Introduction
Resveratrol (trans-3,4,5-trihydroxystilbene) is a
non-flavonoid polyphenolic compound produced naturally in grapes
and nuts (1) that is also present in
Polygonum cuspidatum root (2). Resveratrol is present in the skin of
grapes, and is been used by plants for protection against fungi and
other infections (3). However,
resveratrol has a number of other properties, including
anti-atherogenic, anti-inflammatory and anti-cancer effects
(4). Furthermore, previous research
has demonstrated that using resveratrol as a chemo-preventive agent
results in the inhibition of the cell cycle in tumour cells
(5).
Resveratrol has an anti-tumour potential in
colorectal cancer (6,7). In 2017 colorectal malignancy was the
third most common cancer type in men and women in the United States
that comprises ~9% of all cancer types (8). Colorectal cancer development may be
divided into two main phases, the initiation phase and the
progression phase. The initiation phase includes the inactivation
of the tumour suppressor gene adenomatous polyposis coli, which
controls various cellular processes, including cell division and
proliferation (6,9). In addition, β-catenin is translocated
from the cytoplasm to the nucleus through the Wnt signalling
pathway, in which β-catenin functions as an oncogene. The
progression phase is mostly characterised by the genomic
instability of several proteins including K-Ras, SMAD family member
4 and tumour protein p53 (p53) that results in colorectal carcinoma
progression (10).
p53 is a tumour suppressor that orchestrates the
cellular responses to genotoxic stresses (11–13).
Activation of p53 induces the transcription of its target genes
that are involved in cell cycle arrest, apoptosis and DNA repair
(14). p53 facilitates apoptosis
through transactivating a number of p53-target genes including Bcl2
associated X, apoptosis regulator (BAX) and Bcl2 binding component
3 (PUMA) (15). p53 is mainly
regulated through various post-translational modifications (PTMs),
including acetylation, methylation, neddylation, phosphorylation
and ubiquitination (13,16). Previous studies have demonstrated the
involvement of resveratrol in the cellular function of p53
(17–20). Furthermore, the pro-apoptotic
activity of resveratrol in prostate cancer appeared to be mediated
by the serine-15 phosphorylation of p53 by mitogen-activated
protein kinases (21). However, the
effect of resveratrol on other PTMs of p53 has not been
investigated in colorectal cancer. SET domain containing lysine
methyltransferase 7/9 (SET7/9) is one of the proteins that regulate
p53 through PTMs, which mono-methylates p53 at lysine 372 (K372)
that results in its stabilization and activation (11,16).
In the present study, the investigated the
mechanisms underlying the impact of resveratrol in p53 activation.
To address this question, we examined SET7/9 as a mediator of
Resveratrol-dependent p53 activation. The results of the present
study provide the mechanistic explanation for the regulatory
function of resveratrol in the p53 pathway.
Materials and methods
Plasmid constructs
Scrambled (cat no. shc016:
CCGGGCGCGATAGCGCTAATAATTTCTCGAGAAATTATTAGCGCTATCGCGCTTTTT) and
SET7/9 (cat no. SHCLND-NM_030648:
CCGGGCCAGGGTATTATTATAGAATCTCGAGATTCTATAATAATACCCTGGCTTTTTG) short
hairpin RNA (shRNA) were obtained from Sigma-Aldrich (Merck KGaA,
Darmstadt, Germany). β-galactosidase (cat no. RC200721; Origene
Technologies, Inc., Rockville, MD, USA), pGL3-basic vector
constructs were obtained from Promega Corporation (cat no. E1751;
Madison, WI, USA) and BAX or PUMA promoter regions were synthesized
by GenScript (Piscataway, NJ, USA).
Cell culture and plasmids
All cell lines used in the present study (HCT-116,
CO-115 and SW48) purchased from the American Type Culture
collection (Manassas, VA, USA) were cultured in Dulbecco's modified
Eagle's medium supplemented with 10% foetal bovine serum (Thermo
Fisher Scientific, Inc., Waltham, MA, USA) at 37°C with 5%
CO2. The 293 cells were seeded into 24-well culture
plates (3.5×10−4 cells/ml) and changed the medium
without FBS following an overnight incubation at 37°C with 5%
CO2. Transfections were performed using Lipofectamine
2000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according
to the manufacturer's protocols. Approximately 18–24 h prior to
transfection, plate cells in 1 ml complete growth medium per well
in a 12-well plate. Cells were 70–90% confluent at the time of
transfection. Cells were plated at a density of 1×105
cells/well and incubated at 37°C overnight. The medium was replaced
with 1 ml of fresh complete growth medium and 100 µl of serum-free
medium was placed in a sterile tube. A total of 1 µg plasmid DNA
was added to the medium in the tube and mixed completely by gently
pipetting up and down. A total of 2 µl transfection reagent was
added to the diluted DNA mixture and mixed. The samples was
incubated at room temperature for 20 min to allow sufficient time
for complexes to form. The supernatant of medium was collected to
infect the cancer cells. The viral titer was determined by Virus
drops degree detection; fluorescence/absolute quantitative method
(17). The cancer cells were
infected by the collected supernatant for 48 h and collected for
Western blot analysis. shRNA transfection was performed directly,
without using Lentivirus. Resveratrol or dimethyl sulfoxide (DMSO)
were obtained from Sigma-Aldrich (Merck KGaA).
