Introduction
Tumor metastasis is a major challenge in the
treatment of cancer (1). With the
advances in cancer treatment techniques, including surgery and
targeted therapies, treatment outcomes of patients with
non-metastatic tumors have been improved significantly in the past
decades (2,3). However, the survival of patients with
tumor metastasis is still poor owing to the lack of a radical
treatment strategy (1). Osteosarcoma
is a type of bone cancer that mainly affects children, adolescents,
and young adults (4). Although the
incidence rate is low, osteosarcoma is still a major cause of
cancer-related mortalities owing to its aggressive nature (4). It has been reported that ~20% of
patients with osteosarcoma were diagnosed with tumor metastasis
(5), resulting in a higher mortality
rate.
Beside mRNAs that encode protein products, the human
genome also transcribes functional non-coding RNAs that serve
crucial roles in both physiological and pathological processes
(6). A growing body of literature
has demonstrated that long non-coding RNAs (lncRNAs), a subgroup of
non-coding RNAs that are >200 nucleotides long, are key players
in human diseases (7), including
certain types of cancers (8). Long
intergenic non-coding RNA for kinase activation (LINK-A) lncRNA is
a known oncogenic lncRNA in triple-negative breast cancer (9). As one of the subunits of the
heterodimeric transcription factor hypoxia-inducible factor 1
(HIF1), HIF1α responds to hypoxia during cancer development and
participates in the regulation of tumor invasion of various types
of human malignancies (10). HIF1α
in some cases may participate in cancer biology through
interactions with lncRNAs (11,12). In
the progression of triple-negative breast cancer, LINK-A lncRNA
activates normoxic HIF1α to promote cancer development (9). Therefore, it is reasonable to
hypothesize that the interaction between LINK-A lncRNA and HIF1α
may also participate in osteosarcoma. In the present study, LINK-A
lncRNA was demonstrated to function in the metastasis of
osteosarcoma possibly by upregulating HIF1α.
Materials and methods
Cell line and human materials
MG-63 and U2OS human osteosarcoma cell lines were
bought from the American Type Culture Collection (ATCC; Manassas,
VA, USA). Cells were cultivated with ATCC-formulated Eagle's
Minimum Essential Medium (cat. no. 30–2003; ATCC) supplemented with
10% fetal bovine serum (FBS; Sangon Biotech Co, Ltd., Shangai,
China) in an incubator (37°C; 5% CO2). When required,
cells were treated with HIF1α (cat. no. 776–826; Sigma-Aldrich;
Merck KGaA, Darmstadt, Germany) at 10, 20 and 40 ng/ml for 24 h or
with the HIF inhibitor LW6 (cat. no. S8441; Selleck Chemicals,
Shanghai, China) at 10 ng/ml for 24 h prior to further
experiments.
Plasma samples were obtained from 62 patients with
osteosarcoma and 48 healthy volunteers who were admitted to
Zhongnan Hospital Affiliated to Medical College of Wuhan University
(Wuhan, China) between May 2015 and January 2018. Inclusion
criteria were: i) Patients who were diagnosed with osteosarcoma
through pathological examination; ii) patients who were diagnosed
and treated for the first time; iii) patients who were willing to
join the study. Exclusion criteria were: i) Patients with multiple
diseases; ii) patients who received treatment within 90 days before
admission. Among the 62 patients with osteosarcoma, distant tumor
metastasis was observed in 28 cases, and these patients were
classified into the metastatic osteosarcoma (MO) group based on
imaging findings. The remaining 34 patients were classified into
the non-metastatic osteosarcoma (NMO) group. The patient group
included 34 males and 28 females, aged between 12 and 44 years
(mean, 27.4±4.5 years). The healthy control group included 28 males
and 20 females, aged between 15 and 45 years (mean, 28.8±4.3
years). This study was approved by the Ethics Committee of Zhongnan
Hospital Affiliated to Medical College of Wuhan University; all
participants signed informed consent. Clinicopathological
characteristics of patients included in the study are available in
Table I
 | Table I.Clinicopathological characteristics of
participants. |
Table I.
