Introduction
Pancreatic cancer is one of the most common types of
cancer worldwide and the fourth leading cause of cancer-associated
mortality in developed countries (1,2).
Pancreatic cancer is a malignant disease with a median survival
time of 3–6 months and a 5-year survival rate of less than 5%
(3,4). Chemotherapy is the primary adjuvant
therapy for pancreatic cancer (5).
Dasatinib is an oral inhibitor of dual Bcr/Abl and Src family
kinases (6) commonly used in
hematopoietic tumors (7). Src kinase
was one of the earliest discovered proto-oncoproteins in humans,
which exhibits high activity in a number of human tumors and is
involved in the process of malignant transformation of cells
(8). The Src-mediated tumor cell
signal transduction network serves a crucial role in tumorigenic
processes, such as cell growth (9).
Activated Src kinase promotes cell proliferation through the
Ras/Raf/mitogen-activated protein kinase (MAPK) pathway (10). Dasatinib is also an Src inhibitor
approved by the Food and Drug Administration (FDA) for the
treatment of pancreatic cancer (11). A previous study reported that
Dasatinib could treat epithelial neoplasms, including pancreatic
cancer (12). In addition, dasatinib
can slow down cancer metastasis and the progression of human
pancreatic ductal adenocarcinoma (PDAC) in orthotopic mouse models,
and may be able to stimulate PDAC cell apoptosis in humans and mice
(13–15). Bartscht et al (16) reported that dasatinib inhibits the
function of Src kinases and transforming growth factor β 1
(TGF-β1) in clinical and experimental therapeutics to prevent the
metastatic spread of late-stage PDAC. Dasatinib is a highly
promising treatment of pancreatic cancer; however, most patients
who have a good response to inhibitors typically experience disease
recurrence due to drug-resistance development, which becomes a
severe clinical problem (11).
The mechanism of acquired dasatinib resistance is
unclear. Previous studies reported that SRC/TGF-β alteration and
multiple signals, such as the MAPK signaling pathways, may be
associated with the progression of drug resistance (17,18).
Beauchamp et al (19)
revealed that acquired dasatinib resistance may be related to a
discoidin domain receptor tyrosine kinase 2 gatekeeper mutation and
the loss of neurofibromatosis type 1. These studies demonstrated
that multiple genes participate in the development of
dasatinib-resistance, and that alterations in multiple genes are
often associated with cells resistance to dasatinib. Therefore, it
is not advisable to study the mechanism of drug resistance through
single gene changes or pathways.
Since the precise molecular mechanisms underlying
dasatinib resistance remain unknown, studies on novel treatments
are still in the early stages and their outcomes are not optimal;
the majority of studies focus on specific molecular targets or
genes, ignoring the possibility that dasatinib resistance may be
due to the abnormal expression of multiple genes (20). Traditional treatment methods, which
only consider one gene, may be unable to combat drug resistance
(21). It is therefore crucial to
investigate the resistance-associated gene variations using novel
methods, including genome-wide technologies, which may provide new
understanding of dasatinib resistance and allow the development of
novel treatment strategy.
Microarray is a tool for high-throughput screening,
which is used for the analysis of global gene expression profiles,
particularly for the study of the underlying mechanisms of various
diseases. In the present study, the gene expression profiles of
dasatinib-resistant pancreatic cancer cells were analyzed using
public microarray data to better understand the underlying
mechanisms of dasatinib resistance. Bioinformatics methods were
used to search for differentially expressed genes (DEGs) between
dasatinib-sensitive and dasatinib-resistant pancreatic cancer
cells. The functions of the DEGs were evaluated using gene ontology
(GO) annotation, pathway enrichment and the construction of a
protein-protein interaction (PPI) network. The present study aimed
to understand the mechanisms of drug resistance and to determine
potential tumor therapy targets to prevent dasatinib
resistance.
Materials and methods
DEG identification from public
microarray data
To identify DEGs from acquired dasatinib-resistant
pancreatic cancer cells, the shared gene expression profile
(GSE59357) was obtained from the Gene Expression Omnibus database
(https://www.ncbi.nlm.nih.gov/geo). This
dataset was uploaded by Chien et al (11). The information included the Panc0403,
Panc0504, Panc1005 (dasatinib-sensitive), SU8686, MiaPaCa2 and
Panc1 (dasatinib-resistant) cell lines. The dataset was analyzed
using R software (R version 3.4.1; http://mirrors.tuna.tsinghua.edu.cn/CRAN). Student's
t-test was utilized to screen the dasatinib resistance-associated
DEGs among the cell lines, using a threshold of P<0.05 and a
fold change ≥1.5.
Functional enrichment analysis of
DEGs
The Database for Annotation, Visualization and
Integrated Discovery (DAVID, http://david.ncifcrf.gov) was used to perform the
functional enrichment analysis of the DEGs, including gene ontology
(GO) functional analysis and Kyoto Encyclopedia of Genes and
Genomes (KEGG) pathway analysis. In the GO analysis, the categories
included cellular component (CC), biological process (BP) and
molecular function (MF) terms, and a P-value of <0.01 was
considered to indicate a statistically significant difference. In
the KEGG pathway analysis, enriched pathways were identified based
on their hypergeometric distribution with a P-value of
<0.05.
Construction of a PPI network by
Search Tool for the Retrieval of Interacting Genes (STRING)
The 472 identified DEGs were analyzed by inputting
the identification numbers of the DEGs into the online tool STRING
(https://string-db.org). A combined score of ≥0.7
(high confidence score) was considered significant. The hub
proteins were chosen based on their associations with other
proteins: Proteins coded by DEGs that were associated with a higher
number of other DEGs were considered to serve important roles in
the PPI network.
Analysis of the hub genes and their
association with pancreatic adenocarcinoma prognosis
The Oncolnc website (http://www.oncolnc.org), which uses The Cancer Genome
Atlas database for survival analysis of cancer patients, was used
to analyze the hub genes and their association with the prognosis
of patients with pancreatic cancer.
Results
Identification of the DEGs between
dasatinib-sensitive and acquired dasatinib-resistant pancreatic
cancer cells
The ‘t test’ option in R software was used to
research the gene expression profiles from the GSE59357 public
microarray dataset. It highlighted the DEGs between Panc0403,
Panc0504, Panc1005 (Dasatinib-sensitive), SU8686, MiaPaCa2 and
Panc1 (acquired Dasatinib-resistant) pancreatic cancer cells.
(Fig. 1). A total of 472 DEGs were
identified, which comprised 333 downregulated and 139 upregulated
DEGs. The top 10 upregulated and downregulated DEGs are listed in
Table I.
 | Table I.The top 10 downregulated and
upregulated differentially expressed genes in dasatinib-resistant
cell lines compared with dasatinib-sensitive cell lines. |
Table I.
