2015 and
December 2016. Missing information and loss to follow-up resulted
in the exclusion of 33 patients. Blood samples and clinical data
from 89 patients with identified genes were collected, including
sex, age, smoking status, symptoms, laboratory test results, chest
computed tomography (CT) results, tumor location, pathological
type, Tumor-Node-Metastasis stage (7) and site of metastasis. Among the 89
samples, 50 exhibited KRAS, TP53 and PIK3CA
mutations. The Ethics Committee of Shanghai Changhai Hospital
approved the present study, and written informed consent was
obtained from each participant.
Gene sequencing
Circulating Single-Molecule Amplification and
Resequencing Technology (cSMART; Illumina CN500; Berry Genomics
Co., Ltd., Beijing, China) was used to detect KRAS, PIK3CA
and TP53 mutation in all patients with NSCLC. In brief,
genomic DNA was extracted from the plasma of the patients using
MagMAX Cell-Free DNA Isolation kit, (Thermo Fisher Scientific,
Inc., Waltham, MA, USA; Article no. A29319) DNA was purified using
a DNA purification kit (Berry Genomics Co., Ltd; Article no.
R0037). The libraries were prepared from 10 ng plasma DNA by
ligation of universal sequencing adaptors containing unique 6-bp
barcodes. Modified DNA was denatured and single strands were
circularized by Taq ligase. Bidirectional back-to-back primers, in
either singleplex or multiplex format, were annealed close to the
mutation loci. Inverse PCR was performed to replicate targeted
genes. Amplified products were subjected to massive parallel
sequencing on the MiSeq platform (Illumina, Inc., San Diego, CA,
USA) to generate paired-end reads of 2×200 bp (8).
Treatment
All patients were administered with a first-line
chemotherapy regimen of pemetrexed (500
mg/m2)/paclitaxel (135 mg/m2) and carboplatin
(area under the curve=5). All patients provided written informed
consent.
Survival analysis
Tumors were evaluated every 2 cycles during
chemotherapy treatment or earlier when significant signs of
progression, including aggravation of cough or hemoptysis, were
present. Progression-free survival (PFS) was determined according
to the Response Evaluation Criteria in Solid Tumors guidelines
(version 1.1) (9). The PFS time was
defined as the time from the beginning of chemotherapy to the
presence of objective evidence of progression. The final follow-up
date was June 30, 2017.
Statistical analysis
Survival curves were calculated using the
Kaplan-Meier method from the beginning of chemotherapy to
documented progression or mortality from any cause, differences in
PFS were assessed using the log-rank test. Statistical analysis was
performed with SPSS version 21 (IBM, Corp., Armonk, NY, USA). The
