Introduction
Cervical cancer, one of the most common types of
gynaecological malignancy, is the fourth most frequent cause of
cancer-associated mortality among females worldwide (1). The incidence of cervical cancer is
decreasing each year in developed countries; unfortunately,
however, it continues to increase each year in developing countries
(2). It is estimated that there are
~500,000 novel cases and >250,000 mortalities caused by cervical
cancer per year globally (3). In
recent years, impressive progress has been made in diagnostic
techniques and therapeutic approaches; however, cervical cancer
remains a public health concern worldwide, and the clinical
outcomes of patients with cervical cancer remain unsatisfactory
(4). Multiple factors, such as local
recurrence, distant metastasis and diagnostic delays, are mainly
responsible for the poor prognosis of patients with cervical cancer
(5); however, the specific
underlying molecular mechanism remains largely unknown. Thus,
further studies are required to elucidate the molecular mechanisms
underlying cervical carcinogenesis and progression to develop
promising therapeutic methods for treating patients with cervical
cancer.
MicroRNAs (miRNAs) are a subset of endogenous,
non-coding and small RNA molecules (2). These 17–23-nucleotide-long RNAs are
able to post-transcriptionally modulate the expression of relevant
genes via base-pairing with the 3′-untranslated regions (3′-UTRs)
of their target genes, thereby resulting in either translation
suppression and/or mRNA cleavage (6). To date, >1,500 mature miRNAs have
been identified in the human genome, and these miRNAs can regulate
~30% of human protein-coding genes (6). miRNA aberrations have been identified
in various types of malignancy, including cervical cancer (7). It has been demonstrated that a number
of miRNAs are abnormally expressed during the occurrence and
progression of cervical cancer (8).
For example, miR-143 (9), miR-152
(10) and miR-506 (11) were downregulated in cervical cancer,
whereas miR-21 (12), miR-224
(13) and miR-1297 (14) were expressed at high levels. Recent
evidence has indicated that deregulated miRNAs may serve crucial
roles in cervical cancer initiation and progression by regulating a
wide range of biological functions (15–17).
Therefore, miRNAs may be effective therapeutic targets for the
treatment of patients with cervical cancer.
miR-432 dysregulation has been demonstrated to be
implicated in the carcinogenesis and progression of numerous types
of human cancer, including lung adenocarcinoma (18), neuroblastoma (19), hepatocellular carcinoma (20) and prostate cancer (21). However, the effects and underlying
molecular mechanisms of miR-432 in cervical cancer have yet to be
elucidated. The objectives of the present study were to detect the
expression of miR-432 in cervical cancer and determine its clinical
significance. The effects of miR-432 in cervical cancer cells and
the potential underlying molecular mechanism of these effects were
also investigated.
Materials and methods
Clinical tissue specimens
A total of 47 pairs of cervical cancer tissues and
matched adjacent normal tissues were collected from female patients
(age range, 49–65 years) who received surgery at The Third People's
Hospital of Linyi (Linyi, China) between April 2015 and August
2017. All tissues were obtained using microdissection. None of
these patients had been treated with any pre-operative chemotherapy
or radiotherapy. Once obtained, tissues were immediately frozen in
liquid nitrogen and then stored at −80°C for further study.
Patients that had been treated with pre-operative chemotherapy or
radiotherapy were excluded from the study. All patients with
cervical cancer were divided into either a low miR-432 expression
group or a high miR-432 expression group using the median value of
miR-432 expression (0.45) as the cut-off. The International
Federation of Gynecology and Obstetrics (FIGO) stage is currently
the most commonly used staging system for cancer of the uterine
cervix worldwide (22). Tumours are
classified by roman numerals from I to IV, representing a range
from precancerous or in situ to highly malignant.
Classification subdivisions are indicated by letters and numbers
(22). The present study was
approved by the Ethics Committee of The Third People's Hospital of
Linyi. All patients provided written informed consent and were
apprised of the aims of the study.