The top 10 downregulated and
upregulated differentially expressed genes in dasatinib-resistant
cell lines compared with dasatinib-sensitive cell lines.
| A, Upregulated
genes |
|---|
|
|---|
| Probe set | Gene | logFC | P-value |
|---|
| ILMN_2058251 | VIM | 5.050515 |
6.35×10−5 |
| ILMN_2169966 | TM4SF18 | 3.8899119 |
2.27×10−4 |
| ILMN_2169152 | SRGN | 3.7095162 |
1.77×10−3 |
| ILMN_1804735 | CBS | 3.6128934 |
1.23×10−4 |
| ILMN_1757604 | TPM2 | 3.5489746 |
1.07×10−5 |
| ILMN_1680874 | TUBB2B | 3.5267246 |
7.53×10−4 |
| ILMN_1811468 | IRX3 | 3.3427754 |
3.34×10−5 |
| ILMN_1729117 | COL5A2 | 3.3276338 |
2.21×10−3 |
| ILMN_1703178 | SCG2 | 3.2029857 |
3.70×10−3 |
| ILMN_1790338 | PRRX2 | 3.1810426 |
1.29×10−5 |
|
| B, Downregulated
genes |
|
| Probe
set | Gene | logFC | P-value |
|
| ILMN_1712522 | CEACAM6 | −4.1498441 |
2.22×10−3 |
| ILMN_2188862 | GDF15 | −4.202082 |
6.03×10−4 |
| ILMN_1666222 | PHACTR3 | −4.2059744 |
5.99×10−5 |
| ILMN_2133205 | GPX2 | −4.2320599 |
7.36×10−4 |
| ILMN_2353161 | MSLN | −4.2425188 |
3.35×10−4 |
| ILMN_2160210 | EPCAM | −4.2522758 |
7.48×10−4 |
| ILMN_2163723 | KRT7 | −4.3465465 |
1.70×10−4 |
| ILMN_1692223 | LCN2 | −4.3976553 |
1.98×10−3 |
| ILMN_1739001 | TACSTD2 | −5.0377579 |
9.36×10−4 |
| ILMN_1801216 | S100P | −5.2731601 |
9.94×10−4 |
The functional annotation and pathway
enrichment of DEGs
To investigate the biological functions of the
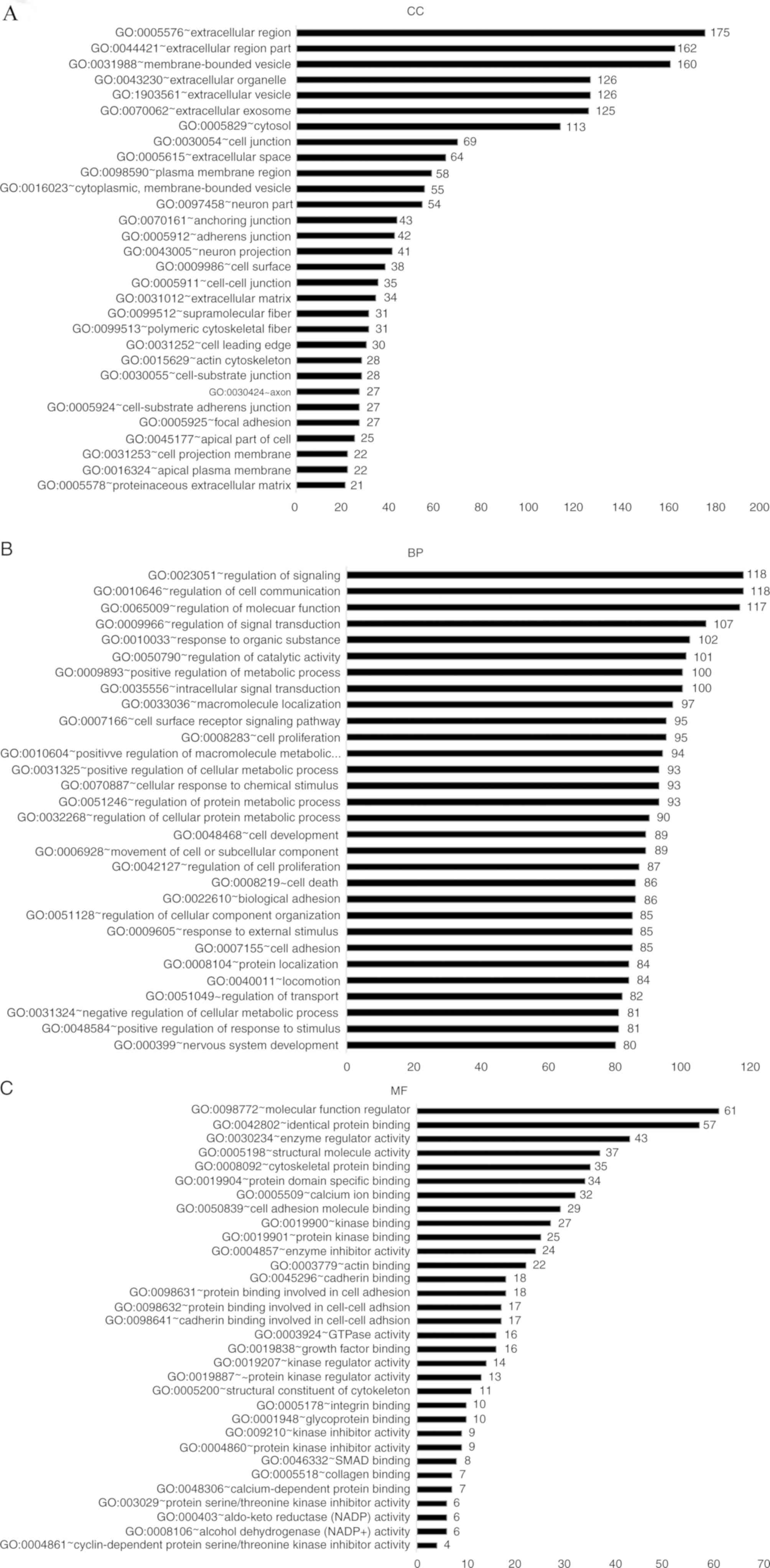
identified DEGs, GO analysis was used in DAVID to cluster the data.
The enriched GO terms were separated into CC, BP and MF ontology
categories (Fig. 2).
In the CC ontology (Fig.