Cell viability
A total of 10,000 (HCT-116, CO-115 and SW480) cells
were cultured in 96-well plates. After 24 h, the cells were treated
with various doses (0, 12.5, 25, 37.5, 50 µM) of resveratrol (cat
no. 34092-100 MG; Sigma-Aldrich, Merck KGaA, Darmstadt, Germany)
and incubated for 24 h at 37°C. Cell viability was measured using a
CellTiter-Glo Luminescent cell viability assay (cat no. G7572;
Promega Corporation) with the Glomax Explorer Microplate Reader
(Promega Corporation) according to the manufacturer's
protocols.
Western blotting
The protein was extracted from the HCT-116, CO-115
and SW480 cells with 10% MSDS (Sigma-Aldrich; Merck KGaA,
Darmstadt, Germany) and the protein quantity was determined using a
bicinchoninic acid assay. The mass of the proteins loaded in per
lane was 60 µg. The proteins were separated using 10% SDS-PAGE, and
then transferred onto polyvinylidene fluoride membranes. The
membranes were blocked in 5% bovine serum albumin (cat. no.
10099141; Gibco; Thermo Fisher Scientific, Inc.) at 37°C for 1 h,
and washed with TBST buffer three times. The membranes were exposed
to primary antibodies (1:1,000 dilution): p53 (Merck KGaA),
methylated-p53 (K372; cat no. ab16033; Abcam, Cambridge, UK),
SET7/9 (cat no. 2813) and poly (ADP-ribose) polymerase (PARP; cat
no. 9542S; Cell Signalling Technology, Inc., Danvers, MA, USA) and
β-actin (cat no. ab8227; Abcam). overnight at 4°C. The membranes
were washed with TBST three times, and incubated with secondary
antibody (anti-rabbit Immunoglobulin G; 1:2,000; cat. no. 7074;
CST) for 2 h at room temperature, and washed with TBST three times.
The bands were visualized with the WEST ZOL Plus system (iNtRON
Biotechnology, Boston, MA, USA) and quantified using ImageJ
software (version 1.44P; National Institutes of Health, Bethesda,
MA, USA).
Luciferase assay
The luciferase assay was performed as previously
described (12). Briefly, 100,000
HCT-116 or CO-115 cells per well were cultured in 24-well plates.
After 24 h, the cells were transfected with luciferase genes
(pGL3-PUMA or pGL3-BAX or pGL3 vectors) and β-galactosidase
constructs using Lipofectamine 2000 at 37°C with 5% CO2.
The following day, the RPMI-1640 medium supplemented with 10% FBS
media was replaced by fresh RPMI-1640 (with 10% FBS) growth media
and 6 samples were treated with different resveratrol doses (0,
12.5, 25, 37.5, 50 µM) at 37°C. A total of 24 h later, the lysates
were collected in lysis buffer (within the kit) and stored at −80°C
for 3 h; and the luciferase signals were assessed using a
luminometer using a Luciferase kit (cat no. K801-200; BioVision,
Inc., Milpitas, CA, USA) according to the manufacturer's
protocol.
Bioinformatics analysis
The publicly available bioinformatics data set
(GSE17537: 244653_at) was downloaded and analysed. The
bioinformatics analyses were obtained from PrognoScan (22,23)
(http://www.abren.net/PrognoScan). 55
patients with colorectal cancer (CRC), included 30 male and 25
female, with a median age of 62 years old (mean ± SD 62.23±3.43,
range 43–76). Tumor stage was determined according to the
tumor-node-metastasis (TNM) classification system of the
International Union against Cancer (2002). The tumors'
differentiation have been graded by the Edmondson-Steiner
classification system. The clinical pathological features of the
patients are summarized in Table
I.
 | Table I.Association of SET7/9 expression and
clinicopathological features in 55 CRCs. |
Table I.
Association of SET7/9 expression and
clinicopathological features in 55 CRCs.
| Clinicopathological
features | Total cases | P-value |
|---|
| Age (years) | 62.23±3.43 |
|
| ≤60 | 21 (38.2) | >0.05 |
|
>60 | 34 (61.6) |
|
| Sex |
|
|
|
Female | 30 (54.5) | >0.05 |
| Male | 25 (45.5) |
|
| Tissue |
|
|
|
Tumour | 55 |
|
|
Normal | 55 |
|
| Depth of
infiltration |
|
|
|
Submucosa, musculeris
propria | 14 (25.5) | >0.05 |
|
Subserosa | 31 (56.4) |
|
| To the
surrounding tissue | 10 (18.2) |
|
| Histological
differentiation |
|
|
| Well
differentiated | 8
(14.5) | <0.01 |
|
Moderately differentiated | 30 (54.5) |
|
| Poorly
differentiated | 17 (31.0) |
|
| TNM
staginga |
|
|
| 0 | 0 | <0.01 |
| I | 25 (45.5) |
|
| II | 22 (40.0) |
|
|
III | 8
(14.5) |
|
| Lymph node
metastasis |
|
|
|
Yes | 32 (58.2) | 0.0293 |
| No | 23 (41.8) |
|
Statistical analyses
Data were compiled in Microsoft Excel 2011 14.7.2
(Microsoft Corporation, Redmond, WA, USA) and the evaluation of
variance between data groups was performed using a one-way analysis
of variance. If significance was detected further comparisons were
performed with a Tukey's post-hoc test. All data are presented as
means ± standard deviation (SD) or median with 95% confidence
interval (95% CI). P<0.05 was considered to indicate a
statistically significant difference. Kaplan-Meier survival curves
with log-rank tests were used in the bioinformatics data
analysis.