χ2 test was used to compare the categorical variables.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Patient characteristics
A total of 122 patients with NSCLC received cSMART
sequencing and 33 patients were excluded due to missing information
or loss to follow-up. A total of 89 patients were therefore
enrolled in the present study, and the baseline demographic
characteristics are shown in Table
I. The study cohort consisted of 52 males and 37 females, with
a median age of 61.0 years and a mean (± standard error) age of
59.4 (±12.2) years. Adenocarcinoma was histologically determined in
75 patients. There were 2 patients with adenosquamous carcinoma and
12 with squamous carcinoma. In total, 41 patients were smokers and
48 had never smoked.
 | Table I.Baseline demographic characteristics
of the 89 patients with non-small cell lung cancer. |
Table I.
Baseline demographic characteristics
of the 89 patients with non-small cell lung cancer.
|
Characteristics | n (%) |
|---|
| Sex |
|
|
Male | 52 (58.4) |
|
Female | 37 (41.6) |
| Age, years |
|
|
<65 | 57 (64.0) |
|
≥65 | 32 (36.0) |
| Surgical
history |
|
|
Yes | 21 (23.6) |
| No | 68 (76.4) |
| Smoking status |
|
|
Former/current | 41 (46.1) |
|
Never | 48 (53.9) |
| First symptom |
|
|
Yes | 60 (67.4) |
| No | 29 (32.6) |
| Tumor site |
|
| Left
lung | 45 (50.6) |
| Right
lung | 44 (49.4) |
| Histology |
|
|
Adenocarcinoma | 75 (84.3) |
|
Adenosquamous carcinoma | 2 (2.2) |
|
Squamous cell carcinoma | 12 (13.5) |
| Invasive
growth |
|
|
Yes | 50 (56.2) |
| No | 39 (43.8) |
| TNM stage |
|
| I | 1 (1.1) |
| II | 3 (3.4) |
|
III | 14 (15.7) |
| IV | 71 (79.8) |
| Metastasis |
|
|
Yes | 71 (79.8) |
| No | 18 (20.2) |
| Metastatic
site |
|
|
Bone | 39 (43.8) |
|
Brain | 20 (22.5) |
|
Adrenal | 8 (9.0) |
|
Liver | 9 (10.1) |
|
Pleura | 27 (30.3) |
| Lymph
nodes | 22 (24.7) |
Gene mutations
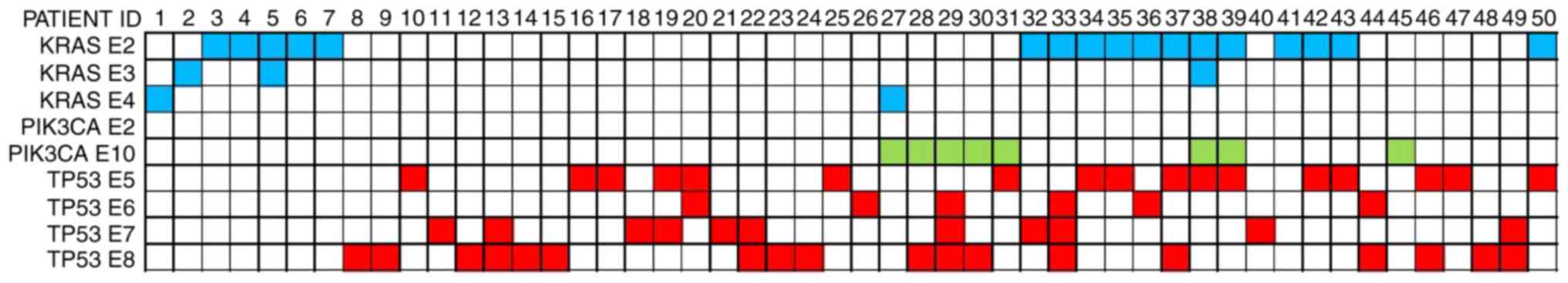
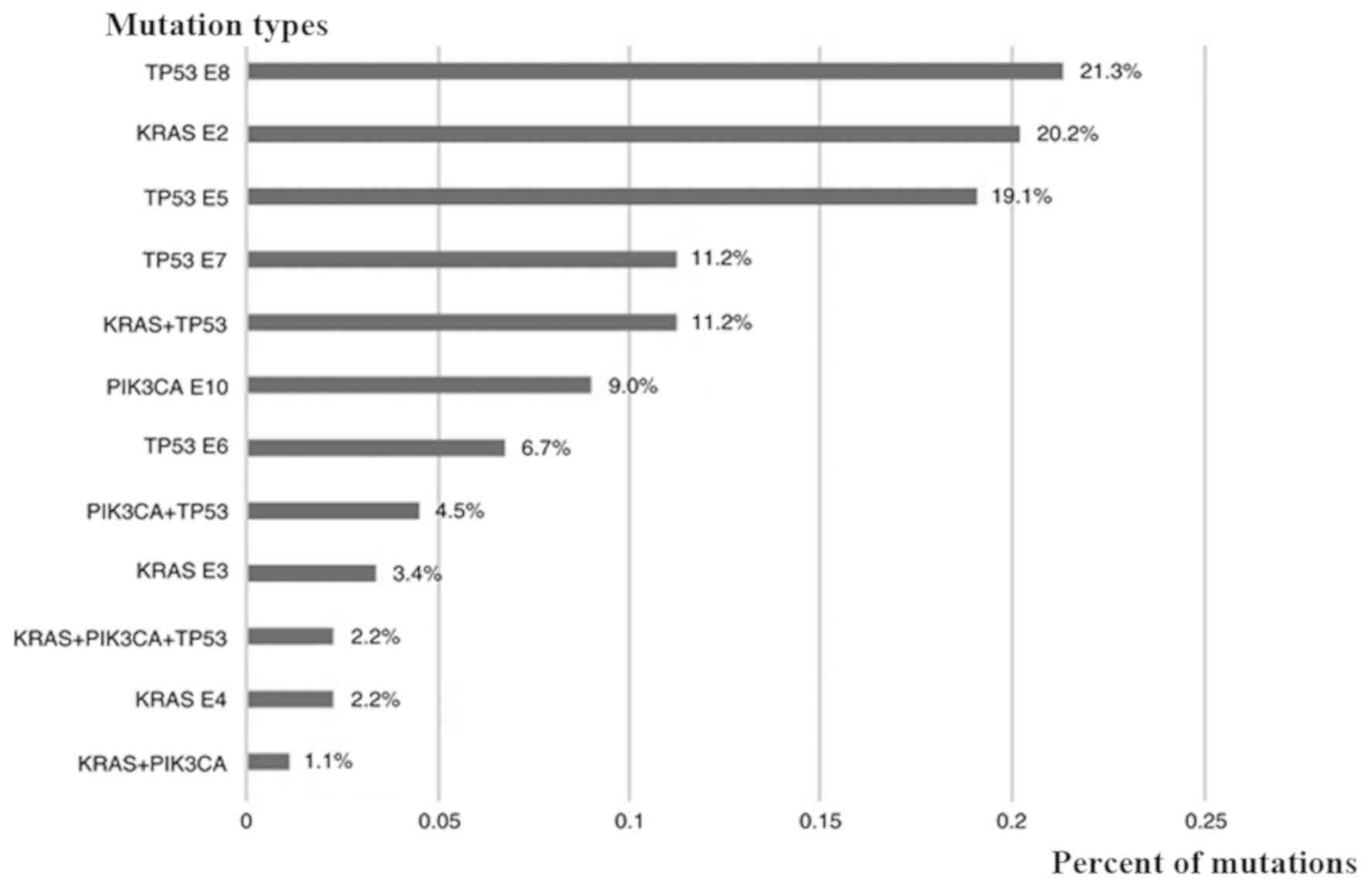
Oncogenic mutations were found in 50 patients,
including KRAS (n=21, 23.6%), PIK3CA (n=8, 9.0%) and
TP53 (n=40, 44.9%). Among the 21 patients with KRAS
mutations, 18 had mutations in exon 2, 3 in exon 3 and 2 in exon 4.
There were 8 patients with a PIK3CA mutation in exon 10. A
total of 17/40 patients had TP53 mutations located in exon
5, 6 in exon 6, 10 in exon 7 and 19 in exon 8. Coexisting mutations
were identified in 17 patients (19.1%), including
KRAS/TP53 (n=10, 11.2%), PIK3CA/TP53
(n=4, 4.5%), KRAS/PIK3CA (n=1, 1.1%) and
KRAS/PIK3CA/TP53 (n=2, 2.2%). There were 32
cases with EGFR mutations (36.0%), 3 cases with the
EMAP-like 4-ALK receptor tyrosine kinase fusion oncogene (3.4%),
and 3 cases of c-MET exon 14 skipping (3.4%). The
KRAS/TP53/PIK3CA mutations and percentage
distribution of the 50 patients are shown in Figs. 1 and 2.