2A), most of the enriched categories were associated with cell
membrane components, including ‘extracellular region’ (175 genes),
‘membrane-bounded vesicles’ (160 genes) and ‘extracellular
vesicles’ (126 genes). In addition, the other highly enriched terms
included ‘extracellular organelles’ (126 genes), ‘the extracellular
region’ (162 genes) and ‘extracellular exosomes’ (125 genes). In
the BP ontology (Fig. 2B), the
highest number of DEGs were enriched in regulation-associated
terms, including ‘signal regulation’ (118 genes), ‘cell
communication regulation’ (118 genes), ‘molecular function
regulation’ (117 genes) and ‘signal transduction regulation’ (107
genes). A number of enriched categories were associated with
transduction, apoptosis and localization, such as ‘cell
proliferation’ (95 genes), ‘intracellular signal transduction’ (100
genes), ‘apoptotic processes’ (73 genes) and ‘macromolecule
localization’ (97 genes). In the MF ontology (Fig. 2C), regulator-associated and
binding-associated components accounted for the majority of the
enriched GO categories, including ‘molecular function regulation’
(61 genes), ‘identical protein binding’ (57 genes), ‘enzyme
regulator activity’ (43 genes) and ‘cytoskeletal protein binding’
(35 genes).
Furthermore, the KEGG pathway analysis identified
dysfunctional enriched pathways, including ‘pathways in cancer’ (26
genes), ‘phosphoinositide 3-kinase (PI3K)-protein kinase B (Akt)
signaling pathway’ (20 genes), ‘MAPK signaling pathway’ (16 genes),
‘transcriptional misregulation in cancer’ (13 genes) and ‘p53
signaling pathway’ (6 genes; Table
II).
 | Table II.Enriched Kyoto Encyclopedia of Genes
and Genomes pathway analysis of the differentially expressed
genes. |
Table II.
Enriched Kyoto Encyclopedia of Genes
and Genomes pathway analysis of the differentially expressed
genes.
| Term | Count | P-value |
|---|
| hsa05200: Pathways
in cancer | 26 |
1.31×10−4 |
| hsa04151: PI3K-Akt
signaling pathway | 20 |
4.54×10−3 |
| hsa04010: MAPK
signaling pathway | 16 |
6.31×10−3 |
| hsa04510: Focal
adhesion | 15 |
2.28×10−3 |
| hsa05202:
Transcriptional misregulation in cancer | 13 |
3.10×10−3 |
| hsa05205:
Proteoglycans in cancer | 12 |
2.83×10−2 |
| hsa04512:
ECM-receptor interaction | 11 |
1.74×10−4 |
| hsa04530: Tight
junction | 10 |
1.66×10−2 |
| hsa04514: Cell
adhesion molecules (CAMs) | 10 |
2.05×10−2 |
| hsa04670: Leukocyte
transendothelial migration | 9 |
1.97×10−2 |
| hsa04550: Signaling
pathways regulating pluripotency of stem cells | 9 |
4.74×10−2 |
| hsa05146:
Amoebiasis | 8 |
3.20×10−2 |
| hsa05212:
Pancreatic cancer | 7 |
1.04×10−2 |
| hsa04610:
Complement and coagulation cascades | 7 |
1.38×10−2 |
| hsa05222: Small
cell lung cancer | 7 |
3.46×10−2 |
| hsa04540: Gap
junction | 7 |
4.00×10−2 |
| hsa05219: Bladder
cancer | 6 |
5.95×10−3 |
| hsa05130:
Pathogenic Escherichia coli infection | 6 |
1.48×10−2 |
| hsa04115: p53
signaling pathway | 6 |
4.24×10−2 |
| hsa00982: Drug
metabolism-cytochrome P450 | 6 |
4.47×10−2 |
| hsa04978: Mineral
absorption | 5 |
4.19×10−2 |
Construction of a PPI network
STRING, which is a database used for the prediction
of protein association, was used to predict the protein
interactions among the identified DEGs. Firstly, the 472 DEGs were
entered in the STRING website to obtain their PPI data. Then, if
the combined score was ≥0.7, the PPIs were chosen to construct a
PPI network. In the PPI network, eight node proteins, including
arrestin β1 (ARRB1), synaptotagmin 1 (SYT1), collagen type I α2
chain (COL1A2), collagen type VI α1 chain (COL6A1), integrin
subunit β4 (ITGB4), integrin subunit α2 (ITGA2), laminin subunit α3
(LAMA3) and Rac family small GTPase 1 (RAC1) had a high association
with other node proteins (>7), which demonstrated their
increased hub degrees (Table III;
Fig. 3). The hub genes may therefore
serve a crucial role in dasatinib resistance within pancreatic
cancer.
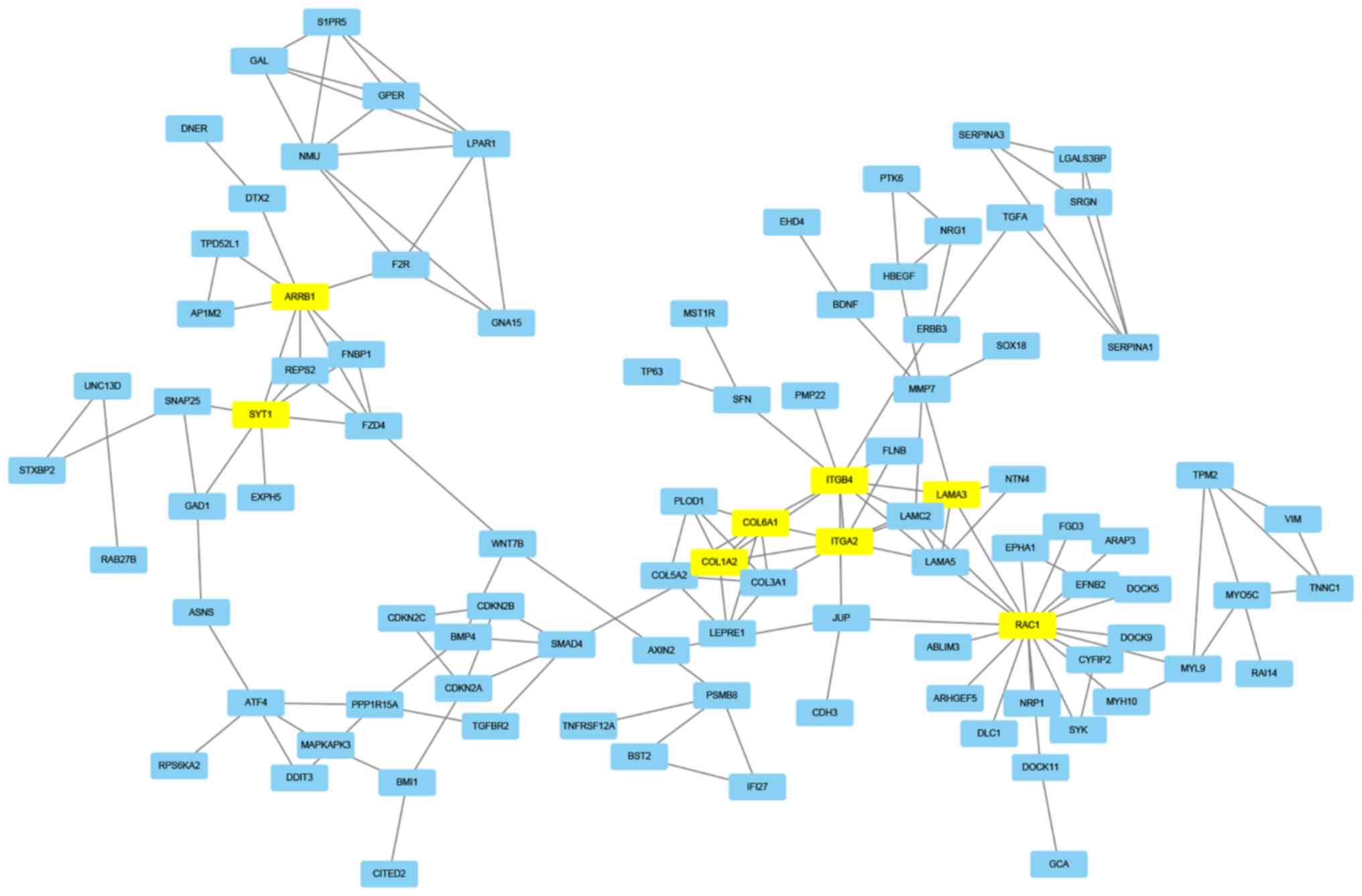 | Figure 3.Protein-protein interaction network.