Results
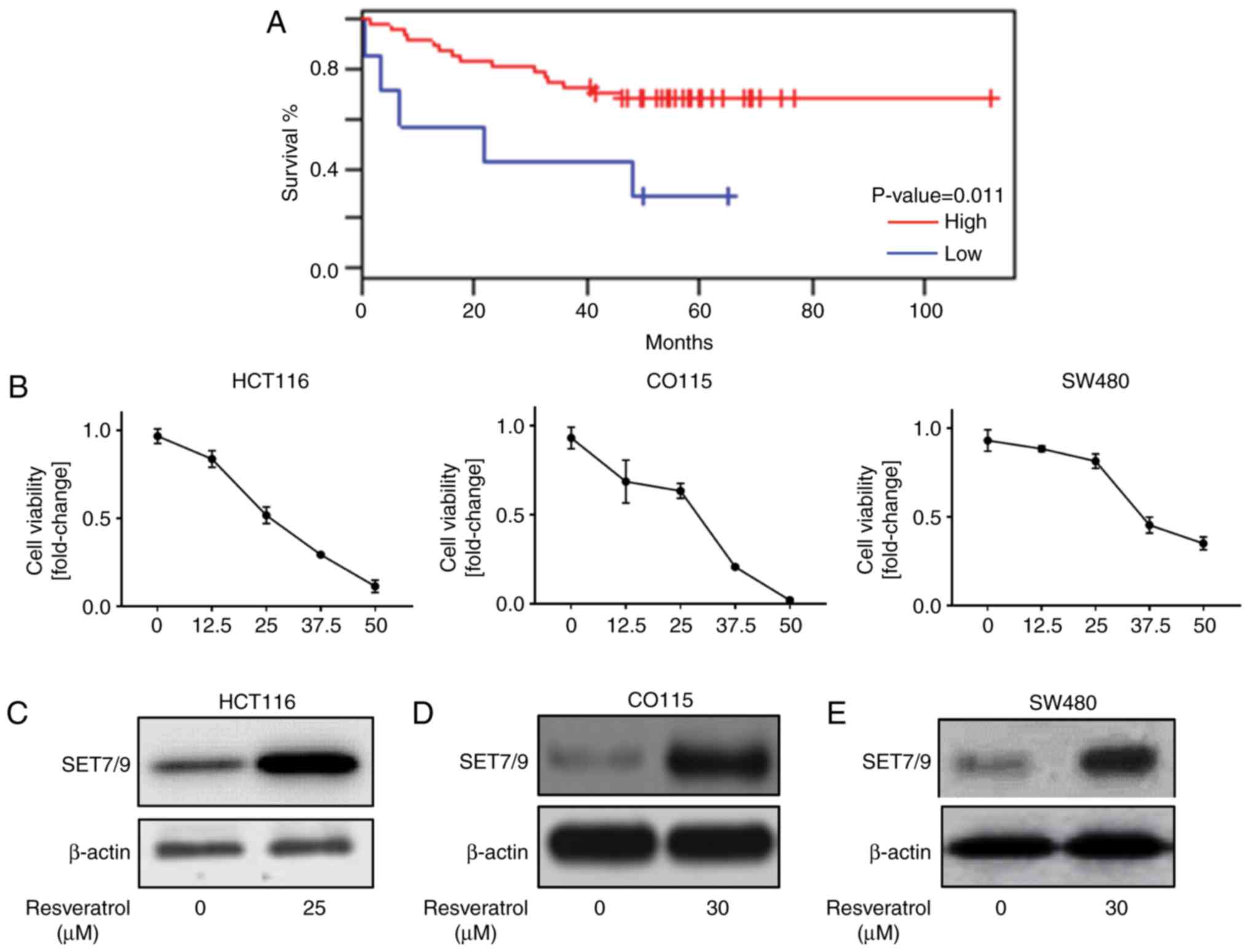
Resveratrol induces SET7/9 expression
in colorectal cancer cells
The expression of SET7/9 is associated with
tumorigenesis in different cancer types, including gastric
(24), breast (25) and colorectal cancer. To understand
the association between SET7/9 expression and the survival rate of
patients with colorectal cancer, the publicly available
bioinformatics data set (GSE17537: 244653_at) was analysed, and the
results suggested a significant positive association between a low
overall survival rate and the downregulation of SET7/9 (P=0.011;
Fig. 1A). Resveratrol has been
suggested as an anti-cancer agent in colorectal cancer (7), which induces the expression of p53
(18). Therefore, it was
hypothesized that resveratrol may regulate p53 through the
induction of SET7/9 expression in colorectal cancer cells. To
address this, initially the concentration (0, 12.5, 25, 37.5, 50
µM) of resveratrol that promoted cell death in HCT116, CO115 and
SW48 cell lines was determined (Fig.
1B). Then, the expression of SET7/9 in response to resveratrol
treatment was assessed. As presented in Fig. 1C-E, the cells treated with
resveratrol demonstrated higher protein expression of SET7/9
compared with those that were treated with DMSO. Collectively,
these data indicate that resveratrol is a positive regulator of
SET7/9.