Clinical characteristics
The clinical characteristics of the 89 patients in
association with the gene mutations are shown in Table II. Patients with KRAS, TP53,
PIK3CA and KRAS/TP53 mutations had a higher
incidence of bone metastasis than those with the wild-type gene
(61.9 vs. 25.6%, P=0.006; 62.5 vs. 25.6%, P=0.024; 62.5 vs. 25.6%,
P=0.042; 70.0 vs. 25.6%, P=0.009). There was also a higher
incidence of adrenal metastasis in the TP53 mutation vs.
wild-type groups (12.5 vs. 5.1%, P=0.017). Patients with
KRAS or KRAS/TP53 mutations had a lower
incidence of pleural metastasis than those with the wild-type gene
(14.3 vs. 43.6%, P=0.022; 0.0 vs. 43.6%, P=0.010). Infiltrative
tumor growth was greater in patients with KRAS, TP53 and
KRAS/TP53 mutations than in the wild-type group (71.4
vs. 51.3%, P=0.039; 67.5 vs. 51.3%, P=0.032; 90.0 vs. 51.3%,
P=0.009).
 | Table II.Association between gene mutation and
clinical features. |
Table II.
Association between gene mutation and
clinical features.
|
Characteristics | All wt, n (%) | KRAS mt, n (%) | χ2 | P-value | PIK3CA mt, n
(%) | χ2 | P-value | TP53 mt, n (%) | χ2 | P-value | KRAS+TP53 mt, n
(%) | χ2 | P-value | PIL3CA+TP53 mt, n
(%) | χ2 | P-value |
|---|
| Sex |
|
| 0.342 | 0.559 |
| 0.219 | 0.640 |
| 0.018 | 0.894 |
| 1.514 | 0.219 |
| 0.120 | 0.729 |
|
Male | 23 (59.0) | 14 (66.7) |
|
| 4 (50.0) |
|
| 23 (57.5) |
|
| 8 (80.0) |
|
| 2 (50.0) |
|
|
|
Female | 16 (41.0) | 7 (33.3) |
|
| 4 (50.0) |
|
| 17 (42.5) |
|
| 2 (20.0) |
|
| 2 (50.0) |
|
|
| Age, years |
|
| 0.115 | 0.694 |
| 0.003 | 0.959 |
| 0.307 | 0.580 |
| 0.245 | 0.620 |
| 0.202 | 0.653 |
|
<65 | 24 (61.5) | 14 (66.7) |
|
| 5 (62.5) |
|
| 27 (67.5) |
|
| 7 (70.0) |
|
| 2 (50.0) |
|
|
|
≥65 | 15 (38.5) | 7 (33.3) |
|
| 3 (37.5) |
|
| 13 (32.5) |
|
| 3 (30.0) |
|
| 2 (50.0) |
|
|
| Tumor site |
|
| 0.012 | 0.914 |
| 0.710 | 0.400 |
| 0.117 | 0.732 |
| 0.122 | 0.727 |
| 0.022 | 0.883 |
| Left
lung | 21 (53.8) | 11 (52.4) |
|
| 3 (37.5) |
|
| 20 (50.0) |
|
| 6 (60.0) |
|
| 2 (50.0) |
|
|
| Right
lung | 18 (46.2) | 10 (47.6) |
|
| 5 (62.5) |
|
| 20 (50.0) |
|
| 4 (40.0) |
|
| 2 (50.0) |
|
|
| Smoking status |
|
| 0.388 | 0.533 |
| 0.004 | 0.947 |
| 0.110 | 0.741 |
| 3.148 | 0.076 |
| 0.002 | 0.961 |
|
Former/current | 19 (48.7) | 12 (57.1) |
|
| 4 (50.0) |
|
| 18 (45.0) |
|
| 8 (80.0) |
|
| 2 (50.0) |
|
|
|
Never | 20 (51.3) | 9 (42.9) |
|
| 4 (50.0) |
|
| 22 (55.0) |
|
| 2 (20.0) |
|
| 2 (50.0) |
|
|
| First symptom |
|
| 0.587 | 0.440 |
| 4.519 | 0.034a |
| 1.654 | 0.198 |
| 1.197 | 0.274 |
| 2.363 | 0.124 |
|
Yes | 24 (61.5) | 15 (71.4) |
|
| 8 (100.0) |
|
| 30 (75.0) |
|
| 8 (80.0) |
|
| 4 (100.0) |
|
|
| No | 15 (38.5) | 6 (28.6) |
|
| 0 (0.0) |
|
| 10 (25.0) |
|
| 2 (20.0) |
|
| 0 (0.0) |
|
|
| CA19-9 |
|
| 1.516 | 0.218 |
| 0.726 | 0.394 |
| 0.748 | 0.387 |
| 5.108 | 0.024a |
| 1.381 | 0.240 |
|
Normal | 30 (76.9) | 13 (61.9) |
|
| 3 (37.5) |
|
| 27 (67.5) |
|
| 6 (60.0) |
|
| 2 (50.0) |
|
|
|
High | 9 (23.1) | 8 (38.1) |
|
| 5 (62.5) |
|
| 13 (32.5) |
|
| 4 (40.0) |
|
| 2 (50.0) |
|
|
| Invasive
growth |
|
| 4.250 | 0.039a |
| 2.621 | 0.105 |
| 4.575 | 0.032a |
| 6.883 | 0.009a |
| 1.439 | 0.230 |
| No | 19 (48.7) | 6 (28.6) |
|
| 2 (25.0) |
|
| 13 (32.5) |
|
| 1 (10.0) |
|
| 1 (25.0) |