The interaction network chart of the eight node proteins containing
ARRB1, SYT1, COL1A2, COL6A1, ITGB4, ITGA2, LAMA3 and RAC1, which
were associated with other node proteins (>7 proteins). ARBB1,
arrestin β1; COL1A2, collagen type I α 2 chain; COL6A1, collagen
type VI α1 chain; ITGA2, integrin subunit α2; ITGB4, integrin
subunit β 4; LAMA3, laminin subunit α 3; RAC1, Rac family small
GTPase 1; SYT1, synaptotagmin 1. |
 | Table III.The eight key hub genes and its
associated genes. |
Table III.
The eight key hub genes and its
associated genes.
| Hub gene | Associated
genes |
|---|
| ARRB1 | FZD4, F2R, DTX2,
SYT1, AP1M2, TPD52L1, REPS2, FNBP1 |
| SYT1 | SNAP25, GAD1,
ARRB1, FZD4, REPS2, FNBP1, EXPH5 |
| COL1A2 | COL3A1, COL5A2,
LEPRE1, COL6A1, SMAD4, ITGA2, PLOD1, ITGB4 |
| COL6A1 | COL3A1, COL1A2,
COL5A2, PLOD1, ITGA2, LEPRE1, ITGB4 |
| ITGA2 | COL3A1, COL1A2,
LAMC2, LAMA3, COL6A1, ITGB4, FLNB, LAMA5 |
| ITGB4 | LAMC2, FLNB,
LAMA3, LAMA5, ERBB3, SFN, PMP22, ITGA2, JUP, COL1A2,
COL6A1 |
| LAMA3 | LAMC2, ITGB4,
NTN4, ITGA2, MMP7, LAMA5, RAC1 |
| RAC1 | MYH10, CYFIP2,
DOCK5, JUP, MYL9, SYK, EPHA1, ARHGEF5, ARAP3, DOCK9, DOCK11,
ABLIM3, EFNB2, DLC1, NRP1, FGD3, LAMA3, LAMC2, LAMA5 |
Association between hub genes and
pancreatic adenocarcinoma prognosis
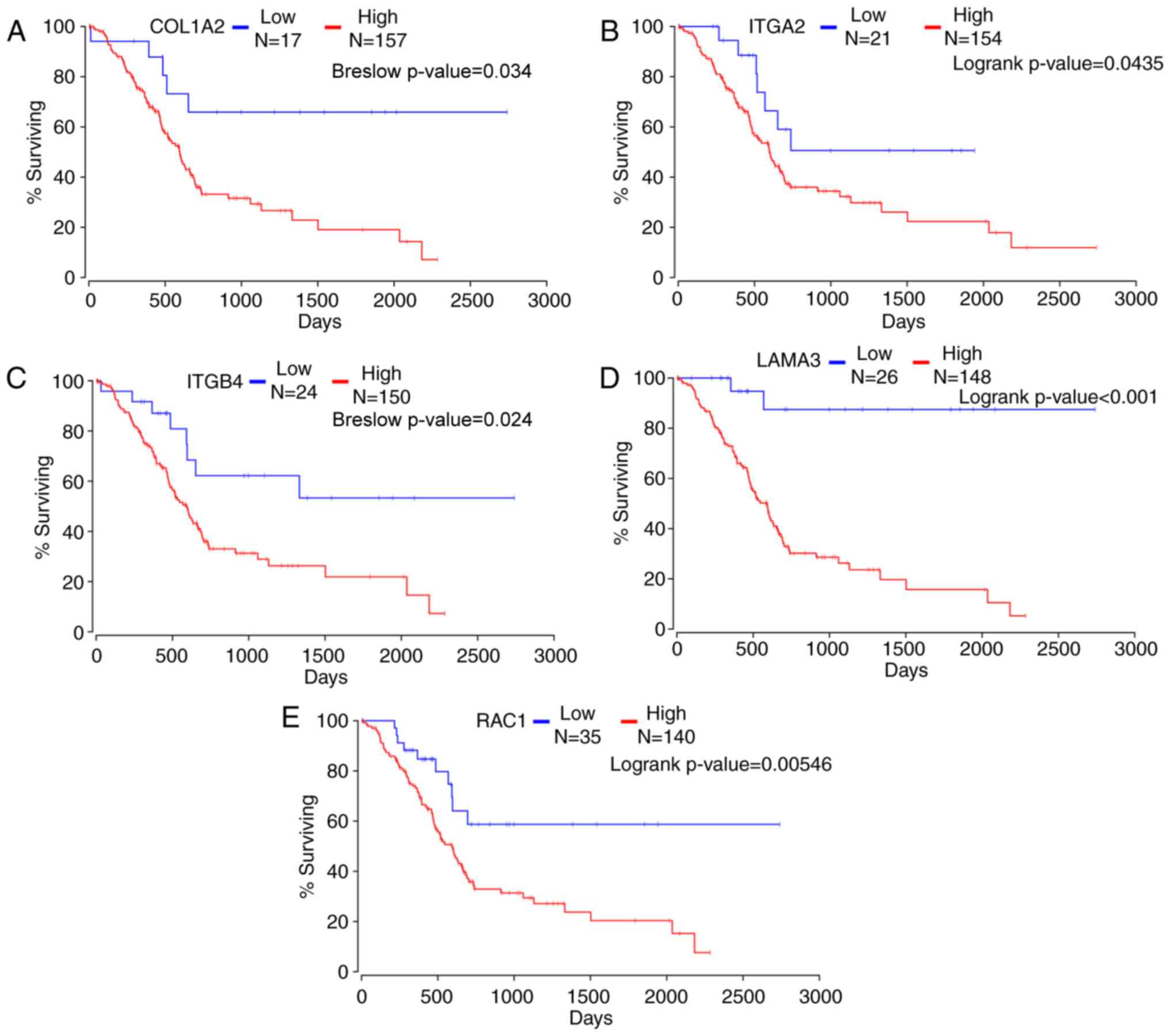
The Oncolnc website was used to analyze the hub
genes and five hub genes were found to have a strong association
with pancreatic adenocarcinoma prognosis in 174 patients (Fig. 4). Kaplan-Meier survival analysis
demonstrated that high expression levels of the hub genes COL1A2,
ITGB4, ITGA2, LAMA3 and RAC1 were associated with a poorer overall
survival in 174 patients with pancreatic adenocarcinoma compared
with patients with low expression levels of each gene (Breslow
P=3.40×10−2, 2.40×10−2, logrank
P=3.09×10−2, 1.27×10−5 and
9.7×10−3, respectively). This suggested that the
selected genes may contribute to dasatinib resistance in pancreatic
cancer.