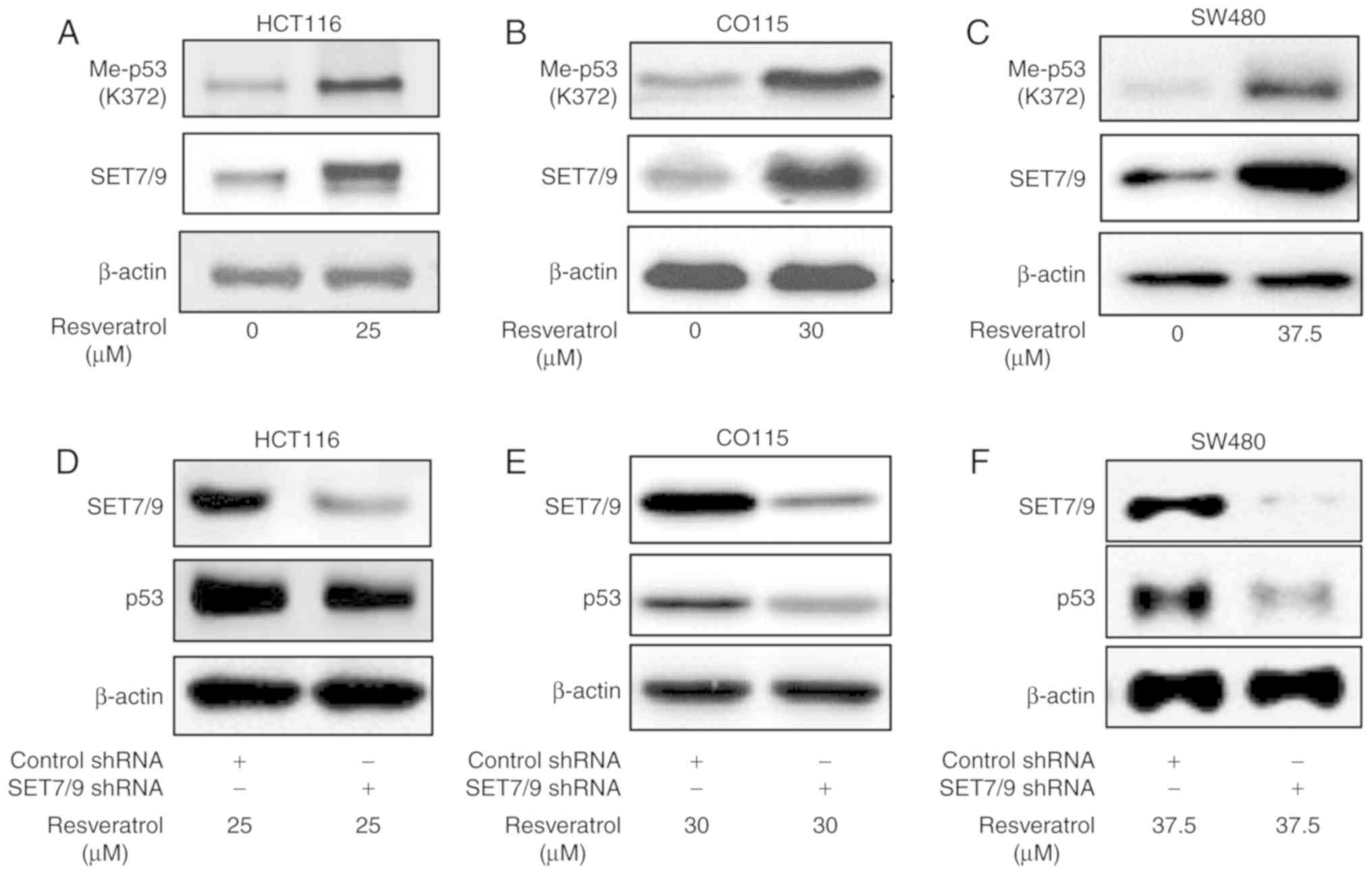
SET7/9 mediates resveratrol-dependent
p53 overexpression
Mono-methylation of p53 at K372 by SET7/9 has a key
role in p53 stability and activity (16). Therefore, the present study
determined the protein expression of methylated p53 K372
(p53K372-Me) in the presence of resveratrol or DMSO
(Fig. 2A-C). The results revealed a
higher protein expression of p53K372-Me in the presence
of resveratrol compared with the DMSO-treated controls.
Importantly, the function of resveratrol towards p53 was attenuated
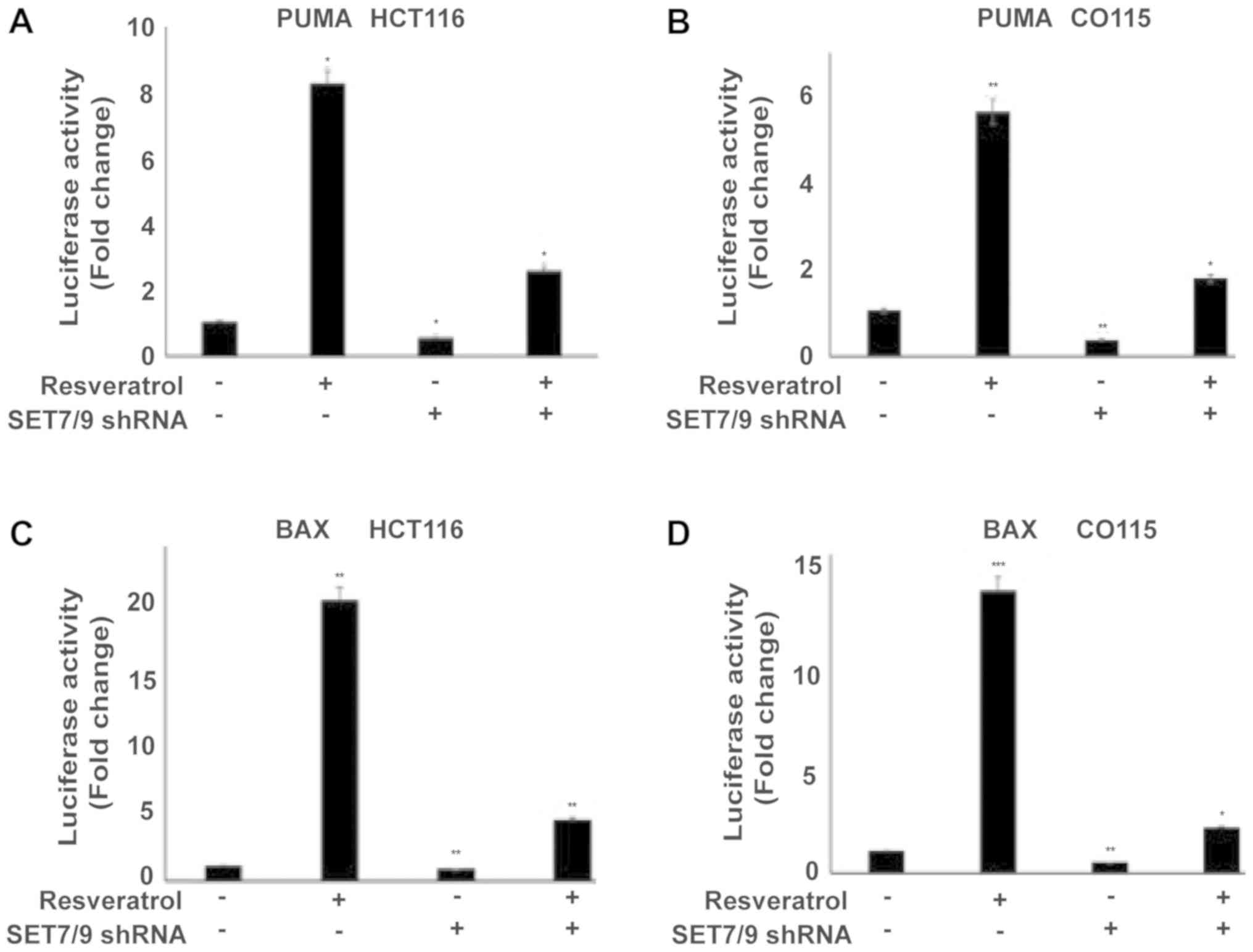
in the presence of shRNA against SET7/9 (Fig. 2D-F). To further confirm the present
results, the expression of p53 target genes (PUMA and BAX) were
examined using a Luciferase assay (Fig.
3A-D). Treating cells with resveratrol significantly induced
PUMA and BAX expression in all cell lines compared with the
untreated control cells (P<0.05), whereas the effect of
resveratrol was significantly abolished in the absence of SET7/9
(P<0.05; Fig. 3A-D). Altogether,
these results suggest that SET7/9 functions as a key mediator of
resveratrol-dependent p53 activation in colorectal cancer
cells.
Resveratrol promotes apoptosis through
the induction of SET7/9 expression
To identify the physiological role of resveratrol in
colorectal cancer cells, the expression of cleaved PARP, an
apoptosis biomarker (26,27), was examined in various colorectal
cancer cell lines (HCT116, CO115 and SW48 As mentioned in Fig. 1B, a viability test was performed to
identify the maximal inhibitory concentration of resveratrol for
HCT116, CO115, and SW480 cells, which were 25, 30, and 37.5 µM
