Introduction
In 2015, cancer-associated mortalities in China were
estimated at 2.8 million, with 4.3 million newly identified cases,
the most common of which being lung cancer (1). Non-small cell lung cancer (NSCLC) is
the most commonly histologically diagnosed lung cancer, accounting
for ~85–90% of cases (2). In the
past decade, genetic and genomic profiling in NSCLC has improved
considerably, and with it, the understanding of underlying
molecular mechanisms of disease pathogenesis and strategies for
targeted therapies (3–5). Potential inhibition targets for 60% of
NSCLC cases in the form of actionable driver alterations have been
identified through molecular profiling (6). Evidence supports the effectiveness of
several of these targeted therapies, including, gefitinib
[epidermal growth factor receptor (EGFR)-tyrosine kinase inhibitor
(TKI)], erlotinib (sensitizing EGFR mutations) and crizotinib
[sensitizing ALK receptor tyrosine kinase (ALK) and ROS
proto-oncogene 1 receptor tyrosine kinase fusion] (7).
Next-generation sequencing (NGS) is used in the
field of genomic research and clinical applications (8). For patients with NSCLC, targeted NGS
provides improved drug target screening and additional potential
avenues of treatment (9–12). Due to the ever-growing number of
discovered predictive markers for therapies and the limited
availability of submitted tissue samples, the long-standing ‘one
test-one drug’ paradigm is gradually being replaced by multiplexed
genotyping platforms (13). One such
platform is the FoundationOne CDx (F1CDx), which received marketing
approval by the Food and Drug Administration (FDA). FICDx performs
NGS-based in vitro diagnostics to detect genetic mutations
in all solid tumor types and serves to identify potential
beneficiaries for several FDA-approved targeted treatment options
(14). With more NGS platforms being
developed, standardization of reports from various test platforms
is warranted (15).
In the present study, a total of 23 NSCLC samples,
containing 21 Formalin-Fixed Paraffin-Embedded (FFPE) and two
cell-free DNA (cf)DNA samples were analyzed. The coding regions of
~180 cancer driver genes were explored using The Roche Nimblegen
(Roche Nimblegen, Inc.) custom-designed panel. Subsequently, the
captured regions were sequenced by NGS to detect various types of
mutations. Whenever available (for a total of 16 patients), the
sequences were compared with exome sequences obtained from white
blood cell (WBC) samples.
Materials and methods
Specimen collection
A total of 23 patients with stage III/IV NSCLC at
the time of diagnosis were included in the present study. Patients
were recruited from The Second Hospital of Anhui Medical University
(Hefei, China) between March 2017 and October 2017. The patients
did not receive preoperative chemotherapy or radiotherapy, and
those with prior TKI therapy were excluded from the study. There
were 21 FFPE specimens and 2 cfDNA specimens in patients who did
not have tissue samples available at presentation. All FFPE samples
were independently assessed by two experienced pathologists and had
a minimum tumor content of 40%. Blood samples from 16 patients were
collected as controls to validate the identified germline
mutations. Patient clinical data are presented in Table I. The samples were obtained from 10
males and 13 females (median age, 62 years; age range, 31–77
years). Among these patients, there was 1 smoker and 22
non-smokers.
 | Table I.Overview of patient and tumor
characteristics in the present study. |
Table I.
Overview of patient and tumor
characteristics in the present study.
|
Characteristics | n (%) |
|---|
| Sex |
|
|
Male | 10 (43.5) |
|
Female | 13 (54.5) |
| Age |
|
|
>60 | 13 (54.5) |
|
≤60 | 10 (43.5) |
| Smoking status |
|
|
Smoker | 1 (4.3) |
|
Non-smoker | 22 (95.7) |
| Stage |
|
| I | 9 (39.1) |
| II | 1 (4.3) |
|
III | 6 (26.1) |
| IV | 3 (13.0) |
|
Unknown | 4 (17.4) |
| Type of
specimen |
|
| FFPE
tissue samples | 21 (91.3) |
|
Cell-free DNA (cfDNA) | 2 (8.7) |
| Genomic
DNA (gDNA) | 16 (69.6) |
Nucleic acid preparation
Deparaffinization of 40 µm tissue sections was
performed with 100% xylene (Sigma Aldrich; Merck KGaA, Darmstadt,
Germany) followed by 100% ethanol as described previously (16). Deparaffinized samples were suspended
in proteinase K-contained buffer (Thermo Fisher Scientific, Inc.,
Waltham, MA, USA). Following extraction using phenol-chloroform
(Sigma Aldrich; Merck KGaA), DNA samples were treated with ethanol
(Sigma Aldrich; Merck KGaA) precipitation and resuspended in
deionized water. cfDNA blood collection tubes (Streck, Inc., Omaha,
NE, USA) were used for the collection of 10 ml blood samples.
Procurement of 1.2 ml of plasma from every patient was performed
prior to cfDNA extraction using centrifugation twice (4°C; 1,600 ×
g for 10 min). cfDNA was isolated with the QIAamp Circulating
Nucleic Acid kit (Qiagen, Inc., Valencia, CA, USA) according to the
manufacturer's protocol. Validation of adopted protocols for the
present study was obtained from previous findings (17). A total of 2 ml of total peripheral
blood was used to process WBC DNA as a control, using a FlexiGene
DNA kit (Qiagen, Inc.). Quantification of isolated DNA samples was
performed using a NanoDrop spectrophotometer (Thermo-Fisher
Scientific, Inc.), and by fluorimetry, which was conducted using
Qubit dsDNA high-sensitivity and/or broad-range assay kits (Thermo
Fisher Scientific, Inc.).
Configuration of the somatic
subpanel
The somatic cancer gene list was selected based on
the following cancer-associated databases such as the Catalogue of
Somatic Mutations in Cancer (COSMIC; cancer.sanger.ac.uk/cosmic), MyCancerGenome
(www.mycancergenome.org), CancerIndex
(www.cancerindex.org), FDA (www.fda.gov), European Medicines Agency (www.ema.europa.eu/ema), National Comprehensive Cancer
Network (NCCN; www.nccn.org), PHARMGKB (www.pharmgkb.org/), MDAnderson Cancer Center
(pct.mdanderson.org), DRUGBACK
(www.drugbank.ca), ClinicalTrial (clinicaltrials.gov), Google Scholar (scholar.google.com) and PubMed (www.ncbi.nlm.nih.gov/pubmed) databases, studies
published between 2000 and 2018.
Configuration of the germline
subpanel
A preliminary cancer gene list was built based on
germline alterations linked with predisposition for hereditary
cancer and sourced from the Human Gene Mutation Database
(www.hgmd.cf.ac.uk), ClinVar (www.ncbi.nlm.nih.gov/clinvar), Online Mendelian
Inheritance in Man (www.omim.org),
Genetics Home Reference (ghr.nlm.nih.gov) and Orphanet (www.orpha.net) databases. Filtering of the list was
performed and genes were removed based on a number of criteria,
including a lack of scientific evidence supporting their
association with cancer. The tumor-associated genes in the FD-180
panel were listed in Table II.
 | Table II.The gene list of FD-180 panel. |
Table II.