Discussion
In pancreatic cancer, vascular endothelial growth
factor receptor (VEGFR), epidermal growth factor receptor (EGFR),
insulin-like growth factor 1 receptor (IGF1R) or matrix
metalloproteinases (MMPs), such as MMP-9, are commonly
overexpressed, which allows treatment of pancreatic cancer using
certain small molecule inhibitors against these proteins (22,23).
Dasatinib is a potent receptor tyrosine kinase/Src inhibitor, which
reduces pancreatic tumor cell growth in vitro (11). Dasatinib is also FDA-approved for the
treatment of pancreatic tumors (11). Previous studies have reported that
increased activation of Src kinases, which are molecular targets of
dasatinib, is associated with poor survival in patients with
pancreatic cancer (15). In these
patients, co-treatment with dasatinib and gemcitabine may
contribute to disease stability (24). However, drug resistance frequently
develops rapidly. Dasatinib resistance is a major challenge for the
treatment of patients with pancreatic cancer (25). It is therefore crucial to determine
the underlying mechanisms of dasatinib resistance in order to
develop appropriate therapeutic strategies against drug resistance.
In the present study, DEGs were identified between
dasatinib-sensitive and dasatinib-resistant pancreatic cancer
cells. GO annotation and pathway enrichment were then performed to
analyze the functions of theses DEGs. The association between these
DEGs was analyzed by constructing a PPI network. In addition,
Kaplan-Meier survival analysis revealed that some DEGs were highly
associated with the prognosis of patients with pancreatic cancer.
Numerous hub genes were confirmed to serve a key role in dasatinib
resistance, and may therefore be considered as potent molecular
targets for the treatment of dasatinib-resistance in pancreatic
cancer.
A total of 472 DEGs between dasatinib-resistant and
dasatinib-sensitive pancreatic cancer cells were identified,
including 139 upregulated and 333 downregulated DEGs. Amongst these
DEGs, some had a significant fold change (>3) in
dasatinib-resistant cancer cells compared with dasatinib-sensitive
cells. Vimentin (VIM), transmembrane 4 l six family member 18,
serglycin, keratin 7, lipocalin 2 (LCN2), tumor associated calcium
signal transducer 2 and S100 calcium binding protein P had 5.05,
3.89, 3.71, −4.35, −4.4, −5.04 and −5.27 fold changes,
respectively. Therefore, these DEGs may represent biomarkers of
dasatinib resistance. However, the molecular mechanisms underlying
dasatinib resistance remain unclear and require further
investigation. Previous studies have reported that these DEGs may
contribute to pancreatic cancer in different ways. For example,
VIM, which is an important factor that coordinates epithelial to
mesenchymal transition and is therefore associated with pancreatic
tumor aggressiveness (26), was an
upregulated DEG in dasatinib-resistant cell lines in the present
study. VIM also participates in colonic neoplastic progression and
resistance to histone deacetylase inhibitors (27). Conversely, methylthioadenosine
phosphorylase (MATP), which has been considered as a potential
therapeutic target for cancer cells with a 9p21 deletion (28), was a downregulated DEG in
dasatinib-resistant cell lines in the present study. A homozygous
deletion of MATP with p16INK/CDKN2A has often been reported in
invasive pancreatic cancer (29). In
addition, BCL2 interacting killer (BIK) was a downregulated DEG in
dasatinib-resistant cells in a previous study (11). ERK1/2 degrades BIK in SRC-, BRAF- or
KRAS-activated tumors (30). In
conclusion, the DEGs identified in the present study may influence
the progression and development of pancreatic cancer, and may
contribute to dasatinib resistance through intricate molecular
mechanisms.
The biological functions of the DEGs identified in
the present study were determined by GO annotation using the DAVID
database. In the CC ontology, the majority of enriched DEGs were
associated with cell membrane components, whereas a number of
enriched DEGs were associated with non-membrane-related components,
including the actin cytoskeleton and extracellular exosome.
Mithraprabhu et al (31),
reported that the actin cytoskeleton may be involved in the drug
resistance of numerous cancers. The results of the CC ontology in
the present study demonstrated that the development of dasatinib
resistance was caused by complex cellular mechanisms and may be
associated with membrane and non-membrane cellular structure. The
results of the BP ontology indicated that the regulation-associated
components were the most enriched, including the regulation of
signaling pathways, such as ‘signal regulation’ (118 genes) and
‘cell communication regulation’ (118 genes). The remaining enriched
categories were associated with cell proliferation, transduction,
apoptosis and localization, including ‘cell proliferation’ (95
genes), ‘intracellular signal transduction’ (100 genes), ‘apoptotic
processes’ (73 genes) and ‘macromolecule localization’ (97
genes).