Clinicopathological characteristics of
participants.
| Characteristics | Osteosarcoma | Controls |
|---|
| Cases (n) | 62 | 48 |
| Sex |
|
|
| Male | 34 | 28 |
|
Female | 28 | 20 |
| Metastasis |
|
|
| Yes | 28 | NA |
| No | 34 | NA |
| Tumor size (cm) |
|
|
|
<2 | 22 | NA |
| 2–4 | 20 | NA |
|
>4 | 20 | NA |
| Age range
(years) | 12–44 | 15–45 |
| Mean age (years) | 27.4±4.5 | 28.8±4.3 |
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted from plasma (1 ml) or MG-63
and U2OS cells (3×104 cells/ml) using RNAzol®
RT RNA Isolation Reagent (GeneCopoeia, Guangzhou, China). RNA
concentrations were measured using a NanoDrop™ 2000
spectrophotometer (Thermo Fisher Scientific, Inc., Waltham, MA,
USA). RNA extraction was repeated until all RNA samples had an
A260/A280 ratio between 1.8 and 2.0. cDNA was synthesized using
RevertAid RT Reverse Transcription kit (Thermo Fisher Scientific,
Inc.) under the following conditions: 25°C for 5 min, 55°C for 30
min and 75°C for 10 min. One-Step PrimeScript RT-PCR kit for Real
Time RT-PCR (Clontech Laboratories, Inc., Mountainview, CA, USA)
was used to prepare all PCR reaction conditions. Primer sequences
were as follows: LINK-A, forward 5′-TTCCCCCATTTTTCCTTTTC-3′,
reverse 5′-CTCTGGTTGGGTGACTGGTT-3′; GAPDH, forward
5′-GAAGGTGAAGGTCGGAGT-3′, reverse 5′-GAAGATGGTGATGGGATTTC-3′. qPCR
reaction conditions were: Initial denaturation at 95°C for 50 sec,
followed by 40 cycles of 95°C for 15 sec and 60.5°C for 34 sec.
Relative expression levels were quantified using the
2−ΔΔCq method (13) and
normalized to GADPH loading control.
LINK-A lncRNA expression vectors and
cell transfection
LINK-A lncRNA overexpression vectors (pcDNA3.1) and
empty vectors were designed and synthesized by Shanghai GenePharma
Co., Ltd., (Shanghai, China). MG-63 and U2OS cells were cultivated
overnight to reach 80–90% confluence. Lipofectamine®
2000 reagent (cat. no. 11668-019; Invitrogen; Thermo Fisher
Scientific, Inc.) was used to perform transfection with vectors at
a dose of 15 mM. Vectors were incubated with 5×105 cells
at 37°C for 6 h. Subsequent experiments were carried out 24 h
post-transfection. Non-transfected cells were used as control
cells; empty vector transfection was used as the negative
transfection control (NC).
Cell migration and invasion assay
Transfected MG-63 and U2OS cells were harvested and
serum-free single cell suspensions with a cell density of
5×104 cells/ml were prepared. Cell migration and
invasion were examined by the following steps: The upper Transwell
chamber (3 µm pore size) was filled with 100 µl serum-free cell
suspension and the lower chamber was filled with ATCC-formulated
Eagle's Minimum Essential Medium containing 20% FBS. Cells were
cultivated in an incubator (37°C; 5% CO2) for 24 h and
0.5% crystal violet (Sigma-Aldrich; Merck KGaA) staining was
performed for 20 min at room temperature. Free cells were removed
using a cotton swab. For the invasion assay, the upper chamber was
pre-coated with Matrigel (cat. no. 356234; EMD Millipore,
Billerica, MA, USA). Membranes were collected and invading or
migrating cells were observed under a light microscope
(magnification, ×40; Olympus Corporation, Tokyo, Japan).
Western blotting
Proteins were extracted from cells (3×104
cells/ml) using ReadyPrep™ Protein Extraction kit (Total Protein;
Bio-Rad Laboratories, Inc., Hercules, CA, USA). Protein
concentration was measured with the bicinchoninic acid assay.
Following denaturing, proteins (35 µg) were separated by 10%
SDS-PAGE. Proteins were transferred onto polyvinylidene difluoride
membrane by semi dry method. Membranes were blocked in 5% milk for
2 h at room temperature. Membranes were then incubated with the
rabbit anti-human primary antibodies against HIF1α (1:1,500; cat.
no. ab2185, Abcam, Shanghai, China) and GAPDH (1:2,000; cat. no.
ab9485; Abcam) at 4°C overnignt, followed by secondary antibody
horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin G
(1:1,000; cat. no. MBS435036; MyBioSource, San Diego, CA, USA) at
room temperature for 2 h. Signals were developed using ECL™
(Sigma-Aldrich; Merck KGaA) and scanned by MYECL™ Imager (Thermo
Fisher Scientific, Inc.). Densitometric analysis was performed
using ImageJ v1.46 software (National Institutes of Health,
Bethesda, MD, USA). GAPDH was used as endogenous control for data
normalization.