The gene list of FD-180 panel.
| APC | ATM | ATR | AXIN2 | RECQL4 | RET | RNF43 | RUNX1 |
|---|
| BAP1 | BLM | BMPR1A | BRCA1 | SBDS | SDHA | SDHAF2 | SDHB |
| BRCA2 | BRIP1 | CDC73 | CDH1 | SDHC | SDHD | SETBP1 | SLC2A2 |
| CDK4 | CDKN1B | CDKN2A | CHEK2 | SLX4 | SMAD4 | SMARCA4 | SMARCB1 |
| CLCN7 | CYLD | DDB2 | DICER1 | SMARCE1 | SPTA1 | STK11 | SUFU |
| EPCAM | ERCC2 | ERCC3 | ERCC4 | TERT | TGFBR2 | TP53 | TP63 |
| ERCC5 | EXT1 | EXT2 | FANCA | TSC1 | TSC2 | VHL | WAS |
| FANCB | FANCC | FANCD2 | FANCE | WRN | WT1 | XPA | XPC |
| FANCF | FANCG | FANCI | FANCL | ABL1 | ABL2 | AKT1 | ALK |
| FANCM | FH | FLCN | GALNT12 | AR | ARAF | ATM | ATR |
| GLA | GPC3 | HNF1A | MAD2L2 | BRAF | BRCA1 | BRCA2 | CDK12 |
| MAX | MEN1 | MLH1 | MLH3 | CDK4 | CDK6 | CRKL | CDKN1A |
| MPL | MSH2 | MSH3 | MSH6 | CDKN2A | CHD4 | CHEK2 | CSF1R |
| MUTYH | NBN | NF1 | NF2 | CTNNB1 | DDR2 | EGFR | ERBB2 |
| PALB2 | PHOX2B | PIK3CA | PMS1 | ERBB3 | ERBB4 | ESR1 | FANCA |
| PMS2 | POLD1 | POLE | PRF1 | FANCB | FANCI | FANCL | FANCC |
| PRKAR1A | PRKCD | PTCH1 | PTEN | FANCD2 | FANCE | FANCF | FANCG |
| RAD50 | RAD51C | RAD51D | RB1 | FANCM | FBXW7 | FGFR1 | FGFR2 |
| TSC1 | TSC2 | XRCC2 | SPOP | FGFR3 | FGFR4 | FRK | FYN |
| MAPK1 | MAPK3 | MET | mTOR | GNA11 | GNAQ | GRM3 | HRAS |
| NF1 | NOTCH1 | NRAS | PDGFRA | IDH1 | IDH2 | KDR | KIT |
| PIK3CA | PIK3CD | PIK3R1 | PTCH1 | KRAS | LYN | MAP2K1 | MAP2K2 |
| PTEN | PTPN11 | RAC1 | RAF1 | RET | ROS1 | SMO |
|
Library construction and
sequencing
The Whole Exome Sequencing (WES) library was
constructed using the Illumina TruSeq DNA Sample Prep kit
(Illumina, Inc., San Diego, CA, USA), and targeted exome enrichment
was performed using the SeqCap EZ Human Exome Library v2.0 kit
(Roche Nimblegen Inc.). Library construction and targeted region
enrichment for FD-180 were completed with the KAPA Hyper Prep kit
for Illumina platforms (Kapa Biosystems; Roche Diagnostics,
Indianapolis, IN, USA) and the SeqCap EZ Choice Library (Roche
Nimblegen, Inc.), respectively. The FD-180 panel was used for the
simultaneous sequencing of all exons of ~180 tumor-associated genes
(data not shown).
Paired-end sequencing was performed with the
Illumina X Ten or NEXT SEQ 500 sequencing instrument (Illumina,
Inc.) according to the manufacturer's protocol, yielding ~150 bp
short sequence reads. An average of ~15G of data was generated for
each WES sample, accounting for 100–200X coverage of the entire
exome. The average data for FD-180 target samples was 3G.
Data analysis
Raw data were processed into a clean FASTQ output
with flexbar (v3.3; http://github.com/seqan/flexbar) through trimming of
adapter sequences and removing reads of insufficient quality
(average quality score <15). Raw reads were checked for the data
quality using FASTQC (v0.11.5) (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/).
Plots of quality scores across all bases in reads indicated that
the majority of positions exhibited a quality score Q≥20. Raw reads
were subsequently trimmed for adapter contamination using
Trimmomatic (v0.32; www.usadellab.org). Leading and trailing low-quality
bases (<3) were removed. Reads were scanned with a
four-base-wide sliding window and the following bases of reads were
cut when the average quality per base dropped below 15. Reads
>50 bases were selected for subsequent analysis. Paired clean
reads following Trimmomatic treatment were aligned against the
human genome (hg19) using Burrows-Wheelers Aligner (18). Genome Analysis Toolkit (19) was used to do base quality score
recalibration, a process in which machine learning is applied to
model quality score errors empirically, and the quality scores are
adjusted accordingly. Analysis of the realigned BAM files, and
detection of somatic single-nucleotide variants and
insertions/deletions was performed using MuTect (20). Normal germline variants were filtered
out using the Single Nucleotide Polymorphism Database (dsSNP)
(21) or the Exome Aggregation
Consortium (ExAC; exac.broadinstitute.org). Default parameter settings
were used for all programs. In-house developed scripts were used to
eliminate erroneous base calls and generate final mutations based
on variant frequency (>0.5%). The frequency of variation is
equal to the ratio of the depth of the variant allele to the total
depth of all alleles at the locus. Remaining variants were
annotated using the ExAC and InterPro database (http://www.ebi.ac.uk/interpro/). Gene ontology
enrichment was analyzed on the website (www.geneontology.org), focusing on the genes which
were mutated in >4 primary tumor samples.
Results
In order to explore the therapeutic relevance of
FD-180 and improve the survival rate of patients with NSCLC, 2
cfDNA and 21 FFPE samples were studied to detect driver mutations
in tumor DNA with (16 samples) or without matched WBC DNA as a
control (mean sequencing depth, 98.96× WES). The average sequencing
depth on the 180 target genes for the cfDNA and FFPE samples was
1,747.7X and 398.4X respectively. Q30 for all data was 86.4%, where
Q scores are defined as a property that is logarithmically
associated with the base calling error probabilities. Q30 means
that the accuracy of base calling is 99.9% (22).