Drug resistance is associated with abnormalities in
signaling pathways, such as the PI3K/AKT/mTOR signaling pathway
(32). A drug is a foreign chemical
capable of damaging tumor cells, and drug resistance is considered
as a cellular adaption to drugs. Regulation of signaling pathways
is therefore a key factor in the development of drug resistance. In
response to dasatinib, tumor cells are able to develop resistance
through mutation or activation of certain pathways. For instance,
the MAPK pathway may contribute to the resistance of dasatinib
(18). During the adaptation
process, numerous gene alterations occur in tumors and may lead to
signaling pathway dysfunction, which may influence cell
proliferation, transduction, apoptosis and localization (33) and result in persistent cell
proliferation, even when tumor cells are exposed to the drug
(34). The results of the present
study indicated that isolated DEGs may affect certain molecular
mechanisms associated with dasatinib resistance in pancreatic
tumors. In the MF ontology, most categories comprised of regulation
and binding-associated components, including ‘molecular function
regulation’ and ‘calcium ion binding’. These results suggested that
the highlighted DEGs may participate in the regulation of molecular
function and ionic pumps. The ‘molecular function regulator’ GO
term is associated with drug resistance (35), and some DEGs identified in the
present study may have significant molecular functions. For
example, a previous study reported that VIM is associated with drug
resistance (36). In addition, LCN2
enhances migration and cisplatin resistance in endometrial cell
carcinoma (37). Ion pumps also
confer drug resistance in various types of cancer, such as gastric,
colorectal and lung cancer (38).
The DEGs highlighted in the present study may therefore serve
crucial roles in dasatinib resistance through regulator-associated
components and/or binding-associated components. However, further
investigation is necessary to confirm these hypotheses.
Pathway analysis may provide additional information
on biological gene function compared with GO analysis. In the
present study, significant pathways were identified, including the
PI3K/Akt and MAPK signaling pathways. Signaling pathways in cancer
may participate in the development of drug resistance; for example,
Guo et al (39) have reported
that the P38/MAPK pathway serves an important role in the
chemotherapy resistance of vincristine in gastric cancer cells. In
addition, the PI3K/Akt and MAPK signaling pathways are crucial for
cancer development, since they are involved in cell proliferation,
maintenance, invasion, drug-resistance and metastasis (40,41).
However, the association between the molecular mechanism of
dasatinib resistance and these signaling pathways remains unknown
and requires further investigation.
Through the construction of a PPI network, the
results from the present study demonstrated that the local network
comprised a series of hub proteins, including ARRB1, SYT1, COL1A2,
COL6A1, ITGB4, ITGA2, LAMA3 and RAC1. It has been previously
reported that these genes are associated with drug resistance and
cancer progression; for example, ARRB1 is an epigenetic regulator
and an important molecule in signal transduction pathways (42). Rosanò et al (43) reported that ARRB1 participates in the
Wnt signaling pathway, which leads to chemotherapy resistance in
ovarian tumor cells. In addition, SYT1 may contribute to cellular
resistance to environmental stresses, such as mechanical stress
(44). Jun et al (45) revealed that SYT1 increases lung
cancer invasiveness through the activation of CD74-ROS, which is a
fusion kinase in lung cancer. COL1A2 and COL6A1 are crucial
molecules associated with drug resistance, and increased expression
of COL1A2 is involved in drug resistance in ovarian cancer cells
(46). In addition, COL6A1 promotes
cancer growth and is upregulated in castration-resistant tumors
(47). Furthermore, integrins are
related to transmembrane glycoprotein receptors in a non-covalent
way; ITGB4 is associated with the progression of taxane-resistant
prostate cancer (48), and Nones
et al (49) reported that
ITGA2 hypomethylation is associated with its high expression and
with poor survival in PDAC. In the present study, COL1A2, COL6A1,
ITGB4, ITGA2 and LAMA3 were associated with the PI3K/AKT signaling
pathway, which indicated that the PI3K/AKT signaling pathway was
enriched in cancer. RAC1 serves key roles in a number of signaling
pathways, including the PI3K/AKT and the MAPK signaling pathways
(50,51). RAC1 is therefore a focal point for
research on tumor resistance (52).
Wang et al (53) reported
that RAC1 can promote chemotherapy resistance in leukemia cells
through the activation of Rac1 GTPase. Previous studies
demonstrated that mutations in RAC1 P29S induce RAF inhibitor
resistance in melanoma cells (54).
RAC1 was highly associated with drug resistance through different
signaling pathways based on the results of the GO and KEGG term
analysis in the present study. Furthermore, the association between
the aforementioned hub genes and the survival curves for pancreatic
adenocarcinoma revealed that five hub genes (COL1A2, ITGB4, ITGA2,
LAMA3 and RAC1) were significantly associated with the overall
survival of patients with pancreatic cancer. These findings
supported the hypothesis that these hub genes may serve crucial
roles in dasatinib resistance. However, whether certain distinct
molecular mechanisms of dasatinib resistance exist, and whether
these genes can be targets for the treatment of dasatinib resistant
pancreatic carcinoma remain unclear.
In conclusion, the results from the present study
revealed certain distinct mechanisms of dasatinib resistance in
pancreatic cancer. Bioinformatics approaches were used to determine
the DEGs in dasatinib-resistant pancreatic cancer cells. The
aberrant signaling pathways were associated with
dasatinib-resistant pancreatic tumors. In addition, five hub genes
were identified as potential targets for the treatment of dasatinib
resistance. The results from the current study may provide a
valuable insight for the determination of dasatinib-resistance in
pancreatic cancer. However, further investigation is required.
Acknowledgements
Not applicable.
Funding
The present study was supported by The National
Natural Science Foundation of China (grant nos. 81572928 and
81772978).
Availability of data and materials
The datasets generated and/or analyzed during the
present study are available in the ‘Dasatinib data’ repository
(https://pan.baidu.com/s/19wCo7dp_Rt2ninY1vGbSMw).
Authors' contributions
JSW and RBH analyzed the data and wrote the
manuscript. XYS, YTC, JFS, XWC, HHZ, YG and XC wrote parts of the
manuscript. JFC revised the manuscript. JSW, RBH, XYS, YTC, JFS,
XWC, HHZ, YG, XC and JFC participated in the statistical analysis
and contributed to the interpretation of the results as well as the
writing of the manuscript. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patients consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2013. CA Cancer J Clin. 63:11–30. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lau MK, Davila JA and Shaib YH: Incidence
and survival of pancreatic head and body and tail cancers: A
population-based study in the United States. Pancreas. 39:458–462.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Quaresma M, Coleman MP and Rachet B:
40-year trends in an index of survival for all cancers combined and
survival adjusted for age and sex for each cancer in England and
Wales, 1971–2011: A population-based study. Lancet. 385:1206–1218.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Reya T, Morrison SJ, Clarke MF and
Weissman IL: Stem cells, cancer, and cancer stem cells. Nature.