|
|
|
Yes | 20 (51.3) | 15 (71.4) |
|
| 6 (75.0) |
|
| 27 (67.5) |
|
| 9 (90.0) |
|
| 3 (75.0) |
|
|
| Margin
lobulation |
|
| 4.290 | 0.038a |
| 0.001 | 0.970 |
| 2.495 | 0.114 |
| 4.273 | 0.039a |
| 1.336 | 0.248 |
|
Yes | 29 (74.4) | 10 (47.6) |
|
| 6 (75.0) |
|
| 23 (57.5) |
|
| 4 (40.0) |
|
| 4 (100.0) |
|
|
| No | 10 (25.6) | 11 (52.4) |
|
| 2 (25.0) |
|
| 17 (42.5) |
|
| 6 (60.0) |
|
| 0 (0.0) |
|
|
| Pleural
traction |
|
| 0.923 | 0.337 |
| 0.710 | 0.400 |
| 0.014 | 0.905 |
| 0.047 | 0.828 |
| 4.210 | 0.040a |
|
Yes | 18 (46.2) | 7 (33.3) |
|
| 5 (62.5) |
|
| 19 (47.5) |
|
| 5 (50.0) |
|
| 4 (100.0) |
|
|
| No | 21 (53.8) | 14 (66.7) |
|
| 3 (37.5) |
|
| 21 (52.5) |
|
| 5 (50.0) |
|
| 0 (0.0) |
|
|
| Vacuolar signs |
|
| 1.187 | 0.276 |
| 0.777 | 0.378 |
| 0.336 | 0.562 |
| 3.921 | 0.048a |
| 0.448 | 0.503 |
|
Yes | 5 (12.8) | 5 (23.8) |
|
| 2 (25.0) |
|
| 7 (17.5) |
|
| 4 (40.0) |
|
| 1 (25.0) |
|
|
| No | 34 (87.2) | 16 (76.2) |
|
| 6 (75.0) |
|
| 33 (82.5) |
|
| 6 (60.0) |
|
| 3 (75.0) |
|
|
| Site of
metastasis |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Bone | 10 (25.6) | 13 (61.9) | 7.594 | 0.006a | 5 (62.5) | 4.150 | 0.042a | 25 (62.5) | 5.081 | 0.024a | 7 (70.0) | 6.912 | 0.009a | 2 (50.0) | 1.070 | 0.301 |
|
Brain | 6 (15.4) | 7 (33.3) | 2.591 | 0.107 | 3 (37.5) | 2.097 | 0.148 | 10 (25.0) | 1.610 | 0.204 | 2 (20.0) | 0.124 | 0.725 | 1 (25.0) | 0.246 | 0.620 |
|
Adrenal | 2 (5.1) | 4 (19.0) | 2.939 | 0.086 | 2 (25.0) | 3.367 | 0.067 | 5 (12.5) | 5.648 | 0.017a | 2 (20.0) | 2.348 | 0.125 | 1 (25.0) | 2.207 | 0.137 |
|
Liver | 4 (10.3) | 3 (14.3) | 0.215 | 0.643 | 1 (12.5) | 0.035 | 0.851 | 5 (12.5) | 2.806 | 0.094 | 2 (20.0) | 0.703 | 0.402 | 0 (0.0) | 0.452 | 0.501 |
|
Pleura | 17 (43.6) | 3 (14.3) | 5.275 | 0.022a | 2 (25.0) | 0.953 | 0.329 | 8 (20.0) | 0.032 | 0.859 | 0 (0.0) | 6.675 | 0.010a | 0 (0.0) | 2.884 | 0.089 |
| Lymph
nodes | 6 (15.4) | 6 (28.6) | 1.484 | 0.223 | 3 (37.5) | 2.097 | 0.148 | 14 (35.0) | 1.610 | 0.204 | 4 (40.0) | 2.969 | 0.085 | 1 (25.0) | 0.246 | 0.620 |
KRAS/TP53 mutations were associated
with elevated carbohydrate antigen 19-9 (CA19-9) expression,
vacuolar signs and margin lobulation in chest CT imaging in
patients. Differences in KRAS mutation were observed in
margin lobulation and invasive growth in chest CT imaging,
meanwhile, first symptoms, including cough and dyspnea, indicated a
statistical significance between wild-type patients and those with
PIK3CA mutation (P=0.034).
Survival analysis
The PFS times of the KRAS mutation and
wild-type group were 8.9±2.3 months (95% CI, 4.3–13.5) and 15.3±1.6
months (95% CI, 12.1–18.4), respectively (P=0.045). Patients with a
single TP53 mutation had a PFS time of 7.8±1.5 months (95%
CI, 4.9–10.7), which was significantly shorter than that of the
wild-type group (P<0.001). Patients with a KRAS/TP53
coexisting mutation had a shorter PFS time of 6.6±1.6 months (95%
CI, 3.5–9.7) compared with the wild-type group (P<0.001). This
result was similar among PIK3CA/TP53 patients (P=0.012). The
difference in the PFS times was not statistically significant
between the single KRAS and KRAS/TP53 mutations, the
single TP53 and KRAS/TP53 mutations or the single
TP53 and PIK3CA/TP53 mutations. (Table III; Fig.
3).
