Introduction
Liver cancer was reported in 2012 as the sixth most
commonly diagnosed cancer, and the second most common cause of
cancer-associated mortality worldwide (1). A total of 782,500 cases of liver cancer
were diagnosed globally, resulting in 745,500 mortalities (1). China has a high incidence of liver
cancer, accounting for ~50% of these cases (2). The main risk factors for the onset of
liver cancer include chronic hepatitis B (CHB) virus (HBV) or
hepatitis C virus infection, aflatoxin exposure, type II diabetes,
liver cirrhosis and smoking (3–5). In
China, 75–85% of cases are caused by CHB infection (6,7). A
number of studies have reported that HBV may result in liver cancer
development by interfering with several cellular processes,
including transcription, signal transduction, cell cycle, protein
breakdown, DNA repair, apoptosis and the maintenance of chromosome
stability (8–10). Additionally, HBV may alter hepatocyte
signaling and modulate the activity of transcription factors and
target proteins involved in hepatocarcinogenesis (11). At present, the diagnosis and
treatment of HBV-associated liver cancer remains inadequate. The
majority of patients with HBV-associated liver cancer are diagnosed
at intermediate or advanced stages (12), and the treatment is typically
surgical resection. However, the aforementioned therapeutic
strategy is unsatisfactory, and the survival rate remains low
(13,14). Therefore, it is necessary to identify
novel molecular markers for HBV-associated liver cancer and to
elucidate their specific underlying molecular mechanisms, for
improved clinical diagnosis and treatment.
Ubiquitin carboxyl-terminal hydrolase 22 (USP22) is
a member of the deubiquitinating enzyme (DUB) family and is located
on chromosome 17p11.2. The 1,578 bp USP22 open reading frame
encodes 525 amino acids, containing highly conserved regions of the
ubiquitin-specific-processing protease family (15). The USP22 protein is predominantly
expressed in the cell nucleus and, through deubiquitinating
modifications, exerts regulatory effects on cellular processes,
including cell differentiation, cell cycle progression,
transcriptional activation and signal transduction (16). Abnormally high USP22 expression has
been detected in various cancer types, and is associated with tumor
differentiation and clinical prognosis (17–21).
However, to the best of our knowledge, whether USP22 is involved in
the development of HBV-associated liver cancer, and how it may
regulate its development, have not yet been reported.
Materials and methods
Patients and samples
Following formalin fixation, a total of 85
paraffin-embedded clinical liver tissue specimens (thickness 4 mm)
from patients [n=85, range, 35–78 years of age, 1.2–20 cm of tumor
size I–IV of Edmonson grade (22),
I–IV of clinic stage (23), 3.7–8736
ng/ml of Alpha-fetoprotein] with liver cancer were obtained from
the Department of Pathology of the Affiliated Hospital of Guilin
Medical University (Guilin, China), and resection specimens (weight
2 g) from patients (n=85) with HBV-associated liver cancer were
obtained following liver cancer resection surgery at this hospital
between February 2013 and February 2017. Inclusion criteria were as
follows: Comply with the diagnostic criteria for hepatitis B
virus-associated liver cancer and all selected subjects were
informed and agreed to the study. Exclusion criteria were as
follows: Patients with non- hepatitis B virus-associated liver
cancer; accompanied by any injury in the human body, including
cardiovascular, liver, kidney, brain and blood system. The
clinicopathological and survival data were collected and analyzed
retrospectively. The present study was approved by the Ethics
Committee of the Affiliated Hospital of Guilin Medical University.
Written informed consent was obtained from all patients, according
to the Declaration of Helsinki.
Cell culture
The human liver L02 cell line, as well as liver
cancer HepG2.2.15, HuH7, HCCLM3 and Hep3B2.1–7 cell lines were
purchased from the Cell Bank of the Chinese Academy of Sciences
(Shanghai, China). The cells were cultured in Dulbecco's modified
Eagle's medium containing 1% penicillin/streptomycin and 10% fetal
bovine serum (all Invitrogen; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) in a 5% CO2 incubator at 37°C.
Cell transfection
The cells were transiently transfected when they
became ~70%-80% confluent. Lipofectamine™ 3000 (5 µl; Invitrogen;
Thermo Fisher Scientific, Inc.) was diluted in 125 µl opti-MEM
(Gibco; Thermo Fisher Scientific, Inc.) medium and the USP22 small
interfering (si)RNA or negative control siRNA (5 µl) was added to
125 µl opti-MEM medium and mixed for 5 min. The prepared
Lipofectamine 3000 and siRNA dilutions were combined and gently
mixed for 20 min. The six-well plates were removed from the
incubator, washed 2–3 times with PBS, and 750 µl serum-free,
antibiotic-free pure medium was added to each well. The
Lipofectamine 3000-siRNA mixture was added to the cells, mixed and
placed into the 5% CO2 incubator at 37°C. After 24 h of
incubation at 37°C, the following experiments were conducted. USP22
siRNA Sequence, sense CAGCAGCCCACGGACAGUCUCAACA, anti-Sense
UGUUGAGACUGUCCGUGGGCUGCUG.
Cell proliferation measurement
The Cell Counting Kit-8 (CCK8; Nanjing KeyGen
Biotech Co., Ltd., Nanjing, China) assay was used to measure cell
proliferation at 24, 48, 72 and 96 h. The cells were seeded in a
96-well culture plate (~5×103 cells/100 µl) following
the addition of 10 µl CCK8 reagent to each well, and the plate was
incubated in a CO2 incubator at 37°C for 1 h. The
optical density value was measured at 450 nm using a microplate
reader. The average value of each group was calculated and a
proliferation curve was plotted.
Apoptosis detection by flow
cytometry
Following digestion with 0.25% trypsin, cells were
resuspended in PBS to prepare a single cell suspension with a
density of ~1×106 cells/ml. The cell suspension from
each group (100 µl) was stained with Annexin V-fluorescein
isothiocyanate and propidium iodide using the Muse Annexin V and
Dead Cell assay reagent (cat. no. MCH100105; Merck KGaA, Darmstadt,
Germany) in equal proportions at room temperature in the dark for
20 min. Flow cytometry (Merck KGaA, Darmstadt, Germany) was used to
determine the rate of apoptosis. Data were analyzed with FlowJo
10.0 software (FlowJo LLC, Ashland, OR, USA).
Colony-formation assay
Cells were seeded into a 6-well plate at
1×103 cells/well, with three replicates for each group.
After 2 weeks, the plate was removed from the incubator, washed
three times with PBS, fixed in 4% paraformaldehyde at room
temperature for 15 min and subsequently stained with 0.1% crystal
violet at room temperature overnight. The plate was washed three
times in PBS and cells were subsequently counted with the image J
1.48 software (National Institutes of Health, Bethesda, MD, USA),
prior to statistical analysis of colony number.