414:105–111. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Shah NP, Tran C, Lee FY, Chen P, Norris D
and Sawyers CL: Overriding imatinib resistance with a novel ABL
kinase inhibitor. Science. 305:399–401. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Keating GM: Dasatinib: A review in chronic
myeloid leukaemia and Ph+ acute lymphoblastic leukaemia. Drugs.
77:85–96. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Summy JM and Gallick GE: Src family
kinases in tumor progression and metastasis. Cancer Metastasis Rev.
22:337–358. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Koon HK, Chan PS, Wong RN, Wu ZG, Lung ML,
Chang CK and Mak NK: Targeted inhibition of the EGFR pathways
enhances Zn-BC-AM PDT-induced apoptosis in well-differentiated
nasopharyngeal carcinoma cells. J Cell Biochem. 108:1356–1363.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang X, Wilson MJ, Slaton JW, Sinha AA,
Ewing SL and Pei D: Increased aggressiveness of human prostate PC-3
tumor cells expressing cell surface localized membrane type-1
matrix metalloproteinase (MT1-MMP). J Androl. 30:259–274. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chien W, Sun QY, Lee KL, Ding LW, Wuensche
P, Torres-Fernandez LA, Tan SZ, Tokatly I, Zaiden N, Poellinger L,
et al: Activation of protein phosphatase 2A tumor suppressor as
potential treatment of pancreatic cancer. Mol Oncol. 9:889–905.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gnoni A, Marech I, Silvestris N, Vacca A
and Lorusso V: Dasatinib: An anti-tumour agent via Src inhibition.
Curr Drug Targets. 12:563–578. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Trevino JG, Summy JM, Lesslie DP, Parikh
NU, Hong DS, Lee FY, Donato NJ, Abbruzzese JL, Baker CH and Gallick
GE: Inhibition of SRC expression and activity inhibits tumor
progression and metastasis of human pancreatic adenocarcinoma cells
in an orthotopic nude mouse model. Am J Pathol. 168:962–972. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Nagaraj NS, Smith JJ, Revetta F,
Washington MK and Merchant NB: Targeted inhibition of Src kinase
signaling attenuates pancreatic tumorigenesis. Mol Cancer Ther.
9:2322–2332. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Morton JP, Karim SA, Graham K, Timpson P,
Jamieson N, Athineos D, Doyle B, McKay C, Heung MY, Oien KA, et al:
Dasatinib inhibits the development of metastases in a mouse model
of pancreatic ductal adenocarcinoma. Gastroenterology. 139:292–303.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Bartscht T, Rosien B, Rades D, Kaufmann R,
Biersack H, Lehnert H, Gieseler F and Ungefroren H: Dasatinib
blocks transcriptional and promigratory responses to transforming
growth factor-beta in pancreatic adenocarcinoma cells through
inhibition of Smad signalling: Implications for in vivo mode of
action. Mol Cancer. 14:1992015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Bartscht T, Rosien B, Rades D, Kaufmann R,
Biersack H, Lehnert H, Gieseler F and Ungefroren H: Dasatinib
blocks transcriptional and promigratory responses to transforming
growth factor-beta in pancreatic adenocarcinoma cells through
inhibition of Smad signalling: Implications for in vivo mode of
action. Mol Cancer. 14:1992015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Beadnell TC, Mishall KM, Zhou Q, Riffert
SM, Wuensch KE, Kessler BE, Corpuz ML, Jing X, Kim J, Wang G, et
al: The mitogen-activated protein kinase pathway facilitates
resistance to the src inhibitor dasatinib in thyroid cancer. Mol
Cancer Ther. 15:1952–1963. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Beauchamp EM, Woods BA, Dulak AM, Tan L,
Xu C, Gray NS, Bass AJ, Wong KK, Meyerson M and Hammerman PS:
Acquired resistance to dasatinib in lung cancer cell lines
conferred by DDR2 gatekeeper mutation and NF1 loss. Mol Cancer
Ther. 13:475–482. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lilian A, Denis S, Johnston JB and Raquel
A: p53 and autophagy contribute to dasatinib resistance in primary
CLL lymphocytes. Leuk Res. 35:99–102. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang L, Huang Y, Zhuo W, Zhu Y, Zhu B and
Chen Z: Identification and characterization of biomarkers and their
functions for Lapatinib-resistant breast cancer. Med Oncol.
34:892017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Andrikou K, Peterle C, Pipitone S, Salati
M and Cascinu S: Emerging antibodies for the treatment of
pancreatic cancer. Expert Opin Emerg Drugs. 22:39–51. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Knapinska AM, Estrada CA and Fields GB:
The roles of matrix metalloproteinases in pancreatic cancer. Prog
Mol Biol Transl Sci. 148:339–354. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hong DS, Choe JH, Naing A, Wheler JJ,
Falchook GS, Piha-Paul S, Moulder SL, George GC, Choe JM, Strauss
LC, et al: A phase 1 study of gemcitabine combined with dasatinib
in patients with advanced solid tumors. Invest New Drugs.
31:918–926. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chen Z, Zhao L, Shi K and Chen B:
Mechanism comparison of gemcitabine and dasatinib-resistant
pancreatic cancer by integrating mRNA and miRNA expression
profiles. Clin Lab. 64:749–757. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Shah AN, Summy JM, Zhang J, Park SI,
Parikh NU and Gallick GE: Development and characterization of
gemcitabine-resistant pancreatic tumor cells. Ann Surg Oncol.
14:36292007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lazarova DL and Bordonaro M: Vimentin,
colon cancer progression and resistance to butyrate and other HDAC
is. J Cell Mol Med. 20:989–993. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Efferth T, Gebhart E, Ross DD and
Sauerbrey A: Identification of gene expression profiles predicting
tumor cell response to L-alanosine. Biochem Pharmacol. 66:613–621.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hustinx SR, Hruban RH, Leoni LM,
Iacobuzio-Donahue C, Cameron JL, Yeo CJ, Brown PN, Argani P, Ashfaq
R, Fukushima N, et al: Homozygous deletion of the MTAP gene in
invasive adenocarcinoma of the pancreas and in periampullary
cancer: A potential new target for therapy. Cancer Biol Ther.