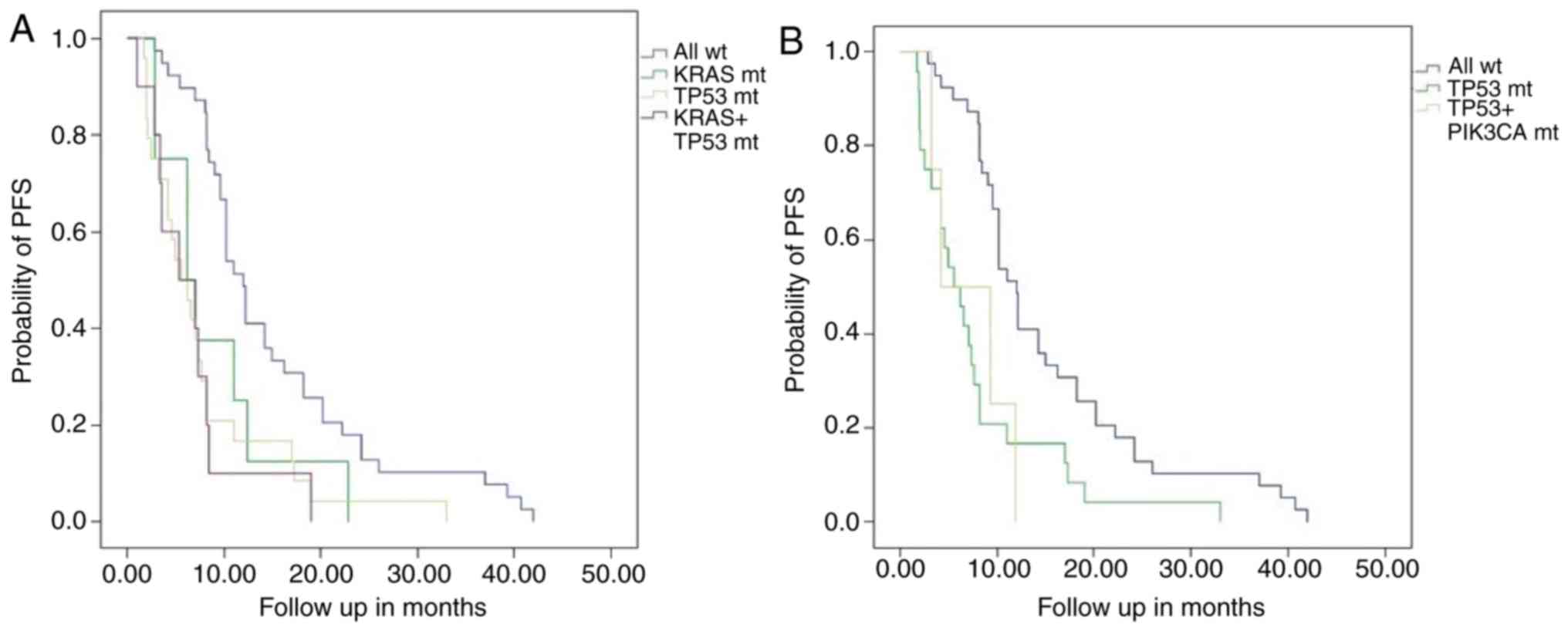 | Figure 3.PFS Kaplan-Meier curves between
different gene mutations group and wild-type group. (A) PFS in
patients with non-small cell lung cancer was compared between
KRAS mutant and all wild-type groups (8.9±2.3 vs. 15.3±1.6
months; P=0.045). PFS was compared between TP53 mutant and
all wild-type groups (7.8±1.5 vs. 15.3±1.6 months; P<0.001), and
between KRAS+TP53 mutant and all wild-type groups
(6.6±1.6 vs. 15.3±1.6 months; P<0.001). Compared with the
KRAS mutant and TP53 mutant, respectively, the PFS
time of patients with the KRAS+TP53 mutant was
shorter, but not statistically different. (B) PFS was also compared
between patients with the PIK3CA+TP53 mutant and all
wild-type groups (7.1±2.1 vs. 15.3±1.6 months; P=0.012). The PFS
time of the patients with the PIK3CA+TP53 mutant was
shorter than that of patients with the TP53 mutant, but was
not statistically different. PFS, progression-free survival; mt,
mutant; wt, wild-type; KRAS, KRAS proto-oncogene GTPase;
PIK3CA, phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic
subunit α; TP53, tumor protein p53. |
 | Table III.Survival prognosis in non-small cell
lung cancer patients with KRAS, PIK3CA and TP53 gene
mutations. |
Table III.
Survival prognosis in non-small cell
lung cancer patients with KRAS, PIK3CA and TP53 gene
mutations.
|
| KRAS mt vs. all
wt | TP53 mt vs. all
wt | KRAS+TP53 mt vs.
all wt |
KRAS+TP53 mt vs. KRAS
mt |
KRAS+TP53 mt vs. TP53
mt |
PIK3CA+TP53 mt vs. all
wt |
PIK3CA+TP53 mt vs.
TP53 mt |
|---|
|
|
|
|
|
|
|
|
|
|---|
| Variables | KRAS mt | All wt | TP53 mt | All wt | KRAS+ TP53 mt | All wt | KRAS+ TP53 mt | KRAS mt | KRAS+ TP53 mt | TP53 mt | PIK3CA+ TP53
mt | All wt | PIK3CA+ TP53
mt | TP53 mt |
|---|
| PFS time,
months | 8.9±2.3 | 15.3±1.6 | 7.8±1.5 | 15.3±1.6 | 6.6±1.6 | 15.3±1.6 | 6.6±1.6 | 8.9±1.3 | 6.6±1.6 | 7.8±1.5 | 7.1±1.5 | 15.3±1.6 | 7.13±11 | 7.8±1.5 |
| 95% CI | 4.3–13.5 | 12.1–18.4 | 4.9–10.7 | 12.1–18.4 | 3.5–9.7 | 12.1–18.4 | 3.5–9.7 | 4.3–13.5 | 3.5–9.7 | 4.9–10.7 | 3.1–11.2 | 12.1–18.4 | 3.1–11.2 | 4.9–10.7 |
| P-value |
| 0.045 |
| <0.001 |
| <0.001 |
| 0.398 |
| 0.873 |
| 0.012 |
| 0.986 |
Discussion
NSCLC accounts for 70–80% of lung cancer cases and
60% of patients are diagnosed at stage III or IV (10). Oncogenes such as EGFR and
ALK have shifted the treatment model of lung cancer from
pathology-guided to molecular-guided precision medicine with
targeted therapy (11). With the
improvement in examination technology and the increase in available
treatment methods, the genetic and clinical characteristics of
NSCLC-related genes, including KRAS, PIK3CA and TP53,
are highly informative.
The present study evaluated 89 cases of NSCLC
patients with KRAS, PIK3CA and TP53 mutations.