respectively. Therefore, each cell line was treated with a specific
concentration of resveratrol followed by Western blotting. The
results suggested the overexpression of p53 and cleaved PARP in the
presence of resveratrol (Fig. 4A-C).
Additionally, the ablation of SET7/9 decreased the expression of
cleaved PARP upon treatment with resveratrol compared with
untransfected cells (Fig. 4D-F).
Similar results were obtained when the expression of cleaved
caspase-3 was examined (Fig. 4D-F).
Altogether, these data confirm that SET7/9 is the mediator of
resveratrol-dependent apoptosis in colorectal cancer.
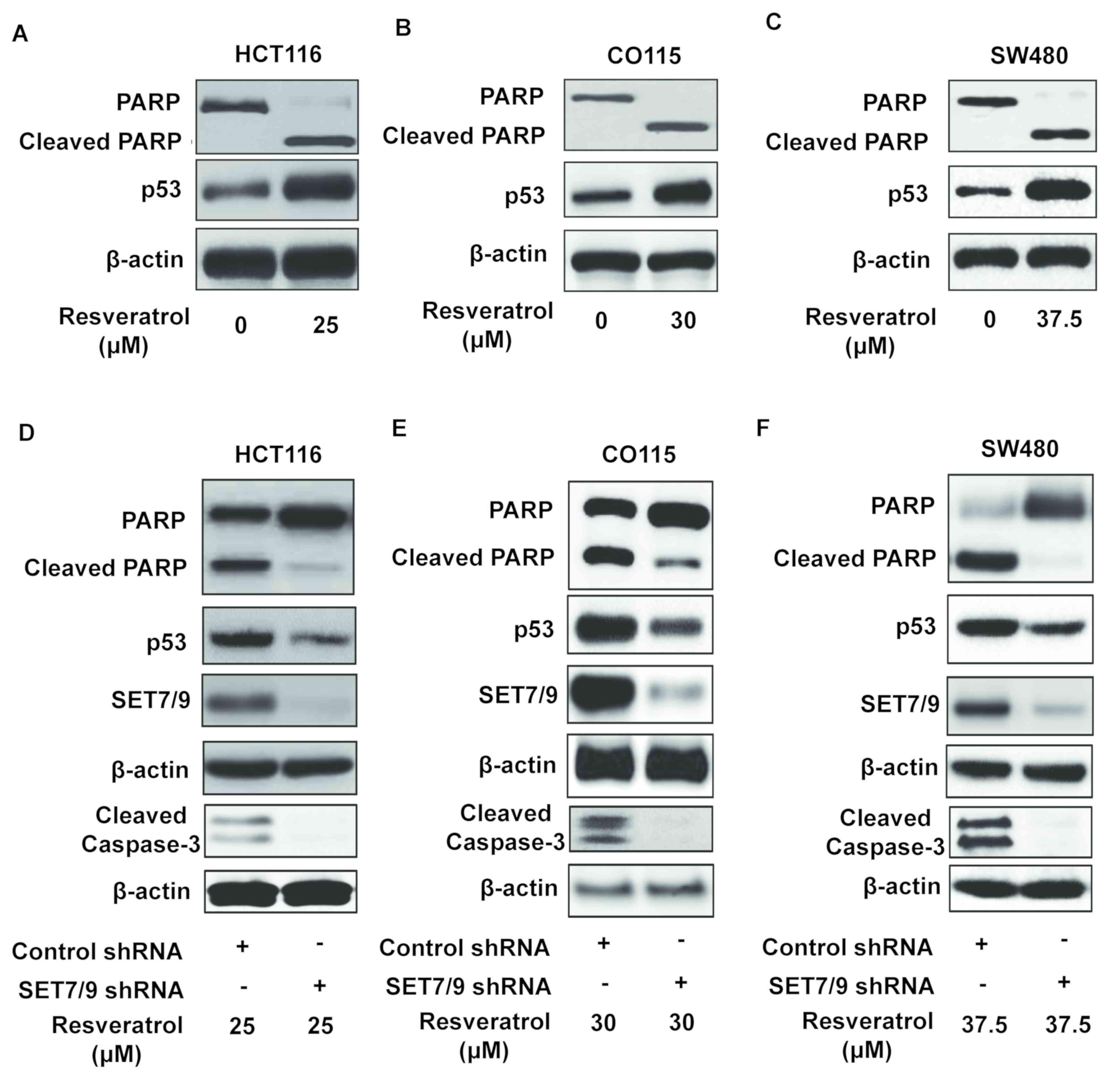 | Figure 4.SET7/9 is required for the induction
of p53-mediated apoptosis by resveratrol. Western blotting was
conducted to evaluate PARP cleavage, apoptosis biomarker, in (A)
HCT116, (B) CO115 and (C) SW480 colorectal cancer cells. Western
blots revealing the response of SET7/9-depleted (D) HCT116, (E)
CO115 and (F) SW480 colorectal cancer cells to resveratrol,
revealing the expression of PARP, cleaved PARP, p53, SET7/9 and
cleaved caspase-3. p53, tumour protein p53; PARP, poly (ADP-ribose)
polymerase; SET7/9, SET domain containing lysine methyltransferase
7/9; shRNA, short hairpin RNA. |
Discussion
SET7/9, a SET domain-containing lysine
methyltransferase, has been proposed to transfer a methyl group to
the target lysine residue of non-histone proteins including p53
(methylated at K372) and histone H3 Lysine 4 (H3-K4) (6). Thus, SET7/9 is involved in various
cellular pathways in cancer cells, including cell proliferation,
cell death, migration and invasion (24). In the present study, SET7/9 was