Statistical analysis
All experiments were performed in triplicates and
data were recorded as the mean ± standard deviation. SPSS 19.0 (IBM
Corp., Armonk, NY, USA) was used to perform all statistical
analyses. Comparisons among multiple groups were performed by
one-way analysis of variance followed by Tukey test. Receiver
operating characteristic (ROC) curve analysis was performed using
patients with osteosarcoma as true positive cases and healthy
controls as true negative cases. P<0.05 was considered to
indicate a statistically significant difference.
Results
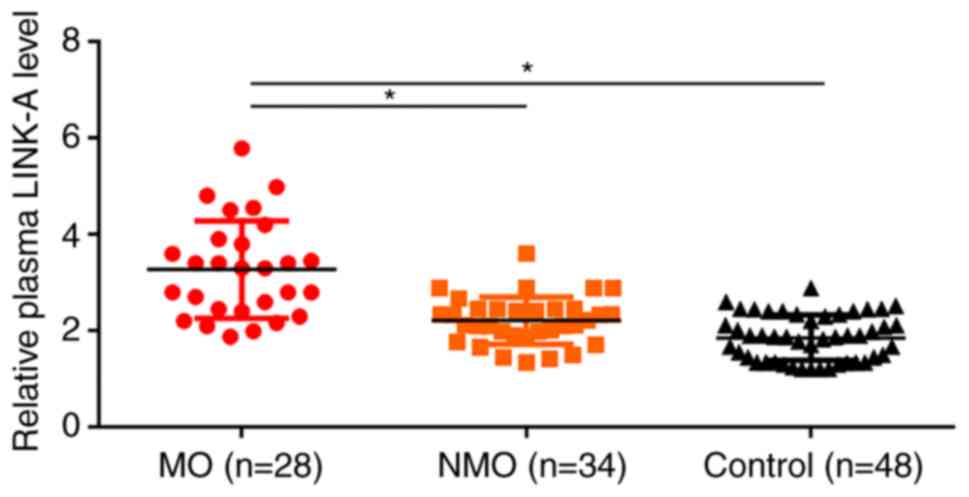
Plasma LINK-A lncRNA is upregulated in
patients with osteosarcoma and is related to tumor metastasis
RT-qPCR results revealed that, compared with the
control and NMO groups, plasma LINK-A lncRNA expression was
significantly upregulated in the MO group (Fig. 1). In addition, plasma levels of
LINK-A lncRNA were also slightly higher in the NMO group than in
control group (Fig. 1).
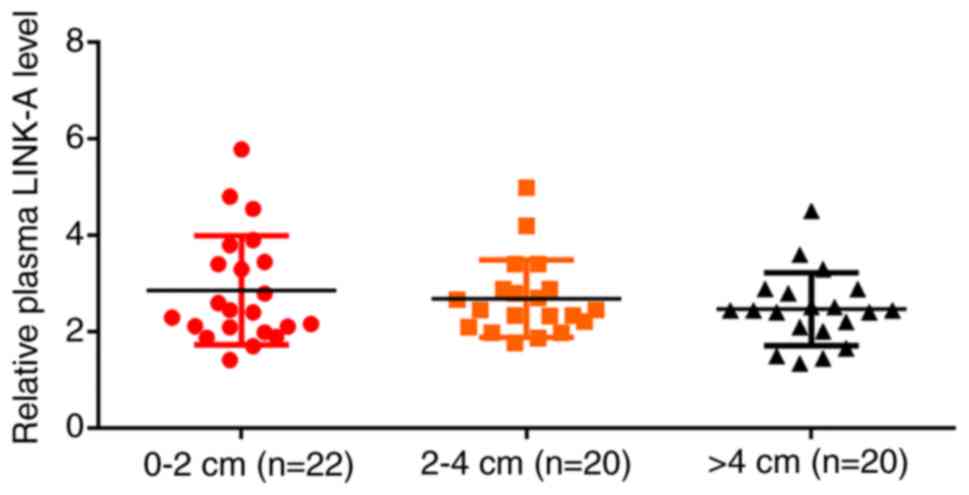
Plasma levels of LINK-A lncRNA in
patients with osteosarcoma are not affected by tumor size
Based on the diameter of the primary tumor, patients
with MO and NMO were divided into 0–2 cm group (n=22), 2–4 cm group
(n=20) and >4 cm group (n=20) (Fig.
2). RT-qPCR results indicated no significant differences in
plasma levels of LINK-A lncRNA among these groups. No significant
differences in plasma levels of LINK-A lncRNA were found in MO
patients with different tumor size (data not shown).