Cell lines
Three human cervical cancer cell lines, HeLa, CaSki
and SiHa, were purchased from the Cell Bank of the Chinese Academy
of Sciences (Shanghai, China). The normal human cervix epithelial
cell line, Ect1/E6E7, was purchased from the American Type Culture
Collection (ATCC; Manassas, VA, USA). All cell lines were cultured
in Dulbecco's modified Eagle's medium (DMEM) containing 10% foetal
bovine serum (FBS), 100 U/ml penicillin and 100 µg/ml streptomycin
(all purchased from Invitrogen; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) and grown in a humidified incubator supplied with
5% CO2 at 37°C.
Oligonucleotide transfection
Synthetic miR-432 mimic and negative control miRNA
mimic (miR-NC) were obtained from Guangzhou RiboBio Co., Ltd.
(Guangzhou, China). For silencing of fibronectin 1 (FN1)
expression, a small interfering RNA (siRNA) against FN1 (FN1 siRNA)
and negative control siRNA (NC siRNA) were chemically synthesized
by GeneCopoeia, Inc. (Rockville, MD, USA). The FN1 expression
plasmid pcDNA3.1-FN1 (pc-FN1) and blank pcDNA3.1 plasmid were
generated by Shanghai GenePharma Co., Ltd. (Shanghai, China). HeLa
and SiHa cells in the exponential growth phase were collected and
seeded into 6-well plates at a density of 6×106
cells/well 1 day before transfection. Cells were transfected with
miRNA mimic (100 pmol), siRNA (100 pmol) or plasmid (4 µg) using
Lipofectamine® 2000 reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol. The
transfected HeLa and SiHa cells were harvested and used for the
detection of miR-432 and FN1 protein expression at 48 and 72 h
after transfection, respectively.
RNA extraction and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was isolated from cell lines and tissues
using TRIzol® reagent (Invitrogen; Thermo Fisher
Scientific, Inc.), and the concentration of isolated total RNA was
determined using a NanoDrop™ 1000 spectrophotometer (Thermo Fisher
Scientific, Inc.). To quantify miR-432 expression, total RNA was
converted into cDNA using the TaqMan MicroRNA RT kit (Applied
Biosystems; Thermo Fisher Scientific, Inc.). The temperature
protocol for reverse transcription was as follows: 16°C for 30 min,
42°C for 30 min and 85°C for 5 min. Next, qPCR was performed using
the TaqMan MicroRNA PCR kit (Applied Biosystems; Thermo Fisher
Scientific, Inc.). The temperature protocol for qPCR was as
follows: 50°C for 2 min, 95°C for 10 min; 40 cycles of denaturation
at 95°C for 15 sec; and annealing/extension at 60°C for 1 min. For
FN1 mRNA expression analysis, cDNA was generated from total RNA
using the PrimeScript RT Reagent kit followed by qPCR using the
SYBR® Premix Ex Taq™ kit (both from Takara Biotechnology
Co., Ltd., Dalian, China). The temperature protocol for reverse
transcription was as follows: 37°C for 15 min and 85°C for 5 sec.
The qPCR was performed with cycling conditions as follows: 5 min at
95°C, followed by 40 cycles of 95°C for 30 sec and 65°C for 45 sec.
U6 small nuclear RNA and GAPDH were used as internal references for
miR-432 and FN1 mRNA expression, respectively. Relative gene
expression was quantified using the 2−ΔΔCq method
(23). The primers were designed as
follows: miR-432 forward, 5′-AACGAGACGACGACAGACT-3′ and reverse,
5′-CTTGGAGTAGGTCATTGGGT-3′; U6 forward,
5′-GCTTCGGCAGCACATATACTAAAAT-3′ and reverse,
5′-CGCTTCACGAATTTGCGTGTCAT-3′; FN1 forward,
5′-CAGTGGGAGACCTCGAGAAG-3′ and reverse, 5′-TCCCTCGGAACATCAGAAAC-3′;
and GAPDH forward, 5′-GCTGGCGCTGAGTACGTCGTGGAGT-3′ and reverse,
5′-CACAGTCTTCTGGGTGGCAGTGATGG-3′.