Somatic variations in primary
tumor
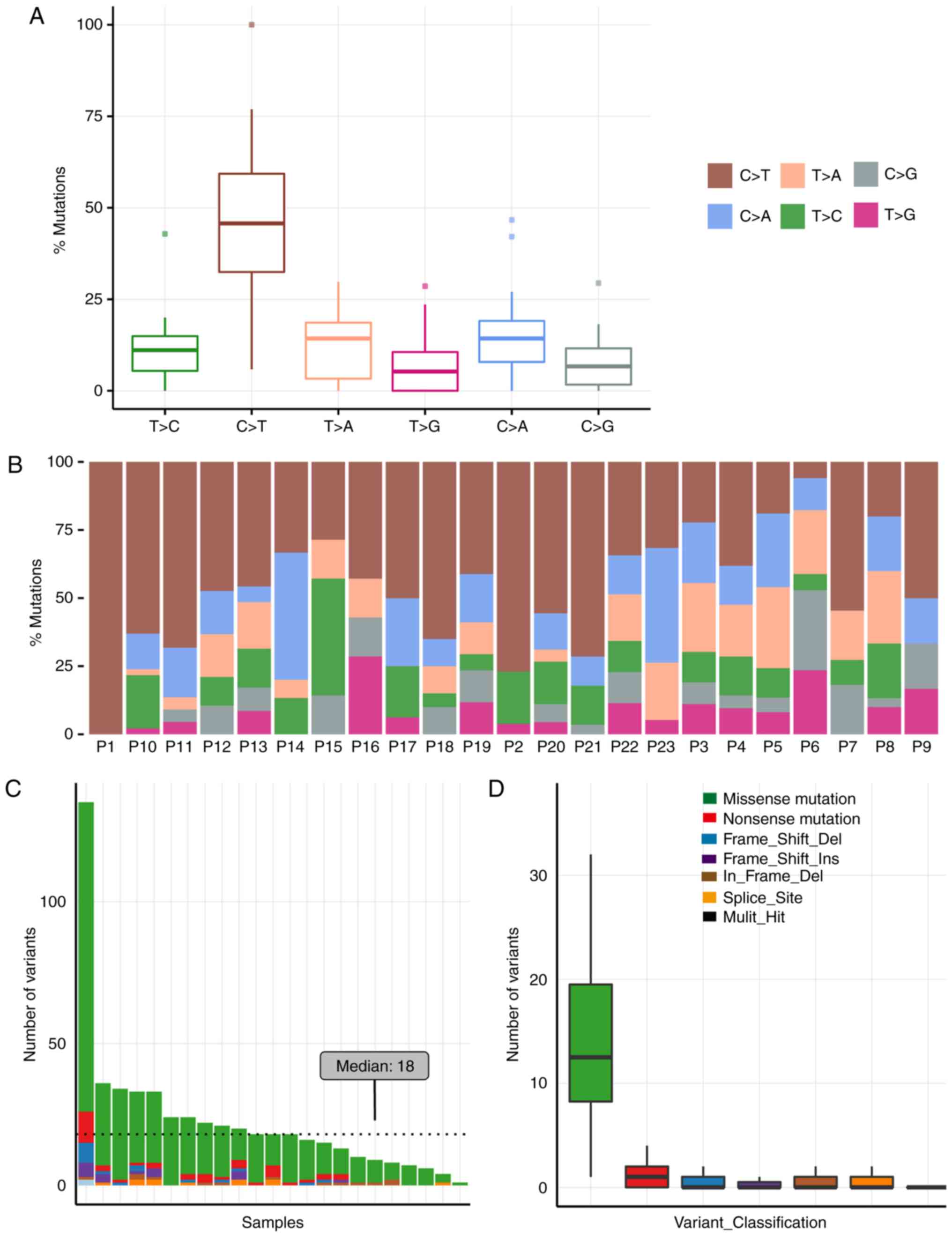
The mutational signature of NSCLC samples was
dissected through inspection of the spectrum of base substitutions.
All 23 samples exhibited similar point mutation spectra. Cytosine
to thymine was the most frequent transversion among somatic
mutations, which accounted for 45.6% in primary tumor tissues
(Fig. 1A and B).
Overall, 525 somatic mutations were detected in the
23 NSCLC samples. The primary type of mutation identified was
missense mutations (82.1%; Fig. 1C and
D). Notably all the mutations reported were within the protein
coding regions, as the capturing probes used in the current study
were restricted to coding sequences. Variations with >5%
frequency, as reported in either the dbSNP or the ExAC database,
were excluded. Mutation types and numbers were compared with the
normal tissue datasets, and the median variant count among the 23
samples was 18 (Fig. 1C and D). The
high variant count in patient 1 remains unclear. Possible causes
include poor living conditions and long cancer progression time
prior to diagnosis.
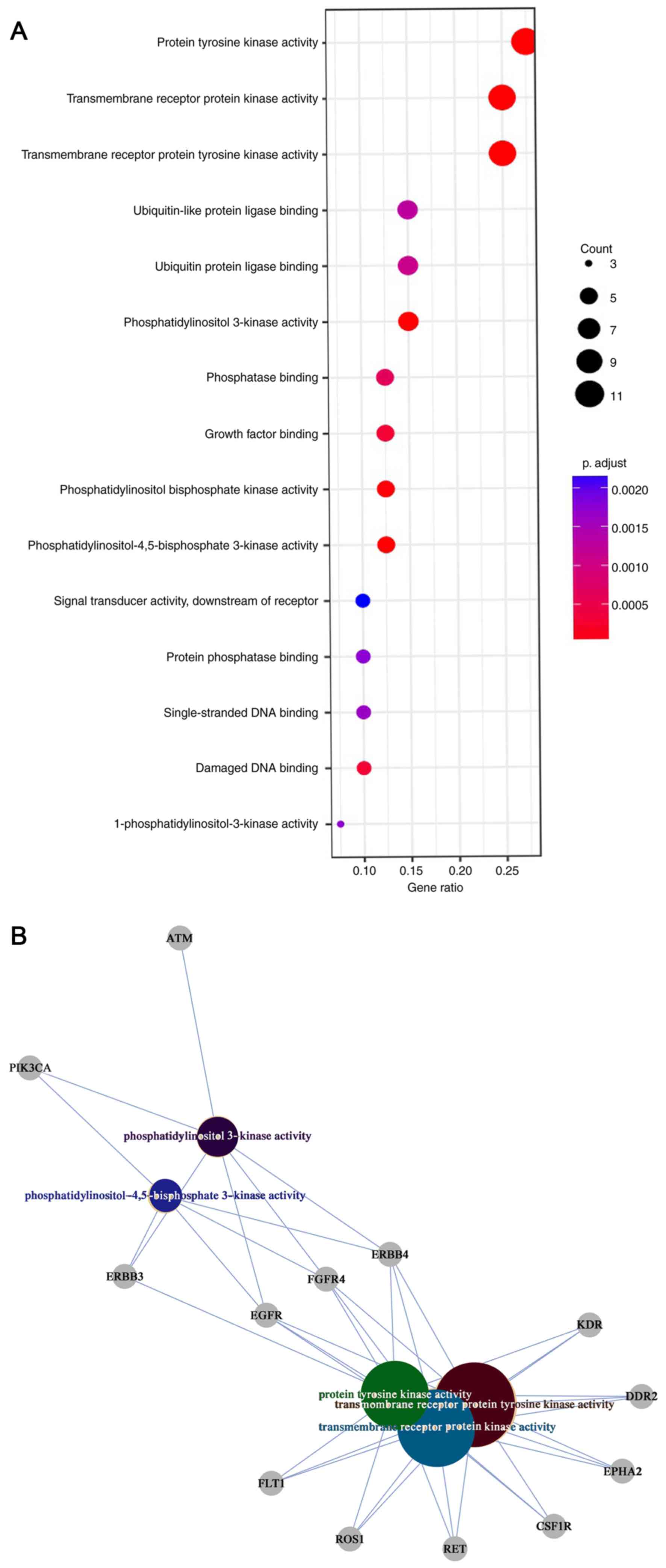
Implicated biological processes and
molecular function
Gene Ontology (www.geneontology.org) enrichment analysis was
performed with 40 frequently mutated genes (genes which were
mutated in >4 primary tumor samples). ‘Peptidyl-tyrosine
phosphorylation’, ‘peptidyl-tyrosine modification’,
‘phosphatidylinositol-mediated signaling’, ‘inositol lipid-mediated
signaling’ and ‘protein autophosphorylation’ were the top five
biological process of these high frequency genes in the 23 NSCLC
samples (Fig. 2A). Notably, the
molecular functions of these genes included ‘protein tyrosine
kinase activity’, ‘transmembrane receptor protein tyrosine kinase
activity’, ‘transmembrane receptor protein kinase activity’,
‘phosphatidylinositol 3-kinase activity’ and
‘phosphatidylinositol-4,5-bisphosphate-3-kinase activity’ (Fig. 2B).