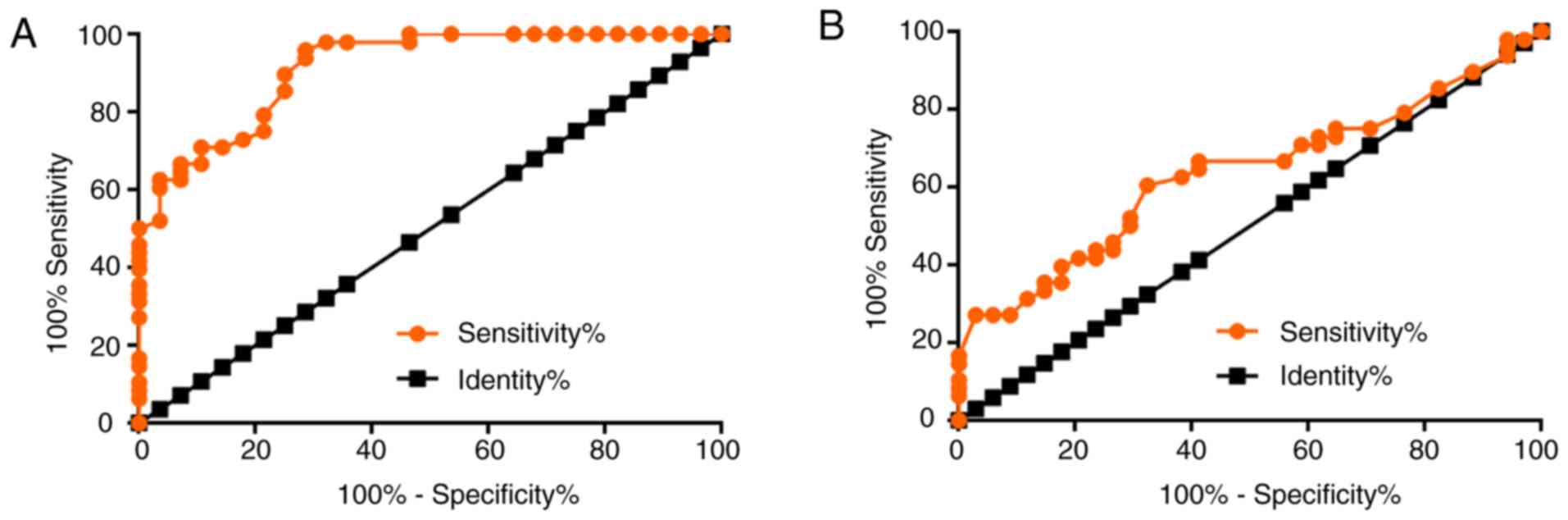
Upregulation of LINK-A lncRNA
distinguishes patients with MO but not those with NMO from healthy
controls
To investigate the diagnostic value of plasma LINK-A
lncRNA for osteosarcoma, ROC curve analysis was performed patients
with NMO or MO as true positive cases and healthy controls as true
negative cases. For metastatic osteosarcoma, the area under the
curve (AUC) was 0.9141, with standard error of 0.03214 and 95%
confidence interval of 0.8511–0.9771 (P<0.0001; Fig. 3A). For non-metastatic osteosarcoma,
the AUC was 0.6351, with standard error of 0.06082 and 95%
confidence interval of 0.5159–0.7543 (P=0.038; Fig. 3B).
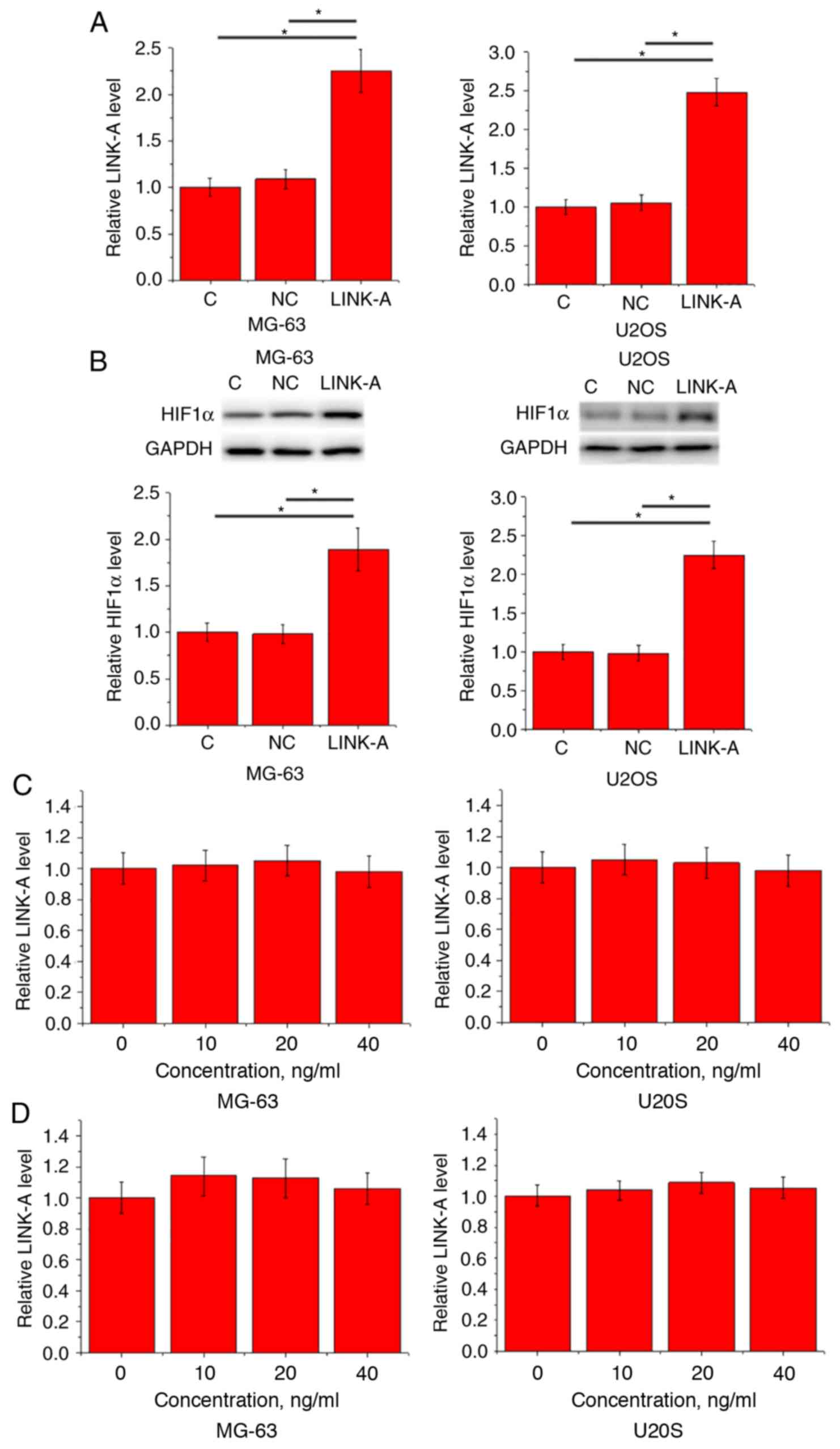
LINK-A lncRNA overexpression mediates
the upregulation of HIF1α in MG-63 and U2OS cell lines
Compared with the Control and NC groups, LINK-A and
HIF1α expression levels were significantly upregulated following
cell transfection (Fig. 4A). LINK-A
lncRNA overexpression led to significantly increased expression of
HIF1α protein in MG-63 and U2OS cells (Fig. 4C). By contrast, treatment with HIF1α
at doses of 10, 20 and 40 ng/ml failed to significantly affect