Immunohistochemistry
The paraffin-embedded tissue sections (thickness, 3
mm) were deparaffinized in xylene at room temperature and
rehydrated in a graded ethanol series. The sections were immersed
in 3% hydrogen peroxide solution to inhibit endogenous peroxidase
activity. Subsequent to being blocked in 5% non-fatty milk at room
temperature in 1 h, they were subsequently incubated with the
primary antibody (dilution, 1:200; Abcam; cat. no. 195289) for 1 h
at 37°C, and washed three times with PBS. An incubation with the
secondary antibody using the MaxVision™ HRP-polymer anti-Rabbit IHC
kit (cat. no. KIT-5006, Fuzhou Maixin Biotech Co., Ltd., Fuzhou,
China), according to the manufacturer's protocols, followed for 30
min at room temperature, and the sections were washed three times
with PBS. The samples were stained with DAB for 5 min,
counterstained with hematoxylin at room temperature and
differentiated with 0.1% HCl alcohol for 1 sec. They were then
dehydrated and dried in a graded alcohol series, hyalinized in
xylene and sealed with neutral gum. The staining intensity score
was calculated by using an Olympus X71 inverted microscope
(magnification, ×200; Olympus Corp., Tokyo, Japan) as follows: 0,
no staining; 1+, mild staining; 2+, moderate staining; 3+, heavy
staining. The staining area scores were as follows: 0, no staining
under the microscope; 1+, positive staining of <30% of tissue;
2+, positive staining of 30–60% of tissue; 3+, positive staining of
>60% of tissue. The sum of the staining intensity and area
scores was used to evaluate the expression of USP22, with the
highest score being 6 and the lowest being 0. The criterion for
positive expression of USP22 was the presence of brown staining in
the nucleus.
RNA extraction and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
TRIzol (Invitrogen; Thermo Fisher Scientific, Inc.)
reagent was added to 0.1 g tissue, which was subsequently cut into
pieces. Chloroform (200 µl) was added and the mixture was
centrifuged in 13,523 × g at 4°C for 15 min. A total of 400 µl
upper layer liquid was collected and mixed with an equal volume of
isopropanol, prior to further centrifugation in 13,523 × g at 4°C
for 15 min. The supernatant was subsequently removed and 1 ml 70%
ice-cold alcohol was added to wash the RNA. Following
centrifugation in 1,127 × g at 4°C for 5 min, the supernatant was
removed, and the RNA was dried and reconstituted in diethyl
pyrocarbonate-treated water. The product was quantified using a
NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Inc.),
and 500 ng was reverse transcribed using the QuantiTect Reverse
Transcription kit (Qiagen, Inc., Valencia, CA, USA). The reaction
cycling conditions were performed as follows: 1 cycle at 95°C for
15 min, followed by 40 cycles at 95°C for 10 sec and at 60°C for 60
sec. The ratio between the target gene and β-actin was used to
calculate the relative quantitation. The amplification products and
the relative expression of RT-PCR were analyzed using the ΔΔCq
method (24). β-actin was used as an
internal control to normalize gene expression levels. The primers
used for amplification were as follows: β-actin, forward primer,
5′-AAGGAAGGCTGGAAGAGTGC-3′, reverse primer,
5′-CTGGGACGACATGGAGAAAA-3′; USP22, forward primer,
5′-GGCGGAAGATCACCACGTAT-3′, reverse primer,
5′-TTGTTGAGACTGTCCGTGGG-3′.
Western blot analysis
Samples were collected and lysis buffer [137 mM
NaCl, 50 mM Tris-HCl, 10% glycerol, 100 mM sodium orthovanadate, 10
mg/ml aprotinin, 1 mM phenylmethylsulfonyl fluoride (PMSF),1%
Nonidet P-40,10 mg/ml leupeptin, and 5 mM protease inhibitor
cocktail; pH 7.4] was added to extract total proteins., and the
concentration was determined using the bicinchoninic acid method.
The proteins (30 ug) were separated by 10% SDS-PAGE and transferred
to polyvinylidene fluoride membranes by wet transfer. The membranes
were blocked in 5% skimmed milk/tris-buffered saline with Tween-20
(TBST) for 1 h at 37°C. The membrane was washed three times in
TBST. Membranes were subsequently incubated with primary antibody
overnight at 4°C. On the following day, the membrane was washed
three times with TBST, and then incubated with secondary antibody
(HRP-labeled goat anti-rabbit IgG; cat. no. A0208; Beyotime
Institute of Biotechnolgy) at a 1:10,000 for 1 h at 37°C.
Subsequently, chemiluminescence was used to display the imprinting,
and the results were analyzed using the Tannon 5200
chemiluminescent imaging system (Tanon Science and Technology,
Shanghai, China). The primary antibodies of USP22 (abcam195289,
concentration is 1:1,000), β-actin, activated caspase-3
(abcam136812, concentration is 1:1,000), caspase-8 (abcam25901,
concentration is 1:1,000), and caspase-9 (abcam2324, concentration
is 1:1,000). The primary antibodies of phosphoinositide 3-kinase
(PI3K) (SC-136298), phosphorylated (p)-PI3K (Cat. No. 20584-1-AP),
protein kinase B (Akt) (SC-135829) and p-Akt (SC-271964) were
purchased from Santa Cruz Biotechnology, Inc. (Dallas, TX, USA) and
proteintech group (Wuhan, China). Protein bands were visualized by
Image J2× (National Institutes of Health, Bethesda, MD, USA).
Microarray analysis
Total RNA quality and quantity were determined using
NanoDrop ND-2000 (Thermo Fisher Scientific, Inc.). GeneChip Human
Genome HU U133 plus 2.0 arrays (Affymetrix; Thermo Fisher
Scientific, Inc.) were used according to the manufacturer's
protocol. The data were initially normalized by robust multiarray
average normalization algorithms in Transcriptome Analysis Console
4.0 software (Affymetrix; Thermo Fisher Scientific, Inc.).
Significantly altered genes between USP22 knockdown and its control
cells were visualized by scatter plots. Clustering analysis was
performed using Gene Cluster v3.0 software. Gene set enrichment
analysis was conducted using ConceptGen (National Center for
Integrative Biomedical Informatics, Ann Arbor, MI, USA).
Statistical analysis
SPSS v22.0 software (IBM Corp., Armonk, NY, USA) was
used to analyze the experimental data. The χ2 test was
used to evaluate the association between USP22 expression and
clinicopathological parameters. Multigroup comparisons of means
were performed by one-way analysis of variance test with post-hoc
comparisons using the Student-Newman-Keuls test. Receiver operating
characteristic curve analysis overall survival (OS) time and
relapse-free survival (RFS) time were also analyzed in the present
study. Student's t-test was used to analyze continuous variables
and the Kaplan-Meier method was used to assess survival
probabilities. P<0.05 was considered to indicate a statistically
significant difference. Each group of experiments was repeated
three times.