Actionable mutations with available
targeted therapies
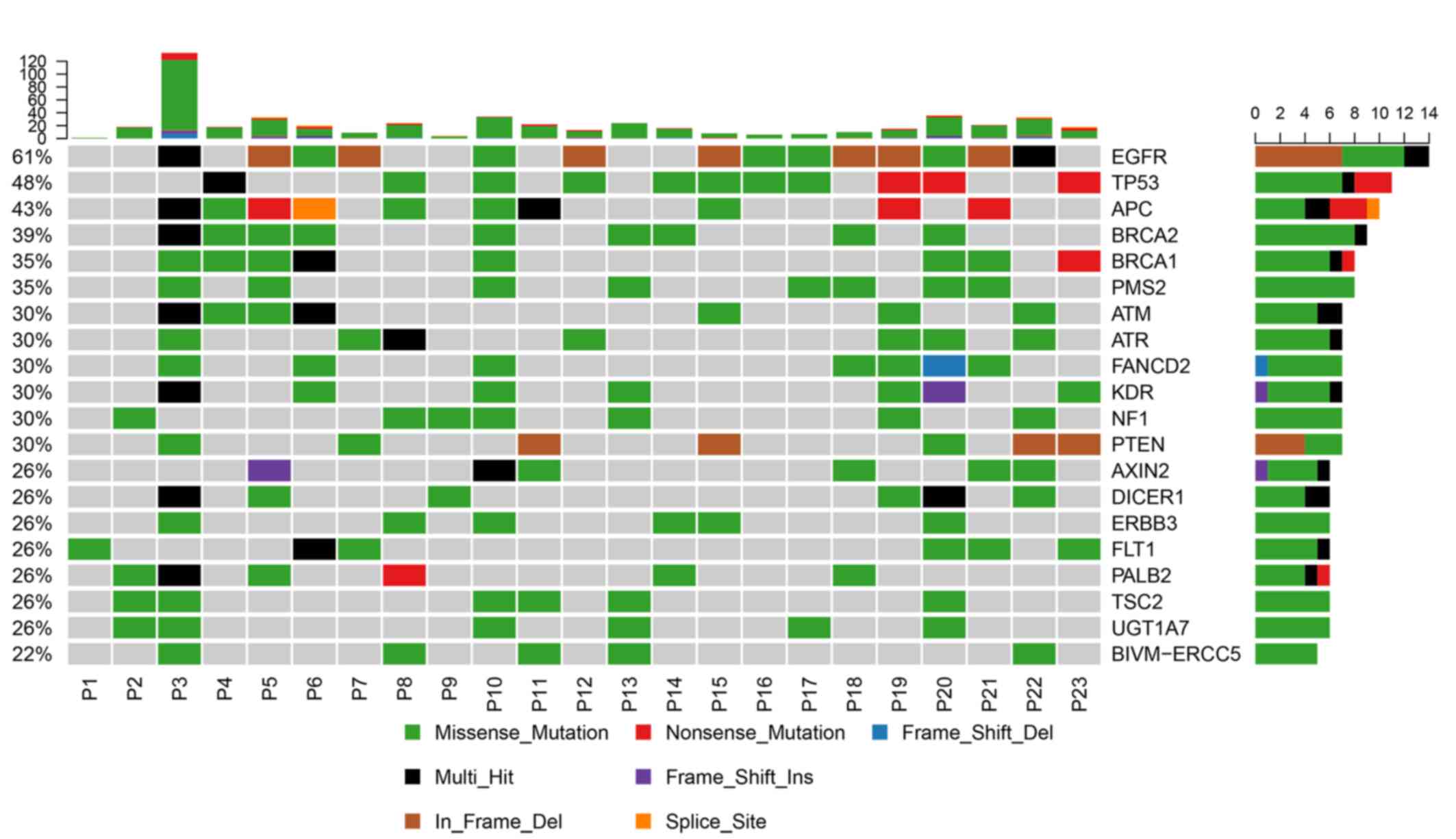
The top 20 mutated genes in the 23 tumor samples
were identified by counting the frequency of mutations in different
genes in the samples (Fig. 3). EGFR,
TP53, and APC were the most conmon mutated gene with a frequency of
61, 48 and 43% respectively. Among the 23 NSCLC samples, 69.57%
(16/23) of the patients carried the mutations with available
matched targeted therapeutic options (Table III). This proportion is consistent
with the previous study (23). In
this study, EGFR mutations which occurred in exon 18–21 were the
most common mutations carried by 14 patients. A total of 13
patients harbored the EGFR sensitizing mutations, which were
recommended for first-line treatment with gefitinib, erlotinib,
afatinib, dacomitinib, Icotinib or osimertinib according to the
NCCN, FDA or China Food and Drug Administration (CFDA) guidelines
(24–26). One of these 13 patients harbored the
acquired resistance mutation EGFR p.T790M, and was amenable to
third-generation EGFR inhibitors, including osimertinib, for
first-line or subsequent treatment (27). Patient 21 carried EGFR p.746_750del
and PIK3CA p.R108H mutations simultaneously (Table III). According to previous studies,
approximately 4–5% of EGFR-mutated lung cancer types with acquired
resistance to EGFR-TKI therapy harbor PIK3CA mutations (28–30). A
total of 2 patients harbored a KRAS p.G12C mutation, which is a
common variant in NSCLC tumors (31). CDKN2A p.W110X was detected
simultaneously with EGFR p.E746_A750del in patient 12 (Table III).
 | Table III.Actionable genomic alterations and
implications for targeted therapeutics. |
Table III.