endogenous LINK-A lncRNA expression in untransfected cells
(Fig. 4D) as well as in cells
overexpressing LINK-A lncRNA (Fig.
4D).
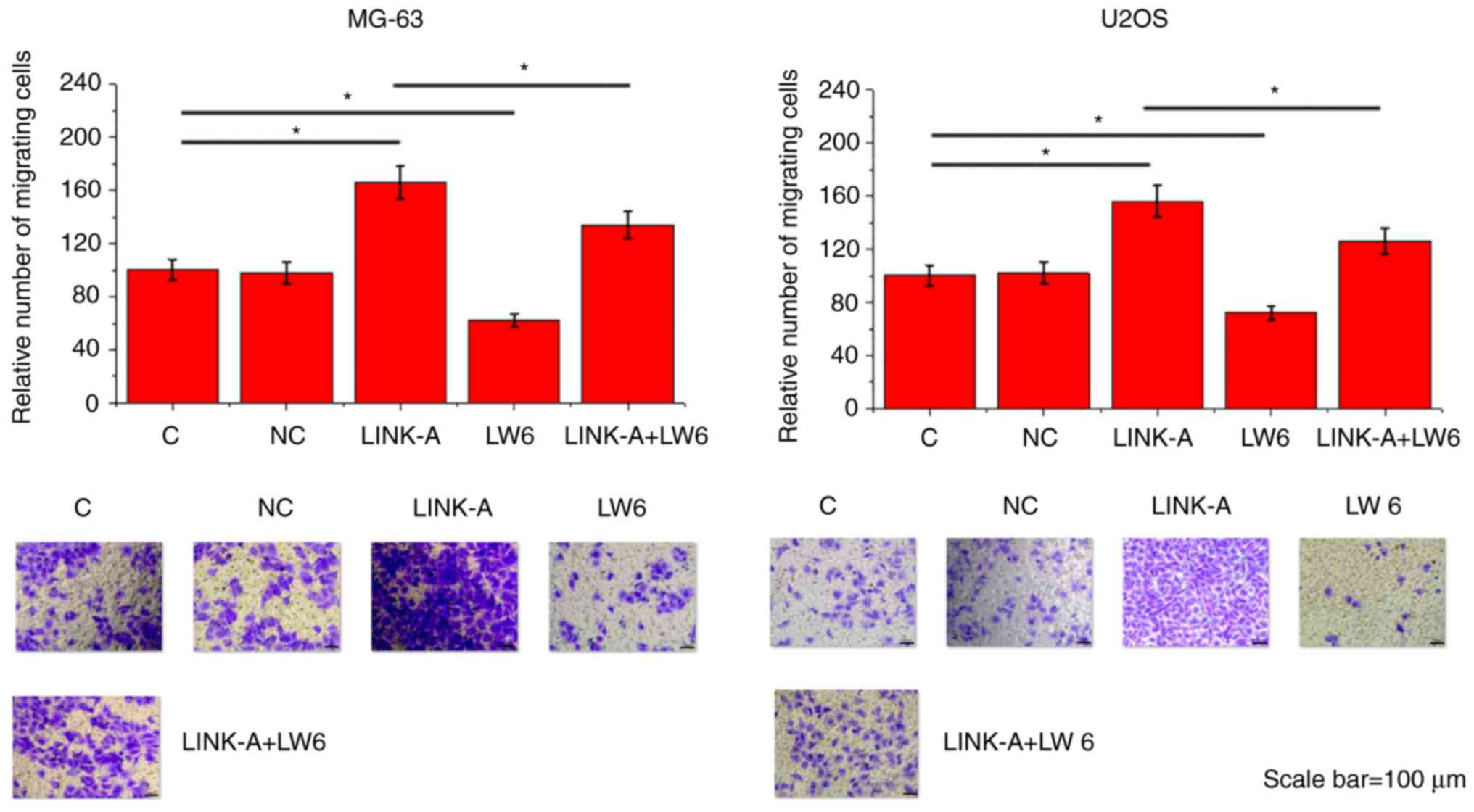
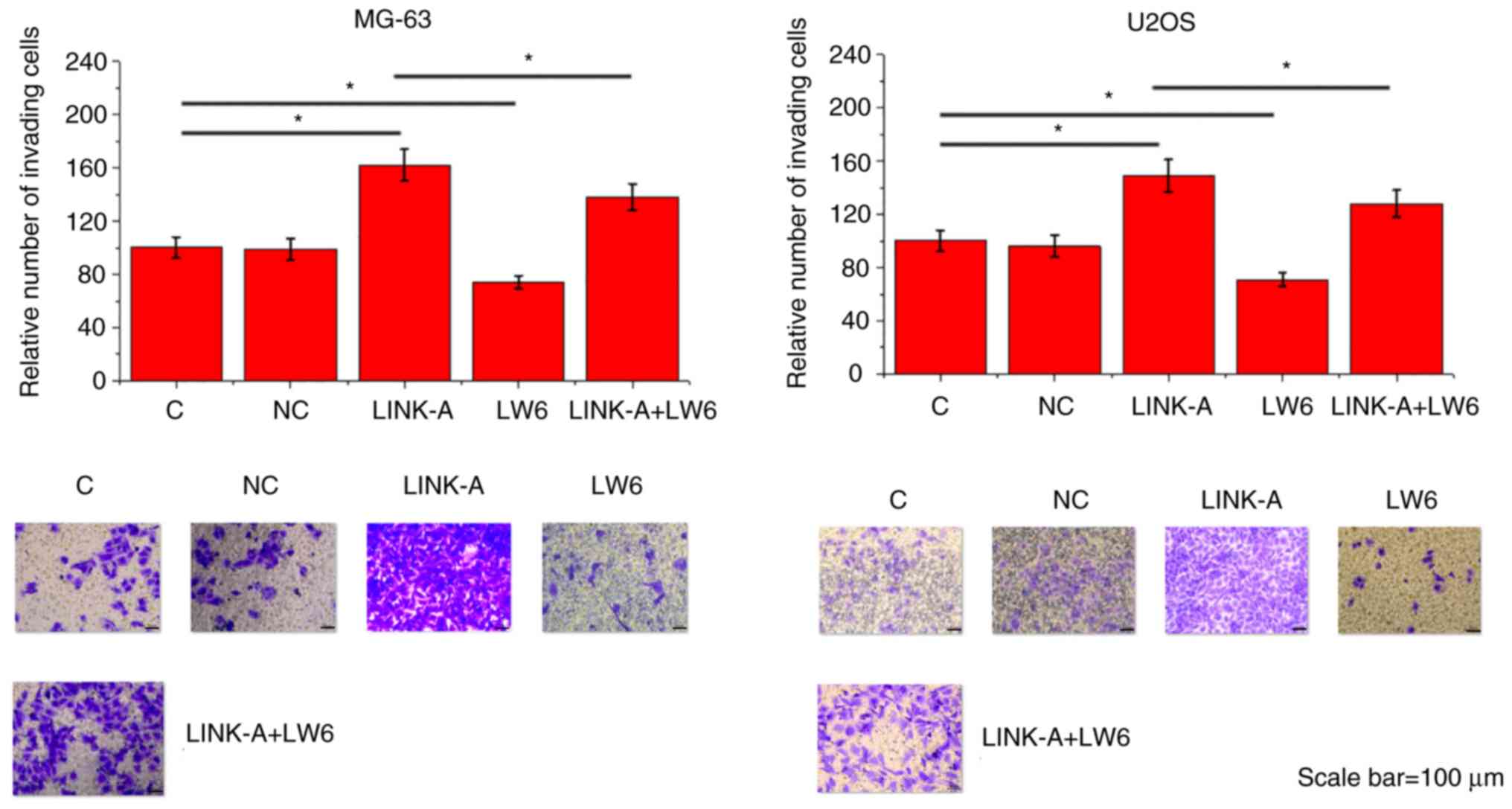
LINK-A lncRNA overexpression promotes
migration and invasion of MG-63 and U2OS cells through HIF1α
Compared with the Control group and the NC group,
LINK-A lncRNA overexpression led to significantly promoted
migration (Fig. 5) and invasion
(Fig. 6) of MG-63 and U2OS cell
lines. In addition, treatment with HIF inhibitor LW6 at a dose of
10 ng/ml significantly attenuated the enhancing effects of LINK-A
overexpression on cancer cell migration (Fig. 5) and invasion (Fig. 6).
Discussion
A recent study reported that LINK-A lncRNA serves an
oncogenic role in triple-negative breast cancer through the
interaction with HIF1α (9). The
present study confirmed the existence of the association between
LINK-A lncRNA and HIF1α in osteosarcoma. Therefore, triple-negative
breast cancer and osteosarcoma may share similar molecular
pathological pathways.
Circulating biomarkers have been widely used in the
diagnosis and prognosis of human diseases (14,15).