Results
USP22 is highly expressed in
HBV-associated liver cancer tissues
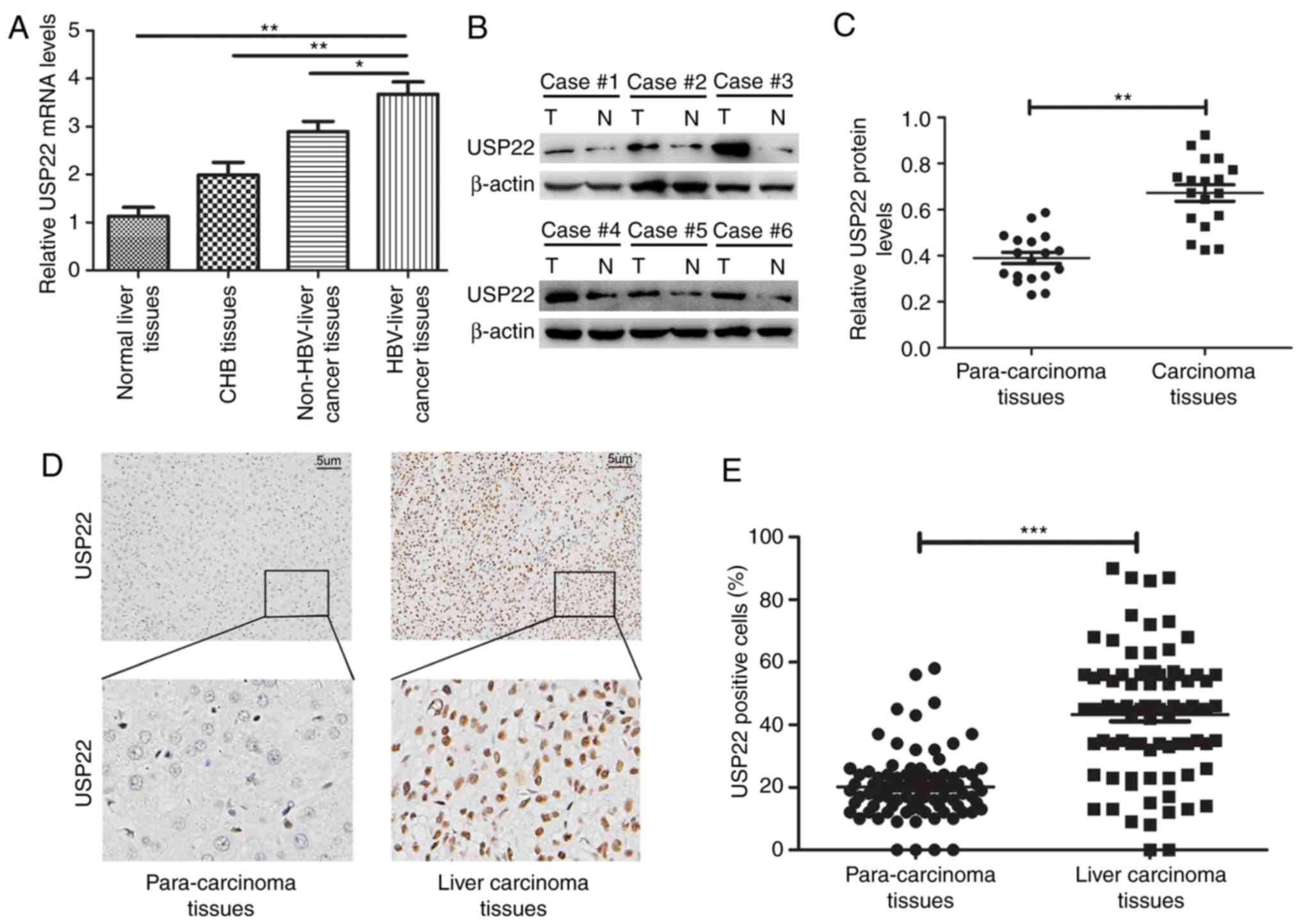
The RT-qPCR results demonstrated that in the 10
normal liver, 15 liver with CHB infection, 23 non-HBV-associated
liver cancer and 28 HBV-associated liver cancer tissues, the
expression levels of USP22 exhibited a gradual increase. The
expression of USP22 in tissues of HBV-associated liver cancer was
higher than in that measured in normal liver, liver with CHB
infection, and non-HBV-associated liver cancer (P<0.01,
P<0.01 and P<0.05, respectively; Fig. 1A). Furthermore, 18 pairs of carcinoma
and para-carcinoma tissues from patients with HBV-associated liver
cancer were randomly selected to analyze the protein expression of
USP22 by western blot analysis (Fig.
1B). It was revealed that the expression level of USP22 in
carcinoma tissues was significantly increased compared with the
para-carcinoma tissues (P<0.01; Fig.
1C). Immunohistochemistry was subsequently performed to detect
USP22 expression in 85 paraffin-embedded sections of HBV-associated
liver cancer tumors (Fig. 1D). The
results revealed that positive USP22 protein staining was
predominantly located in the nucleus, and was more highly expressed
in the cancer tissues compared with the para-carcinoma tissues
(P<0.001; Fig. 1E).
USP22 is associated with poor clinical
prognosis in patients with HBV-associated liver cancer
The expression levels of USP22 in HBV-associated
liver cancer were divided into high- (n=45) and low-expression
(n=40) groups, according to the immunohistochemical scoring system
described above, and compared against clinicopathological data
(Table I). Statistical analysis
determined that the expression of USP22 was associated with tumor
size (P<0.01), Edmonson grade (P<0.01), clinical stage
(clinical staged according to the pTNM (pathologic tumor, node,
metastasis) classification criteria of the International Union
Against Cancer) (P<0.01) and number of tumors (P<0.05).