KRAS mutations were found in 21 cases within exon 2 (n=18),
exon 3 (n=3) and exon 4 (n=2). The total mutation rate of
KRAS was 23.6%, which was similar to the results of a study
undertaken by Mao et al (12), but higher than the mutation rates of
4.4–5.3% reported by Luo et al (13) and Yi et al (14). The mutation rate of PIK3CA was
3% in a study undertaken by Scheffler et al (15), but Liang et al (16) reported a rate of 47.83%. The present
study included 8 cases of PIK3CA exon 10 mutations and the
total mutation rate was 9.0%. TP53 has the highest mutation
rate of all NSCLC-related genes, reported as 39–46% (15,16). The
present study identified 40 cases with TP53 mutations within
exon 5 (n=17), exon 6 (n=6), exon 7 (n=10) and exon 8 (n=19). In
present study the total mutation rate of TP53 was 44.9%,
which is in accordance to previous researches (17,18).
Kris et al (19) found that 3% of patients with NSLCL
exhibited a double gene mutation. The Cancer Genome Atlas
determined that the mutation rate of KRAS/TP53 coexisting
mutation could reach 20% (20). The
present study identified 17 co-mutated samples with a rate of
19.1%, including 15 double-mutations of KRAS/TP53,
PIK3CA/TP53 and KRAS/PIK3CA, and 2 cases of
KRAS/PIK3CA/TP53 co-mutation. This difference may result
from the sensitivity and sequencing depth of next-generation
sequencing by cSMART. The varied sample size between studies may
also contribute toward the discrepancies in gene mutation
rates.
Clinical characteristics, including the baseline
demographics, clinical manifestations, partial laboratory tests,
partial pathological features and certain features of chest CT
imaging, of patients with mutations were not significantly
different from those of wild-type patients (P>0.05). This was
consistent with the results of numerous previous studies (6,21–25). By
contrast, KRAS/TP53 were associated with elevated
CA19-9 expression, vacuolar signs and margin lobulation in chest CT
imaging. However, it is possible that the sample size of each
subgroup resulted in the difference in certain clinical
characteristics to some extent, and further study is required due
to the limited sample size used in the present study.
Invasive growth of the tumor tissue in patients was
associated with KRAS, TP53 and KRAS/TP53,
which was consistent with the clinical features observed. The
incidence of distant metastasis was higher than that of local
metastasis in patients with KRAS and TP53 mutations.
The possible mechanism of this is the activation of the EGFR
downstream Rat sarcoma/Rapidly Accelerated
Fibrosarcoma/mitogen-activated protein kinases signaling pathways
by KRAS mutations to regulate cell differentiation and
proliferation. Prolonged activation of the KRAS signal is
hypothesized to cause tumor cell proliferation and progression
(26). TP53 gene mutations
result in an oncogenic transformation of the tumor suppressor gene
due to a conformational change; therefore, the regulation of cell
growth, apoptosis and DNA repair is disrupted, which allows tumor
cells to proliferate, grow and metastasize (27,28).
The biological significance of these mutations
remains uncertain, but to some extent specific driver genes have
prognostic value. Mascaux et al (29) first reported a poor prognosis in
NSCLC patients with KRAS mutations, and other studies have
confirmed this hypothesis (30).
Recent studies have found that TP53 gene mutations may
generate the same results in patients with NSCLC (31–33).
PIK3CA encodes the type I phosphatidylinositol-3-kinase
p110α catalytic subunit (34)
and is important for the development of NSCLC. PIK3CA
phosphorylates the EGFR bypass pathway, PI3K/AKT/mTOR, to activate
downstream signaling that promotes the proliferation, survival,
adhesion and differentiation of tumor cells (35). Liang et al (16) proposed that PIK3CA gene
mutations are more likely to co-exist with other oncogenic
mutations and that they may weakly induce independent
carcinogenesis.
In the present study, patients with NSCLC who
underwent first-line chemotherapy were divided into groups
according to their genotype. For all patients who have EGFR
mutation in Changhai hospital, targeted therapy is discussed and
anti-EGFR tyrosine kinase inhibitors are recommended as the
first-line treatment. The majority of these patients do receive
targeted therapy. However, due to economic problems or for other
reasons, certain patients cannot afford targeted therapy. For the
baseline balance of the present study, the 89 patients who received
first-line chemotherapy were chosen. Patients with a single KRAS
or TP53 mutation experienced shorter PFS times than the
wild-type patients, which was consistent with the results of the
studies by Molina-Vila et al (36) and Meng et al (37). Shepherd et al (6) hypothesized that a double gene mutation,
such as KRAS/TP53, in NSCLC patients may indicate a
poor prognosis. Patients with KRAS/TP53 or
PIK3CA/TP53 mutations experienced a shorter PFS time
than those patients with the wild-type. The PFS time of the
KRAS/TP53 group was shorter than that in the single
KRAS and single TP53 groups, as was the time in the
PIK3CA/TP53 group compared with the single
TP53 group (P>0.05). We hypothesized that there could be
a ‘gene superposition’ effect in NSCLC patients with a co-mutated
gene, which leads to a shortened PFS compared with a single gene
mutation. However, the trend observed in the present study was not
statistically significant, which was in agreement with the results
of a study by Jao et al (38). The mean PFS time of patients with
KRAS/PIK3CA/TP53 gene co-mutations was 6.2
months, which was shorter than that of the double and single
mutation groups. Only 2 patients had this co-mutation and
therefore, a larger sample size is necessary for further study.
Sampling error may also exist due to the next-generation sequencing
technology and the limited sample size. The subgroups of gene
mutations, as well as the chemotherapy regimen and doses, were not
identical; therefore, further evidence should be obtained in a
large clinical study.