Previous studies have revealed that the development of osteosarcoma
induces altered expression of a large set of human genes, including
lncRNAs (16,17). A number of these differentially
expressed lncRNAs, such as urothelial cancer associated 1 (18) and hepatocellular carcinoma
up-regulated lncRNA (19), have been
demonstrated to have a diagnostic and prognostic potential for
osteosarcoma. As an oncogenic lncRNA, expression of LINK-A lncRNA
is altered in triple-negative breast cancer (9); however, the expression pattern of
LINK-A lncRNA in other human diseases is unknown. The present study
revealed that plasma circulating LINK-A lncRNA was only upregulated
in patients with MO but not in patients with NMO; upregulation of
LINK-A distinguished patients with MO, but not those with NMO, from
healthy controls. These data suggested that LINK-A may be
specifically involved in the distant metastasis of
osteosarcoma.
In the present study, a comparison of patients with
different diameters of primary tumors revealed no significant
differences in plasma levels of LINK-A lncRNA, which suggested that
LINK-A lncRNA was unlikely to be involved in the growth of
osteosarcoma. The in vitro cell proliferation data also
indicated that overexpression of LINK-A lncRNA had no significant
effects on the proliferation of MG-63 and U2OS human osteosarcoma
cells (data not shown).
It has been reported that LINK-A lncRNA in
triple-negative breast cancer activates normoxic HIF1α signalling
(9). The present study confirmed the
existence of the association between LINK-A and HIF1α in
osteosarcoma. LINK-A overexpression had inhibitory effects on HIF1α
expression, whereas HIF1α had no effect on LINK-A. In addition, HIF
inhibitor LW6 reduced the effects of LINK-A overexpression on cell
migration and invasion. Therefore, LINK-A may be an upstream
activator of HIF1α. However, the molecular mechanism of the
regulation of HIF1α by LINK-A remains to be determined.
In conclusion, LINK-A lncRNA is upregulated in
osteosarcoma and may participate in the metastasis of osteosarcoma
by upregulating HIF1α.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
LC and BZ did the experiments, analyzed the data and
were major contributors in writing the manuscript. KL performed a
part of the experimental work. All authors read and approved the
final manuscript and confirmed its accuracy.