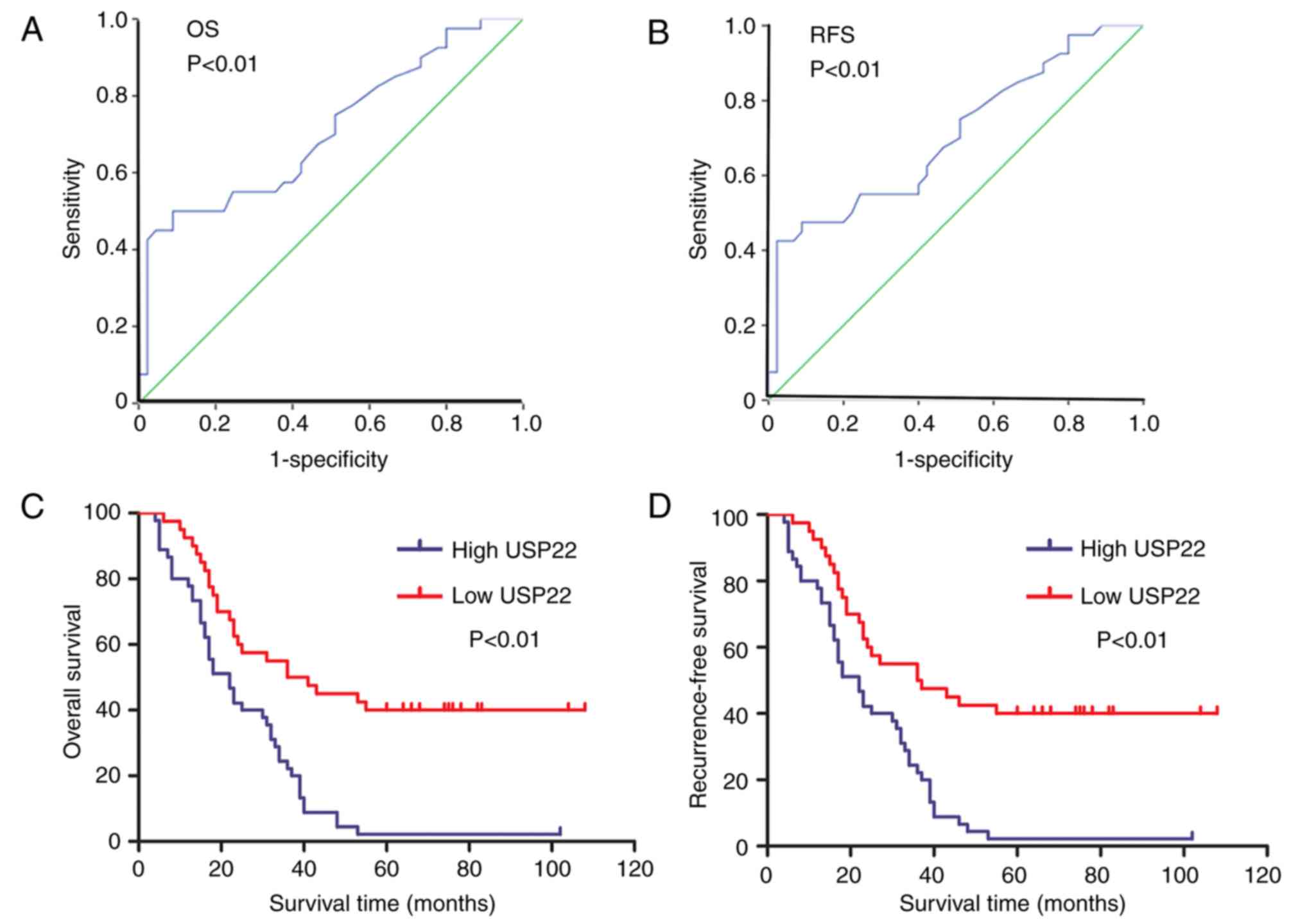
Receiver operating characteristic curve analysis revealed that
USP22 expression was statistically significant in terms of overall
survival (OS) time (P<0.01; Fig.
2A) and recurrence -free survival (RFS) time (P<0.01;
Fig. 2B). The association between
USP22 expression and the clinical prognosis of patients with
HBV-associated liver cancer was further analyzed using Kaplan-Meier
survival curves. The results revealed that the OS time of the USP22
high-expression group was lower than that of the USP22
low-expression group (P<0.01; Fig.
2C), as was the RFS time (P<0.01; Fig. 2D). Together, the results demonstrate
that high USP22 expression in HBV-associated liver cancer is
associated with the degree of malignancy and poor clinical
prognosis of patients.
 | Table I.Association between USP22 staining
results and clinicopathological characteristics of 85 patients with
liver cancer. |
Table I.
Association between USP22 staining
results and clinicopathological characteristics of 85 patients with
liver cancer.
|
| USP22 staining |
|
|
|---|
|
|
|
|
|
|---|
| Variables | Low | High | Total | P-value |
|---|
| Age, years |
|
|
| 0.32 |
|
<50 | 17 | 24 | 41 |
|
|
≥50 | 23 | 21 | 44 |
|
| Sex |
|
|
| 0.18 |
|
Male | 35 | 43 | 78 |
|
|
Female | 5 | 2 | 7 |
|
| AFP, ng/ml |
|
|
| 0.06 |
|
<200 | 26 | 20 | 46 |
|
|
≥200 | 14 | 25 | 39 |
|
| Tumor size (maximum
diameter), cm |
|
|
| <0.01 |
|
<5 | 25 | 12 | 37 |