In conclusion, the treatment strategy for NSCLC
patients with KRAS, PIK3CA and TP53 mutations has not
yet been defined. The present study determined the predictive value
of KRAS, PIK3CA and TP53 mutations in patients with
NSCLC. Additionally, the results of the present study suggested
that patients with NSCLC should undergo routine KRAS, PIK3CA
and TP53 sequencing to determine single or multiple gene
mutations for the analysis of patient clinical characteristics and
prognosis.
Acknowledgements
The authors would like to thank the staff of the
Department of Respiratory and Critical Care Medicine, Shanghai
Changhai Hospital (Shanghai, China) for providing assistance in
data management and statistical analysis.
Funding
The present study was supported by the Shanghai
Scientific Research Projects (grant no. 15411960400).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
JZ, YH, RC and CB conceived and designed the study.
JL analyzed the statistics. CB provided a part of patients'
clinical data and monitored the whole study; JZ and RC wrote the
original draft, YH and CB reviewed and edited the draft. All
authors have read and approved the final version of this
manuscript.
Ethics approval and consent to
participate
The study was approved by the Ethics Committee of
Changhai Hospital affiliated to Second Military Medical University
(Shanghai, China).
Patient consent for publication
Written nformed consent and permission for
publication was obtained for all patients in the present study.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel R, Ma J, Zou Z and Jemal A: Cancer
statistics, 2014. CA Cancer J Clin. 64:9–29. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zeng H, Zheng R, Guo Y, Zhang S, Zou X,
Wang N, Zhang L, Tang J, Chen J, Wei K, et al: Cancer survival in
China, 2003–2005: A population-based study. Int J Cancer.
136:1921–1930. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Moreira AL and Thornton RH: Personalized
medicine for non-small-cell lung cancer: Implications of recent
advances in tissue acquisition for molecular and histologic
testing. Clin Lung Cancer. 13:334–339. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Martin P, Leighl NB, Tsao MS and Shepherd
FA: KRAS mutations as prognostic and predictive markers in
non-small cell lung cancer. J Thorac Oncol. 8:530–542. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Papadimitrakopoulou V: Development of
PI3K/AKT/mTOR pathway inhibitors and their application in
personalized therapy for non-small-cell lung cancer. J Thorac
Oncol. 7:1315–1326. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Shepherd FA, Lacas B, Le Teuff G, Hainaut
P, Jänne PA, Pignon JP, Le Chevalier T, Seymour L, Douillard JY,
Graziano S, et al: Pooled analysis of the prognostic and predictive
effects of TP53 comutation status combined with KRAS or EGFR
mutation in early-stage resected non-small-cell lung cancer in four
trials of adjuvant chemotherapy. J Clin Oncol. 35:2018–2027. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Detterbeck FC, Boffa DJ, Kim AW and Tanoue
LT: The eighth edition lung cancer stage classification. Chest.
151:193–203. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lv W, Wei X, Guo R, Liu Q, Zheng Y, Chang
J, Bai T, Li H, Zhang J, Song Z, et al: Noninvasive prenatal
testing for Wilson disease by use of circulating single-molecule
amplification and resequencing technology (cSMART). Clin Chem.
61:172–181. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Watanabe H, Okada M, Kaji Y, Satouchi M,
Sato Y, Yamabe Y, Onaya H, Endo M, Sone M and Arai Y: New response
evaluation criteria in solid tumours-revised RECIST guideline
(version 1.1). Gan To Kagaku Ryoho. 36:2495–2501. 2009.(In
Japanese). PubMed/NCBI
|
|
10
|
Tsim S, O'Dowd CA, Milroy R and Davidson
S: Staging of non-small cell lung cancer (NSCLC): A review. Respir
Med. 104:1767–1774. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Cooper WA, O'Toole S, Boyer M, Horvath L
and Mahar A: What's new in non-small cell lung cancer for
pathologists: The importance of accurate subtyping, EGFR mutations
and ALK rearrangements. Pathology. 43:103–115. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Mao C, Qiu LX, Liao RY, Du FB, Ding H,
Yang WC, Li J and Chen Q: KRAS mutations and resistance to
EGFR-TKIs treatment in patients with non-small cell lung cancer: A
meta-analysis of 22 studies. Lung Cancer. 69:272–278. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Luo W, Wang H, Xu WJ, et al: Analysis of
KRAS mutation in patients with non- small cell lung cancer.
Guangdong Med J. 35:2025–2028. 2014.(In Chinese).
|
|
14
|
Yi SQ, Zhuang Y, Zhu WD, et al: Analysis
of KRAS gene mutations in non-small cell lung cancer. Zhonghua Lin
Chuan Yi Shi Za Zhi (Electronic Edition). 7:9111–9115. 2013.(In
Chinese).
|
|
15
|
Scheffler M, Bos M, Gardizi M, König K,
Michels S, Fassunke J, Heydt C, Künstlinger H, Ihle M, Ueckeroth F,
et al: PIK3CA mutations in non-small cell lung cancer (NSCLC):
Genetic heterogeneity, prognostic impact and incidence of prior
malignancies. Oncotarget. 6:1315–1326. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Liang NX, Liu YX, Liu L and Li SQ:
Co-mutation of PIK3CA and other oncogenes in patients with
non-small cell lung cancer. Med J PUMCH. 6:186–190. 2015.(In
Chinese).
|
|
17
|
Huang CL, Taki T, Adachi M, Konishi T,
Higashiyama M, Kinoshita M, Hadama T and Miyake M: Mutations of p53
and K-ras genes as prognostic factors for non-small cell lung
cancer. Int J Oncol. 12:553–569. 1998.PubMed/NCBI
|
|
18
|
Kato S, Han SY, Liu W, Otsuka K, Shibata
H, Kanamaru R and Ishioka C: Understanding the function-structure
and function-mutation relationships of p53 tumor suppressor protein
by high-resolution missense mutation analysis. Proc Natl Acad Sci
USA. 100:8424–8429. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kris MG, Johnson BE, Berry LD, Kwiatkowski
DJ, Iafrate AJ, Wistuba II, Varella-Garcia M, Franklin WA, Aronson
SL, Su PF, et al: Using multiplexed assays of oncogenic drivers in
lung cancers to select targeted drugs. Jama. 311:1998–2006. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Cancer Genome Atlas Research Network:
Comprehensive molecular profiling of lung adenocarcinoma. Nature.