Cell Counting Kit-8 (CCK-8) assay
The influence of miR-432 on cervical cancer cell
proliferation was evaluated using the CCK-8 assay. Briefly,
transfected HeLa and SiHa cells were collected after 24 h of
incubation, and 3×103 cells/well were inoculated into
96-well plates in triplicate. Following incubation for 0, 24, 48 or
72 h at 37°C with 5% CO2, a CCK-8 assay was performed by
adding 10 µl CCK-8 solution (Dojindo Molecular Technologies, Inc.,
Kumamoto, Japan) to each well. Cells were then incubated at 37°C
for an additional 2 h. Finally, the optical density at a wavelength
of 450 nm was determined using a spectrophotometer (Victor X;
PerkinElmer, Inc., Waltham, MA, USA).
Transwell Matrigel invasion assay
A Transwell Matrigel invasion assay was used to
identify the invasive capacity of cervical cancer cells. To this
end, transfected cells were harvested after 48 h of incubation and
suspended in FBS-free DMEM. In total, 5×104 cells were
placed in the upper compartment of Transwell inserts that were
coated with Matrigel (both purchased from BD Biosciences, San Jose,
CA, USA). DMEM supplemented with 20% FBS was added to the lower
compartments as a chemoattractant. After 24 h of incubation,
non-invasive cells were gently wiped off using a cotton swab. The
invasive cells on the bottom side of the membrane were fixed with
4% paraformaldehyde at 37°C for 15 min and stained with 0.1%
crystal violet at 37°C for 30 min. The invasive ability of cervical
cancer cells was assessed by counting the number of invasive cells
in five randomly selected visual fields from each insert using a
light microscope (×200 magnification; Olympus Corporation, Tokyo,
Japan).
Bioinformatics analysis
The databases TargetScan (targetscan.org) and miRDB (mirdb.org)
software were used to predicate the putative targets of
miR-432.
Luciferase reporter assay
The wild-type (WT) and mutant (MUT) 3′-UTRs of FN1
were amplified by and purchased from Shanghai GenePharma Co., Ltd.,
and cloned into pMIR (Ambion; Thermo Fisher Scientific, Inc.),
generating the pMIR-WT-FN1-3′-UTR and pMIR-MUT-FN1-3′-UTR plasmids,
respectively. For the reporter assay, cells in the exponential
growth phase were plated on 24-well plates with a density of
1.5×105 cells/well. miR-432 mimic (50 pmol) or miR-NC
(50 pmol) in combination with pMIR-WT-FN1-3′-UTR (0.2 µg) or
pMIR-MUT-FN1-3′-UTR (0.2 µg) were co-transfected into cells using
Lipofectamine 2000 reagent, according to the manufacturer's
protocol.
Luciferase activity was determined using a
dual-luciferase reporter assay system (Promega Corporation,
Madison, WI, USA) 48 h after transfection. The activity of firefly
luciferase was normalized to that of Renilla luciferase.
Western blot analysis
Tissue samples and cells were washed twice with PBS
and lysed with radioimmunoprecipitation assay lysis buffer
(Shanghai Qcbio Science & Technologies Co., Ltd., Shanghai,
China). The concentration of total protein was determined using a
bicinchoninic acid kit (Beyotime Institute of Biotechnology,
Haimen, China). Equal amounts of total protein (30 µg) were
separated by SDS-PAGE (10% gel) and then transferred onto
polyvinylidene difluoride membranes (Beyotime Institute of
Biotechnology). Following blocking with 5% skimmed milk suspended
in Tris-buffered saline-containing Tween-20 (TBST) at room
temperature for 2 h, the membranes were incubated overnight at 4°C
with primary antibodies as follows: Rabbit anti-human FN1 antibody
(catalogue no. 15613-1-AP; 1:200 dilution; ProteinTech Group, Inc.,
Chicago, IL, USA) and rabbit anti-human GAPDH antibody (catalogue
no. ab181603; 1:1,000 dilution; Abcam, Cambridge, UK). Following
extensive washes with TBST, horseradish peroxidase-conjugated goat
anti-rabbit secondary antibody (catalogue no. ab6721; 1:5,000
dilution; Abcam, Cambridge, UK) was added for 2 h at room
temperature. Finally, the immunoreactive proteins were visualized
using an enhanced chemiluminescence protein detection kit (Pierce;
Thermo Fisher Scientific, Inc.). GAPDH was used as a loading
control.
Statistical analysis
Data are presented as the mean ± standard deviation.