|
| ≥5 | 15 | 33 | 48 |
|
| Edmonson grade
(22) |
|
|
| <0.01 |
|
I+II | 34 | 26 | 60 |
|
|
III+IV | 6 | 19 | 25 |
|
| Clinical stage
(23) |
|
|
| <0.01 |
|
I+II | 21 | 5 | 26 |
|
|
III+IV | 19 | 40 | 59 |
|
| Tumor number |
|
|
| <0.05 |
| 1 | 28 | 21 | 49 |
|
|
>1 | 12 | 24 | 36 |
|
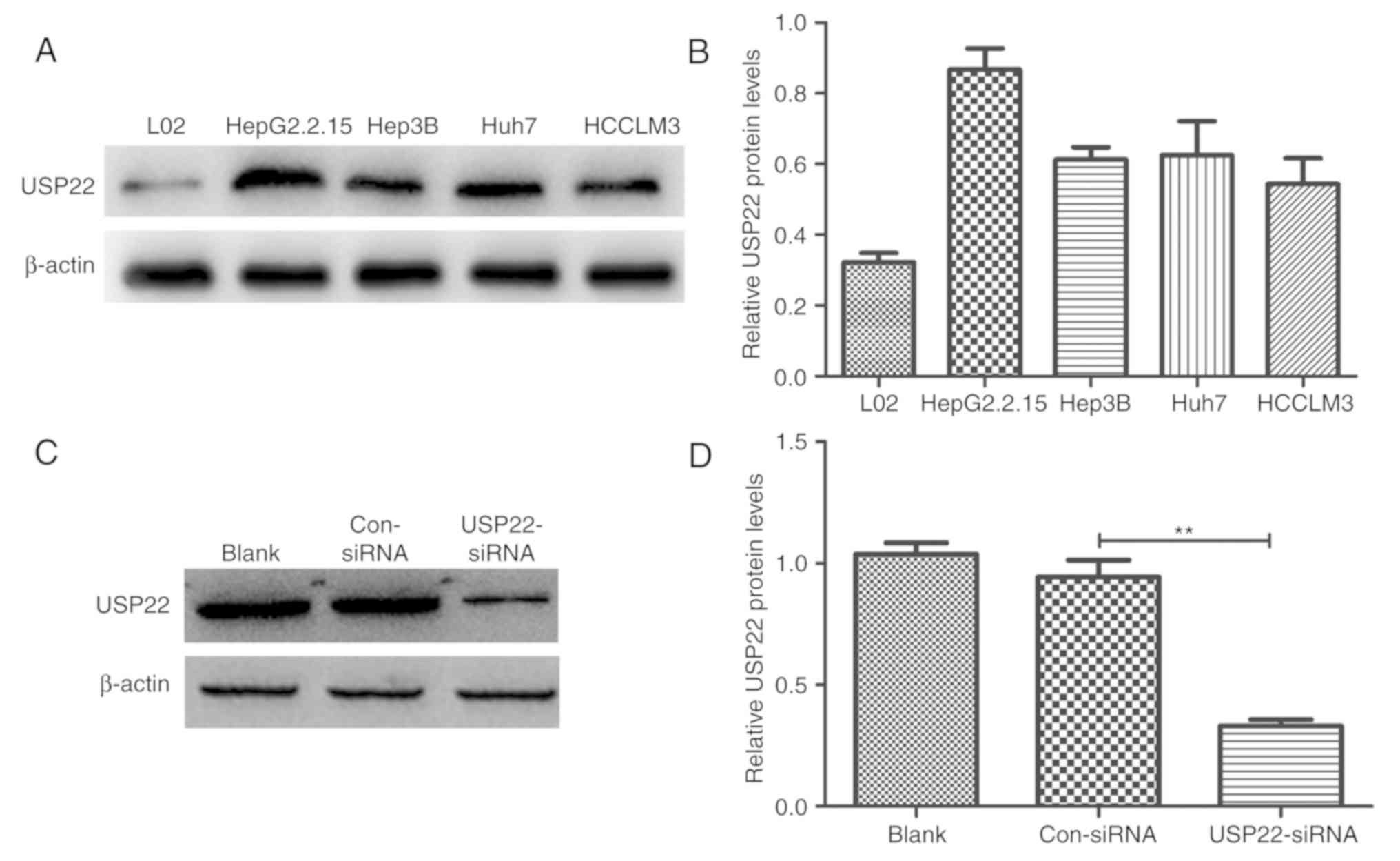
USP22 is highly expressed in
HepG2.2.15 cells, and USP22-siRNA effectively silences the
expression of USP22
Normal liver L02 cells and 4 liver cancer cell lines
(Hep3B, Huh7, HCCLM3 and HepG2.2.15) were used to explore USP22
protein expression by western blot analysis. The USP22 protein
levels in the liver cancer cells was higher than that in the normal
liver cells, and the highest was measured in HepG2.2.15 cells
(Fig. 3A and B). USP22-siRNA was
transiently transfected in HepG2.2.15 cells to silence USP22
expression, and the transfection efficiency was confirmed by
western blotting. The results revealed that USP22-siRNA effectively
decreased USP22 expression (P<0.01 versus control; Fig. 3C and D).
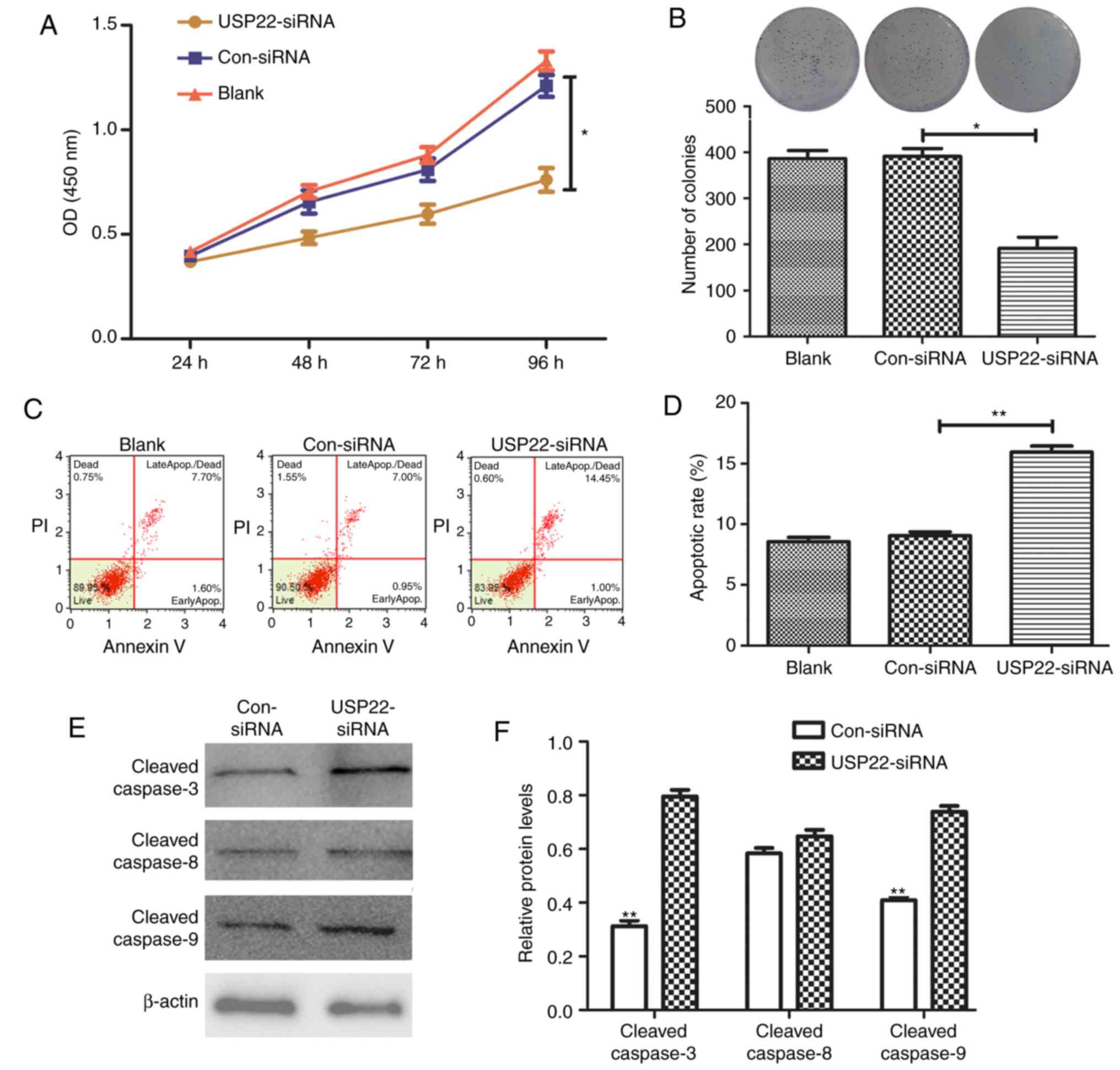
Downregulation of USP22 inhibits the
proliferative ability of HepG2.2.15 cells and promotes
apoptosis
Following knockdown of USP22 expression with siRNA,
the proliferative ability of HepG2.2.15 cells was tested with a
CCK8 assay and colony-formation experiments. The results revealed
that silencing USP22 significantly inhibited HepG2.2.15 cell
proliferation at 96 h (P<0.01; Fig.