Actionable genomic alterations and
implications for targeted therapeutics.
| Patient ID | Genomic
alterations | Allele frequency in
current study | Associated
drugs | Implication for
targeted therapeutics |
|---|
| P3 | EGFR
p.747_753del | 0.249 | Gefitinib,
Erlotinib, Afatinib, Icotinib, Osimertinib | EGFR-TKI
sensitive |
| P5 | EGFR
p.746_750del | 0.205 | Gefitinib,
Erlotinib, Afatinib, Icotinib, Osimertinib | EGFR-TKI
sensitive |
| P6 | EGFR p.L858R | 0.382 | Gefitinib,
Erlotinib, Afatinib, Icotinib, Osimertinib | EGFR-TKI
sensitive |
| P7 | EGFR
p.745_750del | 0.378 | Gefitinib,
Erlotinib, Afatinib, Icotinib, Osimertinib | EGFR-TKI
sensitive |
| P8 | KRAS p.G12C | 0.156 | Trametinib,
Everolimus, Temsirolimus | Possibly
sensitive |
|
|
|
| Gefitinib,
Erlotinib, Afatinib, Icotinib | Decreased EGFR-TKI
sensitivity |
| P10 | EGFR p.L861Q | 0.206 | Gefitinib,
Erlotinib, Afatinib, Icotinib, Osimertinib | EGFR-TKI
sensitive |
|
| EGFR p.G719C | 0.252 | Gefitinib,
Erlotinib, Afatinib, Icotinib, Osimertinib | EGFR-TKI
sensitive |
| P12 | CDKN2A p.W110X | 0.246 | Palbociclib | Possibly
sensitive |
|
| EGFR
p.E746_A750del | 0.102 | Gefitinib,
Erlotinib, Afatinib, Icotinib, Osimertinib | EGFR-TKI
sensitive |
| P15 | EGFR
p.746_750del | 0.248 | Gefitinib,
Erlotinib, Afatinib, Icotinib, Osimertinib | EGFR-TKI
sensitive |
| P16 | EGFR p.L858R | 0.297 | Gefitinib,
Erlotinib, Afatinib, Icotinib, Osimertinib | EGFR-TKI
sensitive |
| P17 | EGFR p.L858R | 0.287 | Gefitinib,
Erlotinib, Afatinib, Icotinib, Osimertinib | EGFR-TKI
sensitive |
| P18 | EGFR
p.747_752del | 0.14 | Gefitinib,
Erlotinib, Afatinib, Icotinib, Osimertinib | EGFR-TKI
sensitive |
| P19 | EGFR
p.E746_A750del | 0.367 | Gefitinib,
Erlotinib, Afatinib, Icotinib, Osimertinib | EGFR-TKI
sensitive |
| P20 | EGFR p.L858R | 0.139 | Gefitinib,
Erlotinib, Afatinib, Icotinib, Osimertinib | EGFR-TKI
sensitive |
| P21 | EGFR
p.746_750del | 0.367 | Gefitinib,
Erlotinib, Afatinib, Icotinib, Osimertinib | Decreased EGFR-TKI
sensitivity |
|
| PIK3CA p.R108H | 0.061 | Everolimus,
Temsirolimus | Possibly
sensitive |
| P22 | EGFR
p.L747_T751del | 0.157 | Gefitinib,
Erlotinib, Afatinib, Icotinib, Osimertinib | EGFR-TKI
sensitive |
|
| EGFR p.T790M | 0.208 | Osimertinib | EGFR-TKI
sensitive |
| P23 | KRAS p.G12C | 0.464 | Trametinib,
Everolimus, Temsirolimus | Possibly
sensitive |
|
|
|
| Gefitinib,
Erlotinib, Afatinib, Icotinib | Decreased EGFR-TKI
sensitivity |
Germline mutations with clinical
implications
A number of cancer types manifest as a result of
inherited defects in key genes within the genome. These genes are a
subclass among the cancer driver genes. Their function may involve
tumor suppressor genes, oncogenes, genes involved in DNA repair,
cell cycle control, apoptosis and angiogenesis factors (32). In the present study, germline
mutations were identified through sequencing of the WBC DNA from
patients and validated with paired tumor sample sequencing for 16
patients. The cancerous germline mutations identified in the 15
patients are presented in Table IV.
A total of 15/16 patients (93.8%) exhibited cancer driver gene
mutations. A total of 57.1% of the mutations were previously
reported in the ExAC database and were described as ‘Uncertain
significance or not provided’ in the NCBI ClinVar database, whereas
42.9% were identified as de novo mutations.
 | Table IV.Germline mutations in cancer
susceptibility genes in 15 patients. |
Table IV.
Germline mutations in cancer
susceptibility genes in 15 patients.