identified as a mediator of the resveratrol-driven overexpression
of p53, while the specific molecular mechanisms underlying the
interaction between resveratrol and SET7/9 remain unclear. Further
experiments are required in order to determine whether resveratrol
directly or indirectly promotes SET7/9 expression.
p53 mono-methylation by SET7/9 is crucial for its
stabilization and promoter occupancy (28). The results revealed and upregulation
of SET7/9 in colorectal cancer (HCT116, CO115, and SW480) cells
upon resveratrol treatment. In agreement with previous studies
(16,29–31),
increasing SET7/9 led to the induction of the expression of
methylated p53 at K372, in addition to the induction of total p53
and p53-target genes. The present study also revealed that
resveratrol-induced p53 overexpression was reduced in cells
expressing shRNA against SET7/9 compared with untransfected cells.
However, resveratrol treatment may still induce the expression of
p53 target genes (BAX and PUMA) in SET7/9-depleted cells implying
that resveratrol regulates p53 activity by other mechanisms.
Further studies are warranted to elucidate the exact molecular
mechanisms by which resveratrol regulates p53 expression and
activity.
Resveratrol has been reported to be one of the most
promising anticancer agents isolated from natural products that
induce apoptosis in certain cancer types in a p53-dependent manner
(17–20,32).
Similarly, the present results propose resveratrol as a positive
regulator of apoptosis in colorectal cancer cells (Fig. 4). Furthermore, the data produced by
the present study indicates that the pro-apoptotic function of
resveratrol depends on SET7/9 in colorectal cancer.