4A). The colony-formation experiments demonstrated that
HepG2.2.15 cells in the USP22-siRNA group formed significantly
fewer colonies compared with the control groups (P<0.01;
Fig. 4B). Flow cytometry was
subsequently used to detect cell apoptosis, and the results
revealed that the apoptotic rate of HepG2.2.15 cells was
significantly increased following USP22 silencing (P<0.01;
Fig. 4C and D). Furthermore, western
blot analysis determined that the expression of
apoptosis-associated proteins, cleaved caspase-3 and −9 increased
in the USP22-siRNA group compared with the control (P<0.01),
whereas no difference was observes in the levels of cleaved
caspase-8 (Fig. 4E and F).
Silencing USP22 inhibits PI3K/Akt
protein expression
In order to further clarify the specific molecular
mechanism of USP22 in liver cancer, microarray analysis was
performed to determine the expression profiles of
HepG2.2.15-USP22-siRNA and HepG2.2.15 control cells. Following
USP22 knockdown, a series of signaling pathways were significantly
changed; the PI3K/Akt signaling pathway was the most enriched
apoptotic pathway (Fig. 5A). The
results of the microarray analysis were verified by western blot
analysis, which revealed that the levels of PI3K, Akt were
significantly decreased (P<0.05; Fig.
5B and C).
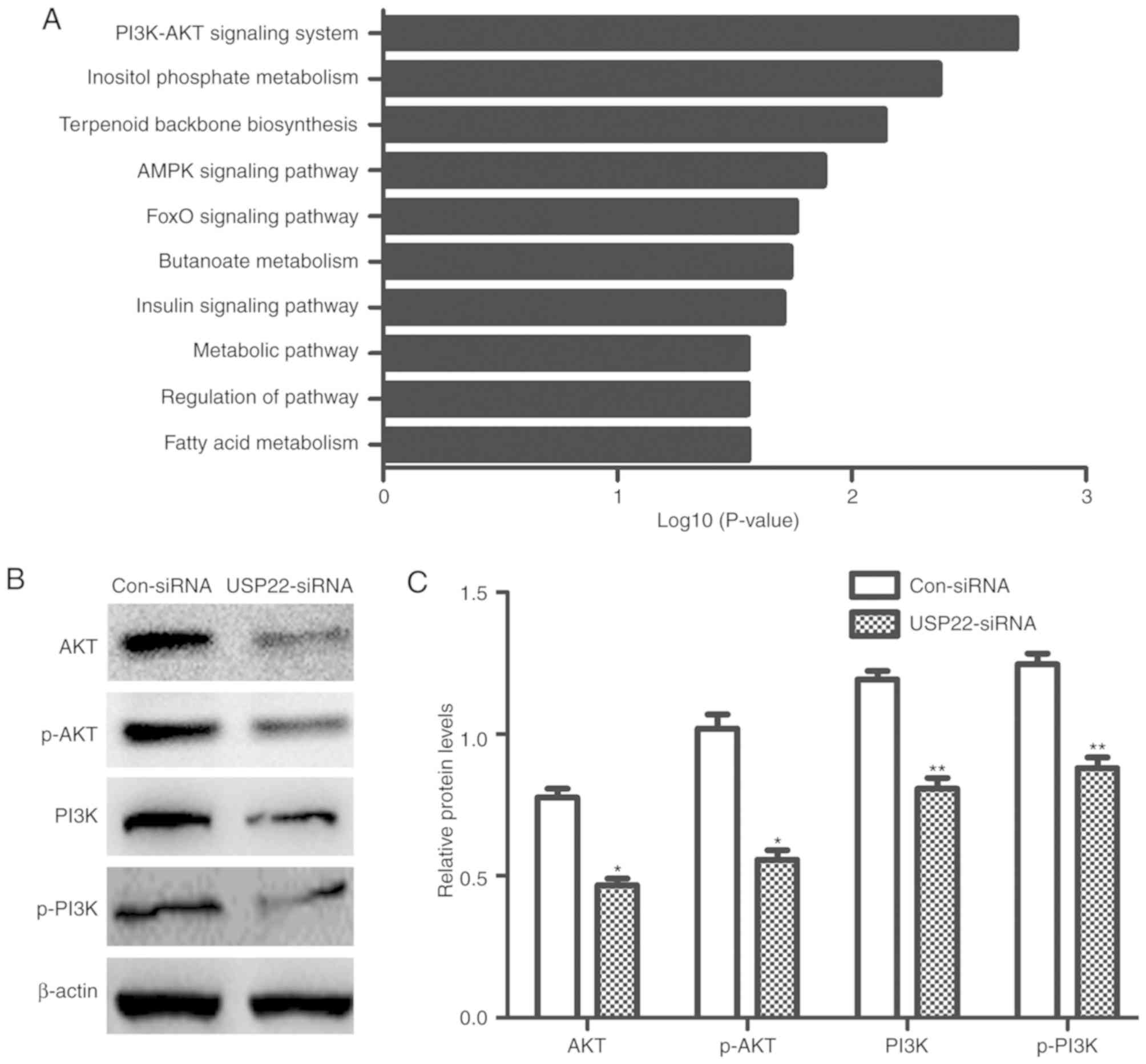 | Figure 5.USP22 silencing inhibits PI3K and Akt
protein expression. (A) Results of the microarray analysis. (B)
Western blotting and (C) relative quantification of PI3K, p-PI3K,
Akt and p-Akt proteins. *P<0.05 and **P<0.01 versus the
equivalent Con-siRNA group. The experiments were repeated three
times. USP22, ubiquitin carboxyl-terminal hydrolase 22; PI3K,
phosphoinositide 3-kinase; Akt, protein kinase B; AMPK,
AMP-activated protein kinase; FOXO, Forkhead box protein O; p-,
phosphorylated; siRNA, small interfering RNA; Con, control. |
Discussion
HBV-associated liver cancer is one of the most
common types of liver cancer, particularly in China (8). Due to rapid tumor progression, it is
difficult to provide an early diagnosis and the associated
mortality rate is high (8). At
present, HBV-associated liver cancer remains a focus of research
for identifying the key regulatory factors controlling the
development and progression of this disease, in order to improve
clinical diagnosis and treatment. Ubiquitination and
deubiquitination are important post-translational modifications
that participate in a variety of biological behaviors (25,26). As
an important member of the DUB family, and as the enzymatic center
of the deubiquitinating module, USP22 participates in a series of
biochemical reactions (16). Studies
have revealed that removal of USP22 leads to a decrease, rather
than an increase, in global monoubiquitination of histone H2B
(H2Bub1). By contrast, depletion of non-enzymatic components
ataxin-7-like protein 3 (ATXN7L3) or transcription and mRNA export
factor ENY2, results in increased H2Bub1 levels. Two new H2Bub1
DUBs, USP27X and USP51, which function independently of the
Spt-Ada-Gcn5 acetyltransferase complex, compete with USP22 for
ATXN7L3 and ENY2 for activity (27).