| Patient ID | Gene | Amino acid
change | InterPro
domain | Reference single
nucleotide polymorphism ID number | Allele frequency in
the Exome Aggregation Consortium | Allele frequency in
current study |
|---|
| P1 | FANCD2 | p.D662V | – | – | – | 0.37 |
|
| SLX4 | p.D1425N | – | rs766448056 |
9.05×10−06 | 0.58 |
| P11 | POLE | p.F2258L | – | – | – | 0.51 |
|
| TSC2 | p.P1158S | – | – | – | 0.47 |
| P14 | FANCM | p.H1103P | – | rs191339110 |
8.28×10−06 | 0.39 |
| P15 | PTCH1 | p.T265S | – | rs201174718 | 0.0001 | 0.51 |
|
| WRN | p.K577T | DEAD/DEAH box
helicase domain; Helicase superfamily 1/2 ATP-binding domain;
P-loop containing nucleoside triphosphate hydrolase | – | – | 0.49 |
| P16 | EXT1 | p.V356L | – | – | – | 0.47 |
| P19 | ATM | p.A1315V | Armadillo-type
fold | – | – | 0.55 |
|
| AXIN2 | p.V619A | – | – | – | 0.45 |
|
| FANCA | p.D694N | – | rs201589909 | 0.0001 | 0.40 |
| P22 | FANCG | p.P590A | – | rs541868979 |
9.07×10−05 | 0.33 |
|
| PMS1 | p.R202K | Histidine kinase
like ATPase C-terminal domain; Ribosomal protein S5 domain 2-type
fold; Ribosomal protein S5 domain 2-type fold subgroup | rs2066459 | 0.012 | 0.58 |
| P23 | MSH3 | p.R574Q | DNA mismatch repair
protein MutS core | rs776668872 |
8.72×10−06 | 0.46 |
| P3 | CLCN7 | p.N104K | Chloride channel
core | – | – | 0.58 |
|
| MLH3 | p.V741F | – | rs28756990 | 0.0145 | 0.44 |
|
| PTEN | c.-326_-324del | – | – | – | 0.18 |
|
| SPTA1 | p.N2057S | – | rs761106571 |
5.80×10−05 | 0.46 |
| P4 | FANCM | p.I552T | Helicase
C-terminal; P-loop containing nucleoside triphos phate
hydrolase | rs200240871 |
8.25×10−06 | 0.58 |
| P5 | BLM | p.R1139Q | RQC domain; Winged
helix turn helix DNA binding domain | rs771776126 |
8.24×10−06 | 0.5 |
| P6 | STK11 | p.P411L | – | – | – | 0.65 |
| P7 | APC | p.A1753P | – | rs587781350 |
1.66×10−05 | 0.49 |
|
| ATM | p.D841Y | – | – | – | 0.41 |
|
| FANCA | p.T1161K | – | rs142833057 |
3.30×10−05 | 0.55 |
|
| SPTA1 | p.D607H | – | rs534906145 |
8.28×10−06 | 0.58 |
| P8 | POLD1 | p.R322H | DNA-directed DNA
poly merase family B exonuclease domain; Ribonuclease H like
domain | – | – | 0.39 |
| P9 | CLCN7 | p.R775H | CBS domain | rs534953229 | 0.0002 | 0.33 |
|
| SLC2A2 | p.G519E | – | rs147959014 | 0.0018 | 0.39 |
Discussion
Several single-institution studies revealed that
targeted NGS permits the detection of actionable mutations in
clinical lung cancer tumor samples, enabling patient selection for
genotype-based therapies (10–12). The
present study demonstrated the clinical utility of FD-180 profiling
in NSCLC. A total of 69.6% (16/23) NSCLC samples in the present
study were identified to harbor mutations with available matched
targeted therapeutic options. Notably, 13 patients harbored
actionable EGFR mutations, with treatment recommended. However, 3
patients may benefit from drug agents, which have not yet been
approved for lung cancer. The available data indicating PIK3CA
mutations that predict the response of solid tumors to
phosphoinositide 3-kinase (PI3K) signaling pathway inhibitors,
including everolimus (33) and
temsirolimus (34,35), remain controversial. Multiple PI3K
inhibitors, including BYL719, buparlisib (BKM120), taselisib
(GDC0032), and GSK2636771, are under investigation in patients with
PIK3CA-mutated or PTEN-mutated solid tumors (36).
KRAS p.G12C has been negatively associated with
progression-free survival (37) and
decreased sensitivity of EGFR-TKIs, including erlotinib and
gefitinib (38,39). Currently, the role KRAS mutations
serve in the selection of anticancer treatment remains unclear;
however, clinical evidence suggests that KRAS-mutant tumors,
specifically in lung adenocarcinoma, respond positively to
selumetinib and trametinib (40). A
phase II randomized trial indicated that a selumetinib and
docetaxel combined therapy increased overall response rates and
progression free survival in patients with KRAS-mutant NSCLC
compared with the placebo control group (41). A mouse model indicated that
serine/threonine kinase 11-deficient KRAS mutant NSCLC tumors were
resistant to mitogen-activated protein kinase (MEK) inhibition,
which may be clinically relevant (42,43). In
a phase I dose escalation trial with the MEK inhibitor trametinib,
reductions in tumor size of 6–52% were observed in 8/22 patients
with KRAS mutation-positive tumors (44). However, the aforementioned studies
require further validation with larger sample sizes and inclusion
of KRAS wild-type tumors for comparison to enhance the evidence for
clinical efficacy. Therefore, the use of MEK inhibitors for
treating KRAS mutation-positive tumors is only recommended in a
clinical trial setting.
Somatic mutations of CDKN2A are present in various
tumor types (45–47). However, currently there are no
existing standard therapy options to treat tumors harboring somatic
CDKN2A alterations. The loss of function mutation of CDKN2A leads
to activation of CDK2/4/6, suggesting that CDK inhibitors may be
beneficial for these patients (48).
Palbociclib is a highly selective CDK4/6 inhibitor, which has
demonstrated sensitivity in in vitro studies of renal cell
carcinoma (49), melanoma (50), ovarian cancer (51) and breast cancer (52) cell lines, suggesting that this drug
may be effective for the treatment of patients included in the
present study.
Even though actionable genomic alterations are
primarily somatically acquired, it has been reported that inherited
cancer predisposition is predominantly associated with germline
mutations (32). In this study a
total of 93.8% of patients (15/16) carried germline mutations which
have been previously associated with cancer. Among these 28
mutations, known and de novo germline mutations were
identified in 57.1% (16/28) and 42.9% (12/28), respectively. Only a
small proportion of NSCLC cases may be attributed to inheritance of
genetic mutations (53). However,
the results obtained in the present study suggested otherwise,
indicating that the association between the identified mutations
and lung cancer requires further investigation with a larger sample
size.
Notably, the current study does not provide
information on the long-term impact of the FD-180 gene panel on
treatment outcomes, including morbidity, quality of life,
disease-free survival and overall mortality. A limitation of the
present study is therefore the lack of long-term follow-up to
monitor these parameters. In conclusion, the present study
demonstrated that the FD-180 gene panel is a robust diagnostic tool
which may be used to identify targeted therapeutics for patients
with NSCLC.
Acknowledgements
The authors would like to thank Mr. Paul Ma [First
Dimension Biosciences (Suzhou) Co., Ltd.,] for his contributions in
revising this manuscript.
Funding
The present study was partly funded by First
Dimension BioSciences (Suzhou), Co., Ltd.
Availability of data and material
The raw sequencing data are not publicly available
due to information that could compromise the privacy of the
research participants.
Authors' contributions
WC and TT designed the study. HFW collected the
data. CY and HLW analyzed the data. DL designed and wrote the
manuscript. All authors have read and approved the final
manuscript.
Ethics approval and consent to
participate
The Anhui Medical University Ethics Committee
approved the study and all patients have given written informed
consent prior to inclusion in the project.
Patient consent for publication
All patients provided a comprehensive consent form
stating that personal data may be used for academic presentation or
paper presentation, while ensuring complete anonymity prior to
receiving treatment.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Li SD, Ma M, Li H, Waluszko A, Sidorenko
T, Schadt EE, Zhang DY, Chen R and Ye F: Cancer gene profiling in
non-small cell lung cancers reveals activating mutations in JAK2
and JAK3 with therapeutic implications. Genome Med. 9:892017.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Díaz-Serrano A, Gella P, Jiménez E,
Zugazagoitia J and Paz-Ares Rodríguez L: Targeting egfr in lung
cancer: Current standards and developments. Drugs. 78:893–911.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ritter DI, Roychowdhury S, Roy A, Rao S,
Landrum MJ, Sonkin D, Shekar M, Davis CF, Hart RK, Micheel C, et
al: Somatic cancer variant curation and harmonization through
consensus minimum variant level data. Genome Med. 8:1172016.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hensing T, Chawla A, Batra R and Salgia R:
A personalized treatment for lung cancer: Molecular pathways,
targeted therapies, and genomic characterization. Adv Exp Med Biol.