Student's t-test was used to compare the difference between two
groups. The differences between multiple groups were evaluated
using one-way analysis of variance, followed by Tukey's post hoc
test. A χ2 test was applied to investigate the
association between miR-432 and the clinical characteristics in
patients with cervical cancer. The correlation between expression
levels of miR-432 and FN1 mRNA in cervical cancer tissues was
assessed using Spearman's correlation analysis. P<0.05 was
considered to indicate a statistically significant difference.
Results
miR-432 expression is decreased and
significantly associated with certain clinicopathological features
of cervical cancer
To investigate the clinical value of miR-432 in
cervical cancer, the expression levels of miR-432 in 47 pairs of
cervical cancer tissues and matched adjacent normal tissues were
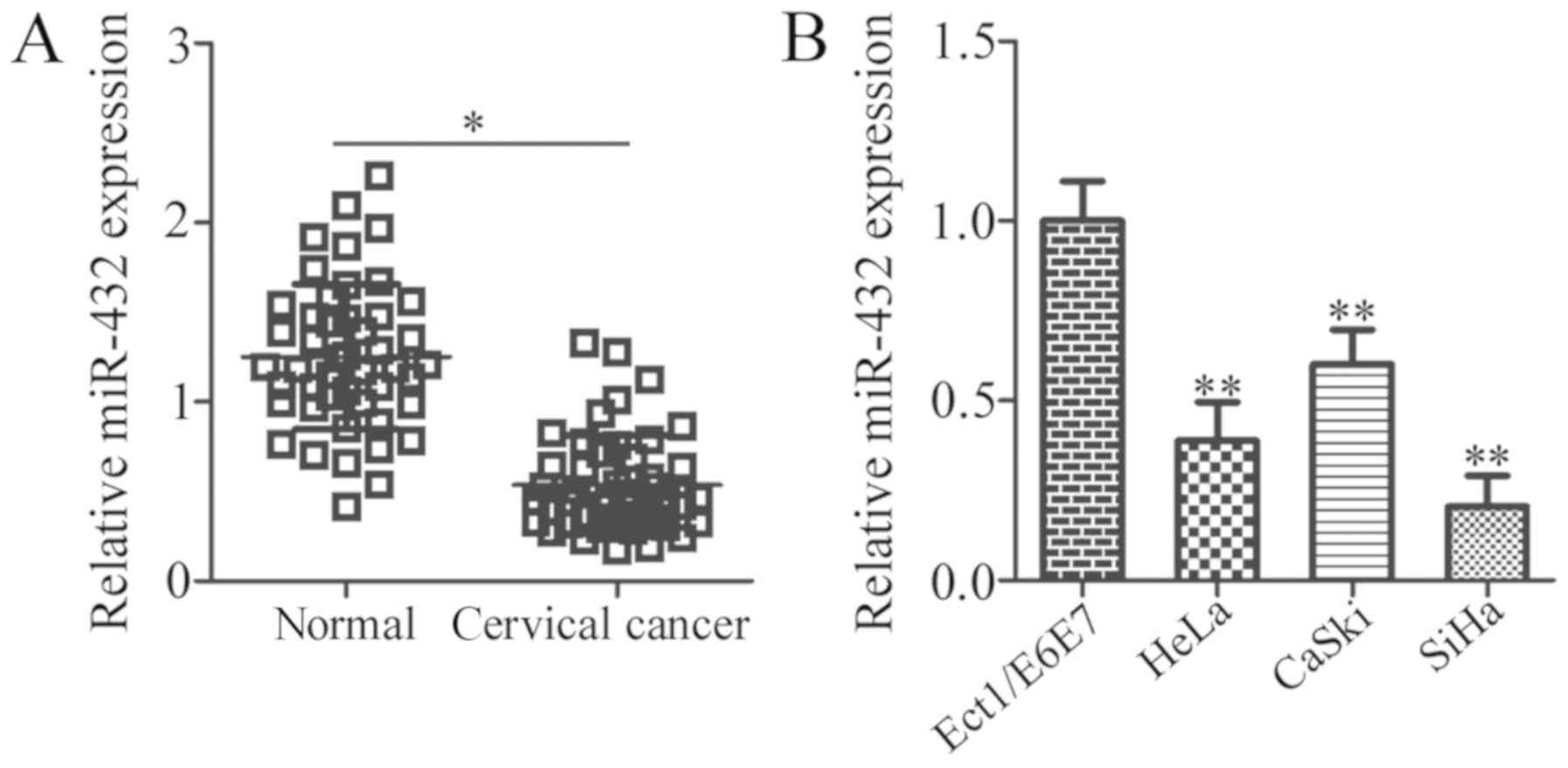
determined using RT-qPCR. The results revealed a significant
decrease in the expression of miR-432 in cervical cancer tissues
compared with in adjacent normal tissues (P<0.05; Fig. 1A). The clinical significance of
miR-432 expression in cervical cancer was then investigated. All
patients with cervical cancer were divided into either a low
miR-432 expression group or a high miR-432 expression group using
the median value of miR-expression (0.45) as the cut-off. Decreased
miR-432 expression was revealed to be significantly associated with