Inhibition of these DUBs suppresses tumor growth, and this has
become a focus of tumor marker research.
A previous study demonstrated that USP22 was highly
expressed in liver cancer tissue and was associated with the
occurrence and development of liver cancer, and poor prognosis
(28). Silencing of USP22 inhibited
the cell proliferation, migration and invasion, and the
chemotherapeutic resistance of liver cancer cells. In addition, it
was demonstrated that USP22 is an independent biomarker for the
prediction of survival and prognosis for patients with liver cancer
(28). The focus of the present
study was HBV-associated liver cancer; the association between this
cancer type and USP22 was verified, and the potential underlying
regulatory mechanism was explored. The results revealed that USP22
expression was varied among normal liver, liver with CHB infection,
non-HBV-associated liver cancer and HBV-associated liver cancer
tissues. However, the levels were significantly higher in
HBV-associated liver cancer tissues, and high expression was
demonstrated to be associated with tumor development and poor
prognosis.
In the in vitro experiments, the expression
of USP22 was higher in liver cancer cells compared with normal
liver cells, and HepG2.2.15 cells exhibited the highest expression
of USP22 among the liver cancer cell lines. Silencing of USP22
inhibited proliferation, promoted apoptosis and increased the
activation levels of the apoptosis-associated proteins caspase-3
and −9 in HepG2.2.15 cells.
Numerous studies have confirmed that USP22 affects
the expression of its target genes, including Myc proto-oncogene
protein (29), polycomb complex
protein BMI-1 (30),
fructose-1,6-bisphosphatase 1 (31),
focal adhesion kinase 1 (32) and
transforming growth factor β (33),
by removing ubiquitin ligase from protein substrates, thereby
regulating a series of biological behaviors, including cell cycle
progression, proliferation and differentiation, and epithelial to
mesenchymal transition. As a major signal transduction pathways,
PI3K/Akt signaling inhibits apoptosis and promotes cell
proliferation by influencing the activation state of a variety of
downstream molecules. It has been demonstrated that the
PI3K/Akt/mammalian target of rapamycin signal transduction pathway
serves a critical role in tumorigenesis and development. Therefore,
it has become a potential novel target for tumor treatment
(21). It has been reported that HBV
X protein promotes the malignant transformation of hepatocytes, by
driving the expression of α-fetoprotein to activate the PI3K/Akt
signaling pathway, which in turn stimulates the expression of
reprogramming-associated proteins and oncogenes (34). HBV large surface proteins are
involved in the development of liver cancer by activating the
proto-oncogene tyrosine-protein kinase Src/PI3K/Akt signaling
pathway and accelerating G1/S cell cycle progression (35). In addition, studies have reported
that serine/threonine-protein kinase PAK1 interacts with the
PI3K/Akt signaling pathway to promote the proliferation and
migration of liver cancer cells (36). In the present study, it was
demonstrated that silencing USP22 in HepG2.2.15 cells modulated the
expression of key proteins in PI3K/Akt pathway, and decreased the
levels of PI3K, Akt,. Therefore, it could be concluded that USP22
may serve an important role in inducing apoptosis and inhibiting
proliferation of liver cancer cells through mechanisms affecting
PI3K/Akt expression levels. In conclusion, it was determined that
USP22 was highly expressed in HBV-associated liver cancer tissues
and was associated with tumor differentiation and poor prognosis.
In addition, it was revealed that USP22 regulated the proliferation
and apoptosis of HepG2.2.15 cells. In terms of molecular mechanism,
microarray and western blot analysis verified that USP22 regulated
the expression of PI3K/Akt pathway-associated proteins, and
therefore may regulate hepatocyte apoptosis. Our next aim is to
further clarify which specific PI3K/Akt signaling molecules are
affected by USP22, by investigating the association between USP22
knockdown and the Akt pathway by means of small molecules that
activate Akt signaling. The present results suggest that USP22 may
be used as an independent predictor of patient survival and
prognosis, as well as a potential molecular target for the
treatment of HBV-associated liver cancer.
Acknowledgements
Not applicable.
Funding
The present study was supported in part by the
Innovation Project of Guangxi Graduate Education (grant. no.
YCSZ2015212), the National Natural Science Foundation of China
(grant no. 81560393), the Guangxi Science Fund for Distinguished
Young Scholars Program (grant no. 2016GXNSFFA380003), the Natural
Science Foundation of Guangxi (grant nos. 2014GXNSFBA118192 and
2015jjDA40010), the Guangxi Health Department Raise Issue (grant
no. Z2013466), the Fund Project in Guangxi Department of Education
(grant no. YB2014265), the Scientific Research and Technology
Development Project for Guilin (grant no. 20140310-2-2), the
Guangxi Department of Education (grant no. YB2014265), Guangxi
Health Department Raise Issue (grant no. Z2013466), the Natural
Science Foundation of Guangxi (grant no. 2014GXNSFBA118192), and
the Guangxi Key Laboratory of Tumor Immunology and
Microenvironmental Regulation (grant no. 2018KF001).
Availability of data and materials
The datasets used during the present study are
available from the corresponding authors upon reasonable
request.
Authors' contributions
BT and ZW conceived and designed the experiments. YL
performed the experiments. XL, WL, ZL, YW, LW and SZ analyzed the
data. BT supervised the experiments and revised the manuscript. WJL
and YL wrote the manuscript. All authors read and approved the
manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Guilin Medical University (Guilin, China). Written
informed consent was obtained from all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bao J, Lu Y, Deng Y, Rong C, Liu Y, Huang
X, Song L, Li S and Qin X: Association between IL-18 polymorphisms,
serum levels, and HBV-related hepatocellular carcinoma in a Chinese
population: A retrospective case-control study. Cancer Cell Int.
15:722015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sun Y, Lu Y, Li T, Xie L, Deng Y, Li S and
Qin X: Interferon gamma +874T/A polymorphism increases the risk of
hepatitis virus-related diseases: Evidence from a meta-analysis.