Ethics approval and consent to
participate
Ethical approval was obtained from The Ethics
Committee of Zhongnan Hospital Affiliated to Medical College of
Wuhan University (Wuhan, China). All procedures performed in
studies involving human participants were in accordance with the
ethical standards of the institutional and national research
committee and with the 1964 Declaration of Helsinki and its later
amendments. Written informed consent was obtained from all patients
and controls following an explanation the nature and possible
consequences of the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Steeg PS: Tumor metastasis: Mechanistic
insights and clinical challenges. Nat Med. 12:895–904. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Miller KD, Siegel RL, Lin CC, Mariotto AB,
Kramer JL, Rowland JH, Stein KD, Alteri R and Jemal A: Cancer
treatment and survivorship statistics, 2016. CA Cancer J Clin.
66:271–289. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
DeSantis CE, Lin CC, Mariotto AB, Siegel
RL, Stein KD, Kramer JL, Alteri R, Robbins AS and Jemal A: Cancer
treatment and survivorship statistics, 2014. CA Cancer J Clin.
64:252–271. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Luetke A, Meyers PA, Lewis I and Juergens
H: Osteosarcoma treatment-where do we stand? A state of the art
review. Cancer Treat Rev. 40:523–532. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Isakoff MS, Bielack SS, Meltzer P and
Gorlick R: Osteosarcoma: Current treatment and a collaborative
pathway to success. J Clin Oncol. 33:3029–3035. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Quinn JJ and Chang HY: Unique features of
long non-coding RNA biogenesis and function. Nat Rev Genet.
17:47–62. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Shi X, Sun M, Liu H, Yao Y and Song Y:
Long non-coding RNAs: A new frontier in the study of human
diseases. Cancer Lett. 339:159–166. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Huarte M: The emerging role of lncRNAs in
cancer. Nat Med. 21:1253–1261. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lin A, Li C, Xing Z, Hu Q, Liang K, Han L,
Wang C, Hawke DH, Wang S, Zhang Y, et al: The LINK-A lncRNA
activates normoxic HIF1α signalling in triple-negative breast
cancer. Nat Cell Biol. 18:213–224. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Semenza GL: Targeting HIF-1 for cancer
therapy. Nat Rev Cancer. 3:721–732. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Cai Q, Wang Z, Wang S, Weng M, Zhou D, Li
C, Wang J, Chen E and Quan Z: Long non-coding RNA LINC00152
promotes gallbladder cancer metastasis and epithelial-mesenchymal
transition by regulating HIF-1α via miR-138. Open Biol.
7:1602472017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Liu L, Zhao X, Zou H, Bai R, Yang K and
Tian Z: Hypoxia promotes gastric cancer malignancy partly through
the HIF-1α dependent transcriptional activation of the long
Non-coding RNA GAPLINC. Front Physiol. 7:4202016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2-ΔΔCT method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Pirola CJ, Fernández Gianotti T, Castaño
GO, Mallardi P, San Martino J, Mora Gonzalez Lopez Ledesma M,
Flichman D, Mirshahi F, Sanyal AJ and Sookoian S: Circulating
microRNA signature in non-alcoholic fatty liver disease: From serum
non-coding RNAs to liver histology and disease pathogenesis. Gut.
64:800–812. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J,
Chen D, Gu J, He X and Huang S: Circular RNA is enriched and stable
in exosomes: A promising biomarker for cancer diagnosis. Cell Res.
25:981–984. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhu KP, Zhang CL, Shen GQ and Zhu ZS: Long
noncoding RNA expression profiles of the doxorubicin-resistant
human osteosarcoma cell line MG63/DXR and its parental cell line
MG63 as ascertained by microarray analysis. Int J Clin Exp Pathol.
8:8754–8773. 2015.PubMed/NCBI
|
|
17
|
Li JP, Liu LH, Li J, Chen Y, Jiang XW,
Ouyang YR, Liu YQ, Zhong H, Li H and Xiao T: Microarray expression
profile of long noncoding RNAs in human osteosarcoma. Biochem
Biophys Res Commun. 433:200–206. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li W, Xie P and Ruan WH: Overexpression of
lncRNA UCA1 promotes osteosarcoma progression and correlates with
poor prognosis. J Bone Oncol. 5:80–85. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sun XH, Yang LB, Geng XL, Wang R and Zhang
ZC: Increased expression of lncRNA HULC indicates a poor prognosis
and promotes cell metastasis in osteosarcoma. Int J Clin Exp
Pathol. 8:2994–3000. 2015.PubMed/NCBI
|