the FIGO stage (P=0.020), myometrium invasion (P=0.001) and lymph
node metastasis (P=0.008). However, no significant association was
identified with age or tumour size (both P>0.05; Table I). In addition, the expression level
of miR-432 in three human cervical cancer cell lines (HeLa, CaSki
and SiHa) and a normal human cervix epithelial cell line
(Ect1/E6E7) was determined. RT-qPCR analysis revealed that miR-432
was expressed at low levels in all three cervical cancer cell lines
relative to Ect1/E6E7 (P<0.05; Fig.
1B). These results suggest that downregulation of miR-432 may
be involved in the development and progression of cervical
cancer.
 | Table I.Association between miR-432 and the
clinicopathological features of patients with cervical cancer. |
Table I.
Association between miR-432 and the
clinicopathological features of patients with cervical cancer.
|
| miR-432 level |
|
|---|
|
|
|
|
|---|
| Clinicopathological
feature | Low | High | P-value |
|---|
| Age, years |
|
| 0.534 |
|
<45 | 9 | 6 |
|
|
≥45 | 15 | 17 |
|
| Tumor size, cm |
|
| 0.380 |
|
<4 | 16 | 12 |
|
| ≥4 | 8 | 11 |
|
| FIGO stage |
|
| 0.020 |
|
I–II | 7 | 15 |
|
|
III–IV | 17 | 8 |
|
| Myometrium
invasion |
|
| 0.001 |
|
<1/2 | 7 | 18 |
|
|
≥1/2 | 17 | 5 |
|
| Lymph node
metastasis |
|
| 0.008 |
|
Negative | 8 | 17 |
|
|
Positive | 16 | 6 |
|
Ectopic miR-432 expression inhibits
the proliferation and invasion of cervical cancer cells