PLoS One. 10:e01211682015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Dastjerdi MN, Kavoosi F, Valiani A,
Esfandiari E, Sanaei M, Sobhanian S, Hakemi MG and Mobarakian M:
Inhibitory effect of genistein on PLC/PRF5 hepatocellular carcinoma
cell line. Int J Prev Med. 6:542015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Peng Q, Yang S, Lao X, Li R, Chen Z, Wang
J, Qin X and Li S: Association of single nucleotide polymorphisms
in VDR and DBP genes with HBV-Related hepatocellular carcinoma risk
in a chinese population. PLoS One. 9:e1160262014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhang T, Zhang J, You X, Liu Q, Du Y, Gao
Y, Shan C, Kong G, Wang Y, Yang X, et al: Hepatitis B virus X
protein modulates oncogene Yes-associated protein by CREB to
promote growth of hepatoma cells. Hepatology. 56:2051–2059. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hodgson AJ, Hyser JM, Keasler VV, Cang Y
and Slagle BL: Hepatitis B virus regulatory HBx protein binding to
DDB1 is required but is not sufficient for maximal HBV replication.
Virology. 426:73–82. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
He P, Zhang D, Li H, Yang X, Li D, Zhai Y,
Ma L and Feng G: Hepatitis B virus X protein modulates apoptosis in
human renal proximal tubular epithelial cells by activating the
JAK2/STAT3 signaling pathway. Int J Mol Med. 31:1017–1029. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Arbuthnot P, Capovilla A and Kew M:
Putative role of hepatitis B virus X protein in
hepatocarcinogenesis: Effects on apoptosis, DNA repair,
mitogen-activated protein kinase and JAK/STAT pathways. J
Gastroenterol Hepatol. 15:357–368. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
El-Serag HB: Hepatocellular carcinoma. N
Engl J Med. 365:1118–1127. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lin S, Hoffmann K and Schemmer P:
Treatment of hepatocellular carcinoma: A systematic review. Liver
Cancer. 1:144–158. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Whittaker S, Marais R and Zhu AX: The role
of signaling pathways in the development and treatment of
hepatocellular carcinoma. Oncogene. 29:4989–5005. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang XY, Pfeiffer HK, Thorne AW and
McMahon SB: USP22, an hSAGA subunit and potential cancer stem cell
marker, reverses the polycomb-catalyzed ubiquitylation of histone
H2A. Cell Cycle. 7:1522–1524. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lee HJ, Kim MS, Shin JM, Park TJ, Chung HM
and Baek KH: The expression patterns of deubiquitinating enzymes,
USP22 and Usp22. Gene Expr Patterns. 6:277–284. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Piao S, Ma J, Wang W, Liu Y, Zhang M, Chen
H and Guo F, Zhang B and Guo F: Increased expression of USP22 is
associated with disease progression and patient prognosis of
salivary duct carcinoma. Oral Oncol. 49:796–801. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li ZH, Yu Y, Du C, Fu H, Wang J and Tian
YU: RNA interference-mediated USP22 gene silencing promotes human
brain glioma apoptosis and induces cell cycle arrest. Oncol Lett.
5:1290–1294. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ning J, Zhang J, Liu W, Lang Y, Xue Y and
Xu S: Overexpression of ubiquitin-specific protease 22 predicts
poor survival in patients with early-stage non-small cell lung
cancer. Eur J Histochem. 56:e462012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Piao S, Liu Y, Hu J, Guo F, Ma J, Sun Y
and Zhang B: USP22 is m as a novel molecular Marker for predicting
disease progression and patient prognosis of oral squamous cell
carcinoma. PLoS One. 7:e425402012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Glinsky GV, Berezovska O and Glinskii AB:
Microarray analysis identifies a death-from-cancer signature
predicting therapy failure in patients with multiple types of
cancer. J Clin Invest. 115:1503–1521. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bosman FT, Carneiro F, Hruban RH and
Theise N: WHO Classification of tumours of the digestive system.
4th. IARC; Lyon: 2010
|
|
23
|
Edge SB and Compton CC: The American Joint
committee on cancer: The 7th edition of the AJCC cancer staging
manual and the future of TNM. Ann Surg Oncol. 17:1471–1474. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hoeller D and Dikic I: Targeting the
ubiquitin system in cancer therapy. Nature. 458:438–444. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tang B, Tang F, Li B, Yuan S, Xu Q,
Tomlinson S, Jin J, Hu W and He S: High USP22 expression indicates
poor prognosis in hepatocellular carcinoma. Oncotarget.
6:12654–12667. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Atanassov BS, Mohan RD, Lan X, Kuang X, Lu
Y, Lin K, McIvor E, Li W, Zhang Y, Florens L, et al: ATXN7L3 and
ENY2 coordinate activity of multiple H2B deubiquitinases important
for cellular proliferation and tumor growth. Mol Cell. 62:558–571.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Tang B, Liang X, Tang F, Zhang J, Zeng S,
Jin S, Zhou L, Kudo Y and Qi G: Expression of USP22 and Survivin is
an indicator of malignant behavior in hepatocellular carcinoma. Int
J Oncol. 47:2208–2216. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhang XY, Varthi M, Sykes SM, Phillips C,
Warzecha C, Zhu W, Wyce A, Thorne AW, Berger SL and McMahon SB: The
putative cancer stem cell marker USP22 is a subunit of the human
SAGA complex required for activator-driven transcription and cell
cycle progression. Mol Cell. 29:102–111. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Liu YL, Jiang SX, Yang YM, Xu H, Liu JL
and Wang XS: USP22 acts as an oncogene by the activation of
BMI-1-mediated INK4a/ARF pathway and Akt pathway. Cell Biochem
Biophys. 62:229–235. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Atanassov BS and Dent SY: USP22 regulates
cell proliferation by deubiquitinating the transcriptional
regulator FBP1. EMBO Rep. 12:924–930. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ning Z, Wang A, Liang J, Xie Y, Liu J, Yan
Q and Wang Z: USP22 promotes epithelial-mesenchymal transition via
the FAK pathway in pancreatic cancer cells. Oncol Rep.
32:1451–1458. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Hu J, Yang D, Zhang H, Liu W, Zhao Y, Lu
H, Meng Q, Pang H, Chen X, Liu Y and Cai L: USP22 promotes tumor
progression and induces epithelial-mesenchymal transition in lung
adenocarcinoma. Lung Cancer. 88:239–245. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhu M, Li W, Lu Y, Dong X, Lin B, Chen Y,
Zhang X, Guo J and Li M: HBx drives alpha fetoprotein expression to
promote initiation of liver cancer stem cells through activating
PI3K/AKT signal pathway. Int J Cancer. 140:1346–1355. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Liu H, Xu J, Zhou L, Yun X, Chen L, Wang
S, Sun L, Wen Y and Gu J: Hepatitis B virus large surface antigen
promotes liver carcinogenesis by activating the Src/PI3K/Akt
pathway. Cancer Res. 71:7547–7557. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Iyer SC, Gopal A and Halagowder D:
Myricetin induces apoptosis by inhibiting P21 activated kinase 1
(PAK1) signaling cascade in hepatocellular carcinoma. Mol Cell
Biochem. 407:223–237. 2015. View Article : Google Scholar : PubMed/NCBI
|