799:85–117. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Vendrell JA, Taviaux S, Béganton B,
Godreuil S, Audran P, Grand D, Clermont E, Serre I, Szablewski V,
Coopman P, et al: Detection of known and novel ALK fusion
transcripts in lung cancer patients using next-generation
sequencing approaches. Sci Rep. 7:125102017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Saarenheimo J, Eigeliene N, Andersen H,
Tiirola M and Jekunen A: The value of liquid biopsies for guiding
therapy decisions in non-small cell lung cancer. Front Oncol.
9:1292019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Cheng Y, Wang S, Han L, Liu P, Li H, Ren
X, Yu J and Hao X: Concurrent somatic mutations in driver genes
were significantly correlated with lymph node metastasis and
pathological types in solid tumors. Oncotarget. 8:68746–68757.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ke EE and Wu YL: Afatinib in the
first-line treatment of epidermal-growth-factor-receptor
mutation-positive non-small cell lung cancer: A review of the
clinical evidence. Ther Adv Respir Dis. 10:256–264. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang S, Cang S and Liu D: Third-generation
inhibitors targeting EGFR T790M mutation in advanced non-small cell
lung cancer. J Hematol Oncol. 9:342016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang YC, Zhou Q and Wu YL: Efficacy of
crizotinib in first-line treatment of adults with ALK-positive
advanced NSCLC. Expert Opin Pharmacother. 17:1693–1701. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gainor JF and Shaw AT: Novel targets in
non-small cell lung cancer: ROS1 and RET fusions. Oncologist.
18:865–875. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Illei PB, Belchis D, Tseng LH, Nguyen D,
De Marchi F, Haley L, Riel S, Beierl K, Zheng G, Brahmer JR, et al:
Clinical mutational profiling of 1006 lung cancers by next
generation sequencing. Oncotarget. 8:96684–96696. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Allegretti M, Fabi A, Buglioni S, Martayan
A, Conti L, Pescarmona E, Ciliberto G and Giacomini P: Tearing down
the walls: FDA approves next generation sequencing (NGS) assays for
actionable cancer genomic aberrations. J Exp Clin Cancer Res.
37:472018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang YC, Zhou Q and Wu YL: The emerging
roles of NGS-based liquid biopsy in non-small cell lung cancer. J
Hematol Oncol. 10:1672017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Miranda E, Destro A, Malesci A, Balladore
E, Bianchi P, Baryshnikova E, Franchi G, Morenghi E, Laghi L,
Gennari L and Roncalli M: Genetic and epigenetic changes in primary
metastatic and nonmetastatic colorectal cancer. Br J Cancer.
95:1101–1107. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Malapelle U, Mayo de-Las-Casas C, Rocco D,
Garzon M, Pisapia P, Jordana-Ariza N, Russo M, Sgariglia R, De Luca
C, Pepe F, et al: Development of a gene panel for next-generation
sequencing of clinically relevant mutations in cell-free DNA from
cancer patients. Br J Cancer. 116:802–810. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li H and Durbin R: Fast and accurate short
read alignment with Burrows-Wheeler transform. Bioinformatics.
25:1754–1760. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
McKenna A, Hanna M, Banks E, Sivachenko A,
Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly
M and DePristo MA: The genome analysis toolkit: A MapReduce
framework for analyzing next-generation DNA sequencing data. Genome
Res. 20:1297–1303. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Cibulskis K, Lawrence MS, Carter SL,
Sivachenko A, Jaffe D, Sougnez C, Gabriel S, Meyerson M, Lander ES
and Getz G: Sensitive detection of somatic point mutations in
impure and heterogeneous cancer samples. Nat Biotechnol.
31:213–219. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
21
|
Sherry ST, Ward MH, Kholodov M, Baker J,
Phan L, Smigielski EM and Sirotkin K: dbSNP: The NCBI database of
genetic variation. Nucleic Acids Res. 29:308–311. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Tassios PT and Moran-Gilad J: Bacterial
next generation sequencing (NGS) made easy. Clin Microbiol Infect.
24:332–334. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Cabanillas R, Diñeiro M, Castillo D,
Pruneda PC, Penas C, Cifuentes GA, de Vicente Á, Durán NS, Álvarez
R, Ordóñez GR and Cadiñanos J: A novel molecular diagnostics
platform for somatic and germline precision oncology. Mol Genet
Genomic Med. 5:336–359. 2017. View
Article : Google Scholar : PubMed/NCBI
|
|
24
|
Forde PM and Ettinger DS: Managing
acquired resistance in EGFR-mutated non-small cell lung cancer.
Clin Adv Hematol Oncol. 13:528–532. 2015.PubMed/NCBI
|
|
25
|
Zhao D, Chen X, Qin N, Su D, Zhou L, Zhang
Q, Li X, Zhang X, Jin M and Wang J: The prognostic role of
EGFR-TKIs for patients with advanced non-small cell lung cancer.
Sci Rep. 7:403742017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kucharczuk CR, Ganetsky A and Vozniak JM:
Drug-drug interactions, safety, and pharmacokinetics of EGFR
tyrosine kinase inhibitors for the treatment of non-small cell lung
cancer. J Adv Pract Oncol. 9:189–200. 2018.PubMed/NCBI
|
|
27
|
Ramalingam SS, Yang JC, Lee CK, Kurata T,
Kim DW, John T, Nogami N, Ohe Y, Mann H, Rukazenkov Y, et al:
Osimertinib as first-line treatment of EGFR mutation-positive
advanced non-small-cell lung cancer. J Clin Oncol. 36:841–849.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Sequist LV, Waltman BA, Dias-Santagata D,
Digumarthy S, Turke AB, Fidias P, Bergethon K, Shaw AT, Gettinger
S, Cosper AK, et al: Genotypic and histological evolution of lung
cancers acquiring resistance to EGFR inhibitors. Sci Transl Med.
3:75ra262011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ludovini V, Bianconi F, Pistola L, Chiari
R, Minotti V, Colella R, Giuffrida D, Tofanetti FR, Siggillino A,
Flacco A, et al: Phosphoinositide-3-kinase catalytic alpha and KRAS
mutations are important predictors of resistance to therapy with
epidermal growth factor receptor tyrosine kinase inhibitors in
patients with advanced non-small cell lung cancer. J Thorac Oncol.
6:707–715. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Li S, Li L, Zhu Y, Huang C, Qin Y, Liu H,
Ren-Heidenreich L, Shi B, Ren H, Chu X, et al: Coexistence of EGFR
with KRAS, or BRAF, or PIK3CA somatic mutations in lung cancer: a
comprehensive mutation profiling from 5125 Chinese cohorts. Br J
Cancer. 110:2812–2820. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Brose MS, Volpe P, Feldman M, Kumar M,
Rishi I, Gerrero R, Einhorn E, Herlyn M, Minna J, Nicholson A, et
al: BRAF and RAS mutations in human lung cancer and melanoma.