4:83–86. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lopez J, Hesling C, Prudent J, Popgeorgiev
N, Gadet R, Mikaelian I, Rimokh R, Gillet G and Gonzalo P: Src
tyrosine kinase inhibits apoptosis through the
Erk1|[sol]|2-dependent degradation of the death accelerator Bik.
Cell Death Differ. 19:1459–1469. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Mithraprabhu S, Khong T and Spencer A:
Overcoming inherent resistance to histone deacetylase inhibitors in
multiple myeloma cells by targeting pathways integral to the actin
cytoskeleton. Cell Death Dis. 5:e11342014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Guerrero-Zotano A, Mayer IA and Arteaga
CL: PI3K/AKT/mTOR: Role in breast cancer progression, drug
resistance, and treatment. Cancer Metastasis Rev. 35:515–524. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Guri Y and Hall MN: mTOR signaling confers
resistance to targeted cancer drugs. Trends Cancer. 2:688–697.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Xu Z, Mei J and Tan Y: Baicalin attenuates
DDP (cisplatin) resistance in lung cancer by downregulating MARK2
and p-Akt. Int J Oncol. 50:93–100. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Kim MH and Kim J: Role of YAP/TAZ
transcriptional regulators in resistance to anti-cancer therapies.
Cell Mol Life Sci. 74:1457–1474. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Xili Y, Yue L, Jianhua Z, Fei G, Dinghong
M, Nianyun W, Guohui L and Guanghua G: Analysis of the mechanism of
drug resistance of VIM-2-type metallo-β-lactamase-producing
Acinetobacter baumannii isolated from burn patients and its
homology. Zhonghua shao shang za zhi (Chinese). 31:205–210.
2015.
|
|
37
|
Miyamoto T, Kashima H, Yamada Y, Kobara H,
Asaka R, Ando H, Higuchi S, Ida K, Mvunta DH and Shiozawa T:
Lipocalin 2 enhances migration and resistance against cisplatin in
endometrial carcinoma cells. PLoS One. 11:e01552202016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Eljack ND, Ma HY, Drucker J, Shen C,
Hambley TW, New EJ, Friedrich T and Clarke RJ: Mechanisms of cell
uptake and toxicity of the anticancer drug cisplatin. Metallomics.
6:2126–2133. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Guo X, Ma N, Wang J, Song J, Bu X, Cheng
Y, Sun K, Xiong H, Jiang G, Zhang B, et al: Increased p38-MAPK is
responsible for chemotherapy resistance in human gastric cancer
cells. BMC Cancer. 8:3752008. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Ersahin T, Tuncbag N and Cetin-Atalay R:
The PI3K/AKT/mTOR interactive pathway. Mol Biosyst. 11:1946–1954.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Xinling Z, Leina M, Jieqiong Q, Hui S,
Wengong Y and Yuchao G: MAPK/ERK signaling pathway-induced
hyper-O-GlcNAcylation enhances cancer malignancy. Mol Cell Biochem.
410:101–110. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Shenoy SK and Lefkowitz RJ:
β-Arrestin-mediated receptor trafficking and signal transduction.
Trends Pharmacol Sci. 32:521–533. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Rosanò L, Cianfrocca R, Tocci P, Spinella
F, Di Castro V, Caprara V, Semprucci E, Ferrandina G, Natali PG and
Bagnato A: Endothelin A receptor/β-arrestin signaling to the Wnt
pathway renders ovarian cancer cells resistant to chemotherapy.
Cancer Res. 74:7453–7464. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Pérez-Sancho J, Vanneste S, Lee E,
McFarlane HE, Esteban Del Valle A, Valpuesta V, Friml J, Botella MA
and Rosado A: The Arabidopsis synaptotagmin1 is enriched in
endoplasmic reticulum-plasma membrane contact sites and confers
cellular resistance to mechanical stresses. Plant Physiol.
168:132–143. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Jun HJ, Johnson H, Bronson RT, de Feraudy
S, White F and Charest A: The oncogenic lung cancer fusion kinase
CD74-ROS activates a novel invasiveness pathway through E-Syt1
phosphorylation. Cancer Res. 72:3764–3774. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Januchowski R, Świerczewska M, Sterzyńska
K, Wojtowicz K, Nowicki M and Zabel M: Increased expression of
several collagen genes is associated with drug resistance in
ovarian cancer cell lines. J Cancer. 7:1295–1310. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Zhu YP, Wan FN, Shen YJ, Wang HK, Zhang GM
and Ye DW: Reactive stroma component COL6A1 is upregulated in
castration-resistant prostate cancer and promotes tumor growth.
Oncotarget. 6:14488–14496. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Kawakami K, Fujita Y, Kato T, Mizutani K,
Kameyama K, Tsumoto H, Miura Y, Deguchi T and Ito M: Integrin β4
and vinculin contained in exosomes are potential markers for
progression of prostate cancer associated with taxane-resistance.
Int J Oncol. 47:384–390. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Nones K, Waddell N, Song S, Patch AM,
Miller D, Johns A, Wu J, Kassahn KS, Wood D, Bailey P, et al:
Genome-wide DNA methylation patterns in pancreatic ductal
adenocarcinoma reveal epigenetic deregulation of SLIT-ROBO, ITGA2
and MET signaling. Int J Cancer. 135:1110–1118. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Zhou G, Peng F, Zhong Y, Chen Y, Tang M
and Li D: Rhein suppresses matrix metalloproteinase production by
regulating the Rac1/ROS/MAPK/AP-1 pathway in human ovarian
carcinoma cells. Int J Oncol. 50:933–941. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Pan Y, Wang N, Xia P, Wang E, Guo Q and Ye
Z: Inhibition of Rac1 ameliorates neuronal oxidative stress damage
via reducing Bcl-2/Rac1 complex formation in mitochondria through
PI3K/Akt/mTOR pathway. Exp Neurol. 300:149–166. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Zhang T and Wang N: miR-135a confers
resistance to gefitinib in non-small cell lung cancer cells by
upregulation of RAC1. Oncol Res. 26:1191–1200. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Wang JY, Yu P, Chen S, Xing H, Chen Y,
Wang M, Tang K, Tian Z, Rao Q and Wang J: Activation of Rac1 GTPase
promotes leukemia cell chemotherapy resistance, quiescence and
niche interaction. Mol Oncol. 7:907–916. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Watson IR, Li L, Cabeceiras PK, Mahdavi M,
Gutschner T, Genovese G, Wang G, Fang Z, Tepper JM, Stemke-Hale K,
et al: The RAC1 P29S hotspot mutation in melanoma confers
resistance to Pharmacological inhibition of RAF. Cancer Res.
74:48452014. View Article : Google Scholar : PubMed/NCBI
|