HeLa and SiHa cell lines exhibited the greatest
decrease in miR-432 expression among the three cervical cancer cell
lines, therefore these two cervical cancer cell lines were selected
for subsequent functional assays. To identify the specific roles of
miR-432 in cervical cancer cells, miR-432 mimic or miR-NC were
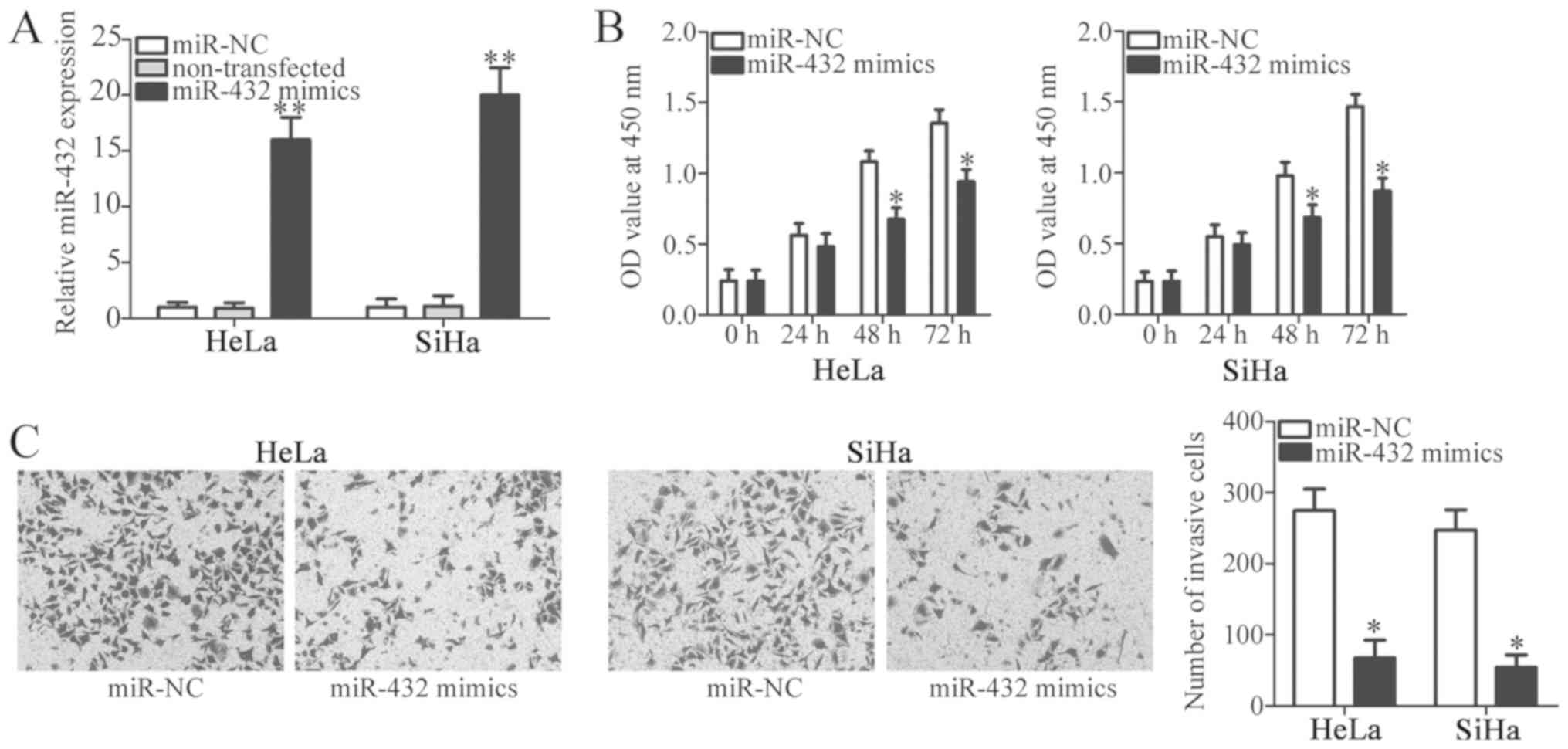
introduced into HeLa and SiHa cells. RT-qPCR analysis verified that
miR-432 was markedly overexpressed in miR-432 mimic-transfected
HeLa and SiHa cells compared with cells transfected with miR-NC and
non-transfected cells (P<0.05; Fig.
2A). In addition, no significant difference in miR-432
expression was observed between the miR-NC and non-transfected
groups (P<0.05). A CCK-8 assay was performed in order to
investigate the effect of miR-432 overexpression on the
proliferation of cervical cancer cells. The results indicated that
when HeLa and SiHa cells were transfected with miR-432 mimic, the
proliferative ability was significantly decreased compared with
that of miR-NC groups at 48 and 72 h (P<0.05; Fig. 2B). Furthermore, a Transwell Matrigel
invasion assay was performed to investigate the effect of miR-432
on cervical cancer cell invasion. It was revealed that enforced
miR-432 expression significantly inhibited the invasion of HeLa and
SiHa cells (P<0.05; Fig. 2C).
These results suggest that miR-432 may have anti-proliferative and
anti-invasive roles in cervical cancer cells.