Cancer Res. 62:6997–7000. 2002.PubMed/NCBI
|
|
32
|
Cheng DT, Prasad M, Chekaluk Y, Benayed R,
Sadowska J, Zehir A, Syed A, Wang YE, Somar J, Li Y, et al:
Comprehensive detection of germline variants by MSK-IMPACT, a
clinical diagnostic platform for solid tumor molecular oncology and
concurrent cancer predisposition testing. BMC Med Genomics.
10:332017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wheler JJ, Moulder SL, Naing A, Janku F,
Piha-Paul SA, Falchook GS, Zinner R, Tsimberidou AM, Fu S, Hong DS,
et al: Anastrozole and everolimus in advanced gynecologic and
breast malignancies: Activity and molecular alterations in the
PI3K/AKT/mTOR pathway. Oncotarget. 5:3029–3038. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Moroney J, Fu S, Moulder S, Falchook G,
Helgason T, Levenback C, Hong D, Naing A, Wheler J and Kurzrock R:
Phase I study of the antiangiogenic antibody bevacizumab and the
mTOR/hypoxia-inducible factor inhibitor temsirolimus combined with
liposomal doxorubicin: Tolerance and biological activity. Clin
Cancer Res. 18:5796–5805. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Mackay HJ, Eisenhauer EA, Kamel-Reid S,
Tsao M, Clarke B, Karakasis K, Werner HM, Trovik J, Akslen LA,
Salvesen HB, et al: Molecular determinants of outcome with
mammalian target of rapamycin inhibition in endometrial cancer.
Cancer. 120:603–610. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Lovly C, Horn L and Pao W: PIK3CA
c.1633G>A (E545K) mutation in non-small cell lung cancer. my
cancer genome. https://www.mycancergenome.org/content/disease/lung-cancer/pik3ca/8/(Updated
January 13). 2017
|
|
37
|
Fiala O, Pesek M, Finek J, Benesova L,
Belsanova B and Minarik M: The dominant role of G12C over other
KRAS mutation types in the negative prediction of efficacy of
epidermal growth factor receptor tyrosine kinase inhibitors in
non-small cell lung cancer. Cancer Genet. 206:26–31. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Riely GJ and Ladanyi M: KRAS mutations: An
old oncogene becomes a new predictive biomarker. J Mol Diagn.
10:493–495. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Riely GJ, Marks J and Pao W: KRAS
mutations in non-small cell lung cancer. Proc Am Thorac Soc.
6:201–205. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Bhattacharya S, Socinski MA and Burns TF:
KRAS mutant lung cancer: Progress thus far on an elusive
therapeutic target. Clin Transl Med. 4:352015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Jänne PA, Shaw AT, Pereira JR, Jeannin G,
Vansteenkiste J, Barrios C, Franke FA, Grinsted L, Zazulina V,
Smith P, et al: Selumetinib plus docetaxel for KRAS-mutant advanced
non-small-cell lung cancer: A randomised, multicentre,
placebo-controlled, phase 2 study. Lancet Oncol. 14:38–47. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Carretero J, Shimamura T, Rikova K,
Jackson AL, Wilkerson MD, Borgman CL, Buttarazzi MS, Sanofsky BA,
McNamara KL, Brandstetter KA, et al: Integrative genomic and
proteomic analyses identify targets for Lkb1-deficient metastatic
lung tumors. Cancer Cell. 17:547–559. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Chen Z, Cheng K, Walton Z, Wang Y, Ebi H,
Shimamura T, Liu Y, Tupper T, Ouyang J, Li J, et al: A murine lung
cancer co-clinical trial identifies genetic modifiers of
therapeutic response. Nature. 483:613–617. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Infante JR, Fecher LA, Falchook GS,
Nallapareddy S, Gordon MS, Becerra C, DeMarini DJ, Cox DS, Xu Y,
Morris SR, et al: Safety, pharmacokinetic, pharmacodynamic, and
efficacy data for the oral MEK inhibitor trametinib: a phase 1
dose-escalation trial. Lancet Oncol. 13:773–781. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Xing X, Cai W, Shi H, Wang Y, Li M, Jiao J
and Chen M: The prognostic value of CDKN2A hypermethylation in
colorectal cancer: A meta-analysis. Br J Cancer. 108:2542–2548.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Padhi SS, Roy S, Kar M, Saha A, Roy S,
Adhya A, Baisakh M and Banerjee B: Role of CDKN2A/p16 expression in
the prognostication of oral squamous cell carcinoma. Oral Oncol.
73:27–35. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Kim N, Song M, Kim S, Seo Y, Kim Y and
Yoon S: Differential regulation and synthetic lethality of
exclusive RB1 and CDKN2A mutations in lung cancer. Int J Oncol.
48:367–375. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Gopalan PK, Pinder MC, Chiappori A, Ivey
AM, Villegas AG and Kaye FJ: A phase II clinical trial of the
CDK4/6 inhibitor palbociclib (PD0332991) in previously treated,
advanced non-small cell lung cancer (NSCLC) patients with
inactivated CDKN2A. J Clin Oncol. 32:80772014. View Article : Google Scholar
|
|
49
|
Logan JE, Mostofizadeh N, Desai AJ, VON
Euw E, Conklin D, Konkankit V, Hamidi H, Eckardt M, Anderson L,
Chen HW, et al: PD-0332991, a potent and selective inhibitor of
cyclin-dependent kinase 4/6, demonstrates inhibition of
proliferation in renal cell carcinoma at nanomolar concentrations
and molecular markers predict for sensitivity. Anticancer Res.
33:2997–3004. 2013.PubMed/NCBI
|
|
50
|
Young RJ, Waldeck K, Martin C, Foo JH,
Cameron DP, Kirby L, Do H, Mitchell C, Cullinane C, Liu W, et al:
Loss of CDKN2A expression is a frequent event in primary invasive
melanoma and correlates with sensitivity to the CDK4/6 inhibitor
PD0332991 in melanoma cell lines. Pigment Cell Melanoma Res.
27:590–600. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Konecny GE, Winterhoff B, Kolarova T, Qi
J, Manivong K, Dering J, Yang G, Chalukya M, Wang HJ, Anderson L,
et al: Expression of p16 and retinoblastoma determines response to
CDK4/6 inhibition in ovarian cancer. Clin Cancer Res. 17:1591–1602.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Finn RS, Dering J, Conklin D, Kalous O,
Cohen DJ, Desai AJ, Ginther C, Atefi M, Chen I, Fowst C, et al: PD
0332991, a selective cyclin D kinase 4/6 inhibitor, preferentially
inhibits proliferation of luminal estrogen receptor-positive human
breast cancer cell lines in vitro. Breast Cancer Res. 11:R772009.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Kanwal M, Ding XJ and Cao Y: Familial risk
for lung cancer. Oncol Lett. 13:535–542. 2017. View Article : Google Scholar : PubMed/NCBI
|