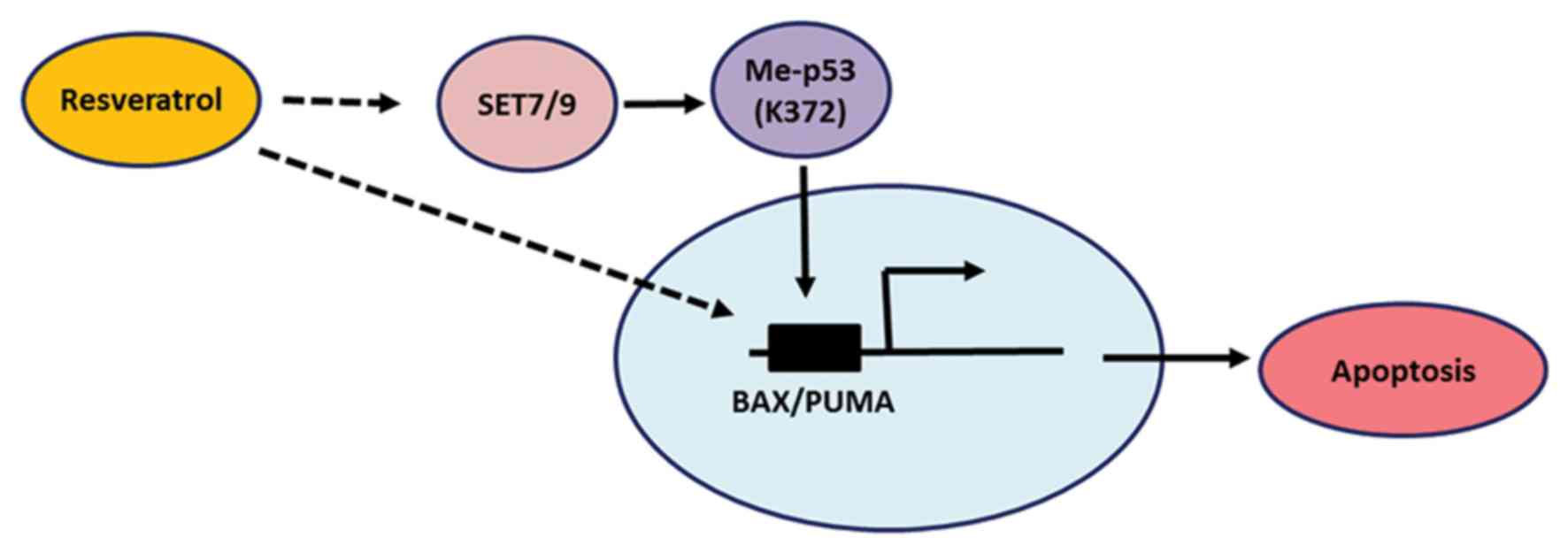
From the present study, it was concluded that SET7/9
functions as a mediator of resveratrol-dependent p53 activation,
and the results confirm that resveratrol modulates p53 through its
mono-methylation at K372 (Fig. 5).
In agreement with the present study, other studies have reported
the induction of p53 phosphorylation at serine 15 by resveratrol
(21), which is vital for the
stabilization and activation of p53 (11,14,16). The
results of the present study add another layer of complexity to the
anti-cancer function of resveratrol and further studies are
required to identify the effect of resveratrol-induced SET7/9 in
p53 mutant cancer cells.
Acknowledgments
The authors would like to thank Dr Chao Gao (Albany
Medical College, Albany, NY, USA) for assistance with the
bioinformatics analysis
Funding
The present study was supported by the Lianyungang
Health and Family Planning Commission (grant no. 201623).
Availability of data and materials
The publicly available bioinformatics data set
(GSE17537: 244653_at) was downloaded on prognoscan website by this
article (22,23) and made publicly available (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE17537),
which shows overall survival rate.
Authors' contributions
ZL, XW and JL performed experiments, analyzed data,
and wrote the manuscript. HS performed the experiments and wrote
the manuscript. FZ supervised the project, designed the
experiments, and wrote the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Singh UP, Singh NP, Singh B, Hofseth LJ,
Price RL, Nagarkatti M and Nagarkatti PS: Resveratrol
(trans-3,5,4′-trihydroxystilbene) induces silent mating type
information regulation-1 and down-regulates nuclear transcription
factor-kappaB activation to abrogate dextran sulfate sodium-induced
colitis. J Pharmacol Exp Ther. 332:829–839. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kimura Y and Okuda H: Resveratrol isolated
from Polygonum cuspidatum root prevents tumor growth and metastasis
to lung and tumor-induced neovascularization in Lewis lung
carcinoma-bearing mice. J Nutr. 131:1844–1849. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Schmidlin L, Poutaraud A, Claudel P,
Mestre P, Prado E, Santos-Rosa M, Wiedemann-Merdinoglu S, Karst F,
Merdinoglu D and Hugueney P: A stress-inducible resveratrol
O-methyltransferase involved in the biosynthesis of pterostilbene
in grapevine. Plant Physiol. 148:1630–1639. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Das DK and Maulik N: Resveratrol in
cardioprotection: A therapeutic promise of alternative medicine.
Mol Interv. 6:36–47. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Colin D, Gimazane A, Lizard G, Izard JC,
Solary E, Latruffe N and Delmas D: Effects of resveratrol analogs
on cell cycle progression, cell cycle associated proteins and
5fluoro-uracil sensitivity in human derived colon cancer cells. Int
J Cancer. 124:2780–2788. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bai Q, Shen Y, Yao X, Wang F, Du Y, Wang
Q, Jin N, Hai J, Hu T and Yang J: Modeling a new water channel that
allows SET9 to dimethylate p53. PLoS One. 6:e198562011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ji Q, Liu X, Fu X, Zhang L, Sui H, Zhou L,
Sun J, Cai J, Qin J, Ren J and Li Q: Resveratrol inhibits invasion
and metastasis of colorectal cancer cells via MALAT1 mediated
Wnt/β-catenin signal pathway. PLoS One. 8:e787002013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Siegel RL, Miller KD, Fedewa SA, Ahnen DJ,
Meester RGS, Barzi A and Jemal A: Colorectal cancer statistics,
2017. CA Cancer J Clin. 67:177–193. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Aoki K and Taketo MM: Adenomatous
polyposis coli (APC): A multi-functional tumor suppressor gene. J
Cell Sci. 120:3327–3335. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Losi L, Luppi G and Benhattar J:
Assessment of K-ras, Smad4 and p53 gene alterations in colorectal
metastases and their role in the metastatic process. Oncol Rep.