511:543–550. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gao J, Chen JQ, Zhang L and Liang ZY:
Relationship between EGFR and KRAS mutations and prognosis in
Chinese patients with non-small cell lung cancer: A mutation
analysis with real-time polymerase chain reaction using scorpion
amplification refractory mutation system. Zhonghua Bing Li Xue Za
Zhi. 41:652–656. 2012.(In Chinese). PubMed/NCBI
|
|
22
|
Kim HR, Ahn JR, Lee JG, Bang DH, Ha SJ,
Hong YK, Kim SM, Nam KC, Rha SY, Soo RA, et al: The impact of
cigarette smoking on the frequency of and qualitative differences
in KRAS mutations in Korean patients with lung adenocarcinoma.
Yonsei Med J. 54:865–874. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Li Y, Li Y, Yang T, Wei S, Wang J, Wang M,
Wang Y, Zhou Q, Liu H and Chen J: Clinical significance of EML4-ALK
fusion gene and association with EGFR and KRAS gene mutations in
208 Chinese patients with non-small cell lung cancer. PLoS One.
8:e520932013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhang Q, Wang J, Li X, Zhang H, Nong J,
Qin N, Zhang X, Wu Y, Yang X, Lv J and Zhang S: Clinical Analysis
of 107 NSCLC Patients Harboring KRAS Mutation. Zhongguo Fei Ai Za
Zhi. 19:257–262. 2016.(In Chinese). PubMed/NCBI
|
|
25
|
Hu W, Liu Y and Chen J: Concurrent gene
alterations with EGFR mutation and treatment efficacy of EGFR-TKIs
in Chinese patients with non-small cell lung cancer. Oncotarget.
8:250462017.PubMed/NCBI
|
|
26
|
Korpanty GJ, Graham DM, Vincent MD and
Leighl NB: Biomarkers that currently affect clinical practice in
lung cancer: EGFR, ALK, MET, ROS-1, and KRAS. Front Oncol.
4:2042014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Deben C, Van den Bossche J, Van Der Steen
N, Lardon F, Wouters A, de Beeck KO, Hermans C, Jacobs J, Peeters
M, Van Camp G, et al: Deep sequencing of the TP53 gene reveals a
potential risk allele for non-small cell lung cancer and supports
the negative prognostic value of TP53 variants. Tumour Biol.
39:10104283176943272017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Vanderlaan PA, Rangachari D, Mockus SM,
Spotlow V, Reddi HV, Malcolm J, Huberman MS, Joseph LJ, Kobayashi
SS and Costa DB: Mutations in TP53, PIK3CA, PTEN and other genes in
EGFR mutated lung cancers: Correlation with clinical outcomes. Lung
Cancer. 106:17–21. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Mascaux C, Iannino N, Martin B, Paesmans
M, Berghmans T, Dusart M, Haller A, Lothaire P, Meert AP, Noel S,
et al: The role of RAS oncogene in survival of patients with lung
cancer: A systematic review of the literature with meta-analysis.
Br J Cancer. 92:131–139. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lin EY, Rupani R and Gitlitz BJ: Markers
in lung cancer. Springer. (New York, NY). 2013. View Article : Google Scholar
|
|
31
|
Scoccianti C, Vesin A, Martel G, Olivier
M, Brambilla E, Timsit JF, Tavecchio L, Brambilla C, Field JK and
Hainaut P;: European Early Lung Cancer Consortium: Prognostic value
of TP53, KRAS and EGFR mutations in nonsmall cell lung cancer: The
EUELC cohort. Eur Respir J. 40:177–184. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ma X, Rousseau V, Sun H, Lantuejoul S,
Filipits M, Pirker R, Popper H, Mendiboure J, Vataire AL, Le
Chevalier T, et al: Significance of TP53 mutations as predictive
markers of adjuvant cisplatin-based chemotherapy in completely
resected non-small-cell lung cancer. Mol Oncol. 8:555–564. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lee SY, Jeon HS, Hwangbo Y, Jeong JY, Park
JY, Lee EJ, Jin G, Shin KM, Yoo SS, Lee J, et al: The influence of
TP53 mutations on the prognosis of patients with early stage
non-small cell lung cancer may depend on the intratumor
heterogeneity of the mutations. Mol Carcinog. 54:93–101. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Mcgowan M, Hoven AS, Lund-Iversen M,
Solberg S, Helland Å, Hirsch FR and Brustugun OT: PIK3CA mutations
as prognostic factor in squamous cell lung carcinoma. Lung Cancer.
103:52–57. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Kang S, Bader AG and Vogt PK:
Phosphatidylinositol 3-kinase mutations identified in human cancer
are oncogenic. Proc Natl Acad Sci USA. 102:802–807. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Molina-Vila MA, Bertran-Alamillo J, Gascó
A, Mayo-de-las-Casas C, Sánchez-Ronco M, Pujantell-Pastor L,
Bonanno L, Favaretto AG, Cardona AF, Vergnenègre A, et al:
Nondisruptive p53 mutations are associated with shorter survival in
patients with advanced non-small cell lung cancer. Clin Cancer Res.
20:4647–4659. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Meng D, Yuan M, Li X, Chen L, Yang J, Zhao
X, Ma W and Xin J: Prognostic value of K-RAS mutations in patients
with non-small cell lung cancer: A systematic review with
meta-analysis. Lung Cancer. 81:1–10. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Jao K, Tomasini P, Kamel-Reid S and Tsao
MS: Prognostic effect of single versus multiple somatic mutations
in non-small cell lung cancer (NSCLC). J Clin Oncol.
33:75212015.
|