12:1221–1225. 2004.PubMed/NCBI
|
|
11
|
Zilfou JT and Lowe SW: Tumor suppressive
functions of p53. Cold Spring Harb Perspect Biol. 1:a0018832009.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Rada M, Althubiti M, Ekpenyong-Akiba AE,
Lee KG, Lam KP, Fedorova O, Barlev NA and Macip S: BTK blocks the
inhibitory effects of MDM2 on p53 activity. Oncotarget.
8:106639–106647. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Rada M, Vasileva E, Lezina L, Marouco D,
Antonov AV, Macip S, Melino G and Barlev N: Human EHMT2/G9a
activates p53 through methylation-independent mechanism. Oncogene.
36:922–932. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bieging KT, Mello SS and Attardi LD:
Unravelling mechanisms of p53-mediated tumour suppression. Nat Rev
Cancer. 14:359–370. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lee DH, Kim C, Zhang L and Lee YJ: Role of
p53, PUMA, and Bax in wogonin-induced apoptosis in human cancer
cells. Biochem Pharmacol. 75:2020–2033. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Marouco D, Garabadgiu AV, Melino G and
Barlev NA: Lysine-specific modifications of p53: A matter of life
and death? Oncotarget. 4:1556–1571. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Cong P, Yi C and Wang XY: Expression of
Smo in pancreatic cancer CD44+CD24+ cells and
construction of a lentiviral expression vector to silence Smo.
Oncology Lett. 16:4855–4862. 2018.
|
|
18
|
Hsieh TC, Wang Z, Hamby CV and Wu JM:
Inhibition of melanoma cell proliferation by resveratrol is
correlated with upregulation of quinone reductase 2 and p53.
Biochem Biophys Res Commun. 334:223–230. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Oi N, Yuan J, Malakhova M, Luo K, Li Y,
Ryu J, Zhang L, Bode AM, Xu Z, Li Y, et al: Resveratrol induces
apoptosis by directly targeting Ras-GTPase-activating protein SH3
domain-binding protein 1. Oncogene. 34:2660–2671. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li B, Hou D, Guo H, Zhou H, Zhang S, Xu X,
Liu Q, Zhang X, Zou Y, Gong Y and Shao C: Resveratrol sequentially
induces replication and oxidative stresses to drive p53-CXCR2
mediated cellular senescence in cancer cells. Sci Rep. 7:2082017.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Shih A, Davis FB, Lin HY and Davis PJ:
Resveratrol induces apoptosis in thyroid cancer cell lines via a
MAPK- and p53-dependent mechanism. J Clin Endocrinol Metab.
87:1223–1232. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Mizuno H, Kitada K, Nakai K and Sarai A:
PrognoScan: A new database for meta-analysis of the prognostic
value of genes. BMC Med Genomics. 2(18)2009.PubMed/NCBI
|
|
23
|
Smith JJ, Deane NG, Wu F, Merchant NB,
Zhang B, Jiang A, Lu P, Johnson JC, Schmidt C, Bailey CE, et al:
Experimentally derived metastasis gene expression profile predicts
recurrence and death in patients with colon cancer.
Gastroenterology. 138:958–968. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Akiyama Y, Koda Y, Byeon SJ, Shimada S,
Nishikawaji T, Sakamoto A, Chen Y, Kojima K, Kawano T, Eishi Y, et
al: Reduced expression of SET7/9, a histone mono-methyltransferase,
is associated with gastric cancer progression. Oncotarget.
7:3966–3983. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Montenegro MF, Sánchez-Del-Campo L,
González-Guerrero R, Martínez-Barba E, Piñero-Madrona A,
Cabezas-Herrera J and Rodríguez-López JN: Tumor suppressor SET9
guides the epigenetic plasticity of breast cancer cells and serves
as an early-stage biomarker for predicting metastasis. Oncogene.
35:6143–6152. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Los M, Mozoluk M, Ferrari D, Stepczynska
A, Stroh C, Renz A, Herceg Z, Wang ZQ and Schulze-Osthoff K:
Activation and caspase-mediated inhibition of PARP: A molecular
switch between fibroblast necrosis and apoptosis in death receptor
signaling. Mol Biol Cell. 13:978–988. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Althubiti M, Rada M, Samuel J, Escorsa JM,
Najeeb H, Lee KG, Lam KP, Jones GD, Barlev NA and Macip S: BTK
modulates p53 activity to enhance apoptotic and senescent
responses. Cancer Res. 76:5405–5414. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
West LE and Gozani O: Regulation of p53
function by lysine methylation. Epigenomics. 3:361–369. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Chuikov S, Kurash JK, Wilson JR, Xiao B,
Justin N, Ivanov GS, McKinney K, Tempst P, Prives C, Gamblin SJ, et
al: Regulation of p53 activity through lysine methylation. Nature.
432:353–360. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kurash JK, Lei H, Shen Q, Marston WL,
Granda BW, Fan H, Wall D, Li E and Gaudet F: Methylation of p53 by
Set7/9 mediates p53 acetylation and activity in vivo. Mol Cell.
29:392–400. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Nishioka K, Chuikov S, Sarma K,
Erdjument-Bromage H, Allis CD, Tempst P and Reinberg D: Set9, a
novel histone H3 methyltransferase that facilitates transcription
by precluding histone tail modifications required for
heterochromatin formation. Genes Dev. 16:479–489. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kai L, Samuel SK and Levenson AS:
Resveratrol enhances p53 acetylation and apoptosis in prostate
cancer by inhibiting MTA1/NuRD complex. Int J Cancer.
126:1538–1548. 2010.PubMed/NCBI
|