FN1 is a direct target gene of miR-432
in cervical cancer cells
To further illustrate the molecular mechanisms by
which miR-432 inhibits cervical cancer cell proliferation and
invasion, a bioinformatics analysis was performed in order to
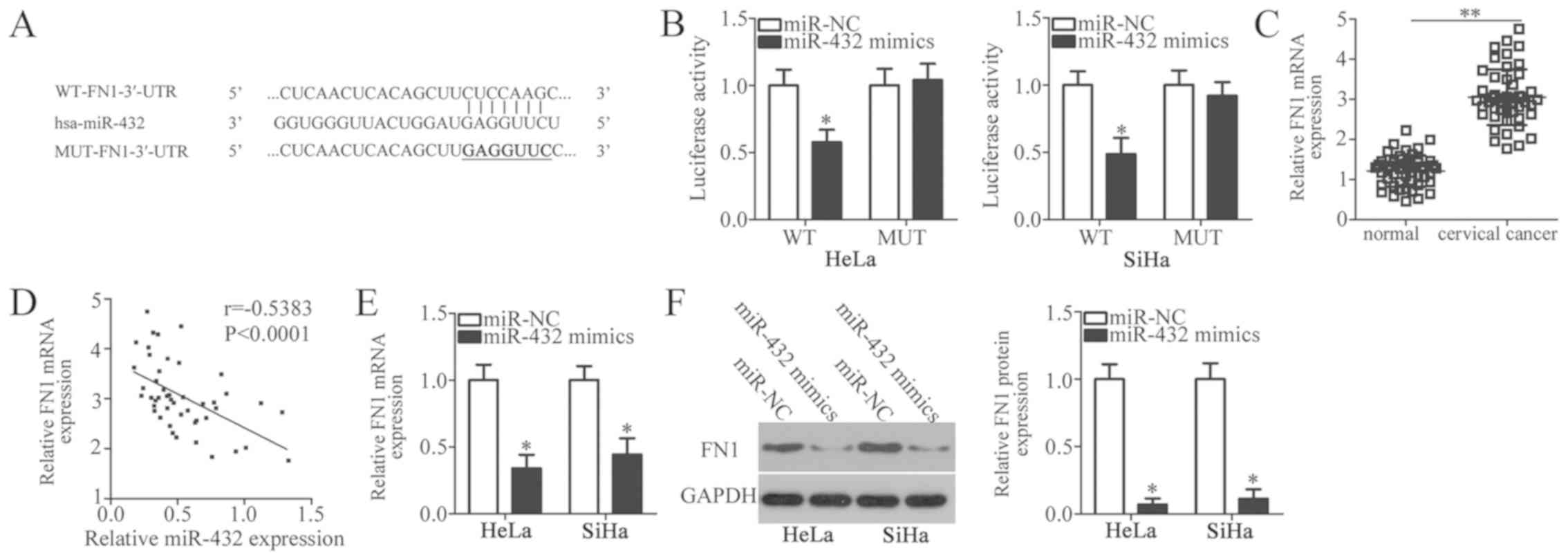
predict the potential targets of miR-432. A highly conserved
putative binding site was revealed at 154-160 bp of the FN1-3′-UTR
(Fig. 3A). FN1 was selected for
further investigation as this gene was considered to have a role in
the formation and progression of cervical cancer (24–27). To
confirm this prediction, luciferase reporter plasmids were
chemically synthesized and co-transfected with miR-432 mimic or
miR-NC into HeLa and SiHa cells. The results of the luciferase
reporter assays in the present study indicated that miR-432
overexpression notably decreased the luciferase activity of the
plasmid harbouring WT-FN1-3′-UTR (P<0.05); however, the
suppression of luciferase activity was eliminated when HeLa and
SiHa cells were co-transfected with miR-432 mimic and luciferase
plasmid containing MUT-FN1-3′-UTR (Fig.
3B).
RT-qPCR was performed to determine FN1 expression in
47 pairs of cervical cancer tissues and matched adjacent normal
tissues. The data revealed that the expression level of FN1 mRNA
was significantly higher in cervical cancer tissues compared with
in adjacent normal tissues (P<0.05; Fig. 3C). Notably, FN1 mRNA expression
levels in cervical cancer tissues were inversely correlated with
miR-432 expression levels (r=−0.5383, P<0.0001; Fig. 3D). Furthermore, re-expression of
miR-432 resulted in a significant decrease in FN1 expression in
HeLa and SiHa cells at the mRNA (P<0.05; Fig. 3E) and protein (P<0.05; Fig. 3F) levels. Collectively, these results
indicate that FN1 is a direct target gene of miR-432 in cervical
cancer cells.
Inhibition of FN1 stimulates the
tumour suppressor activity of miR-432 in cervical cancer cells
To clarify the biological roles of FN1 in cervical
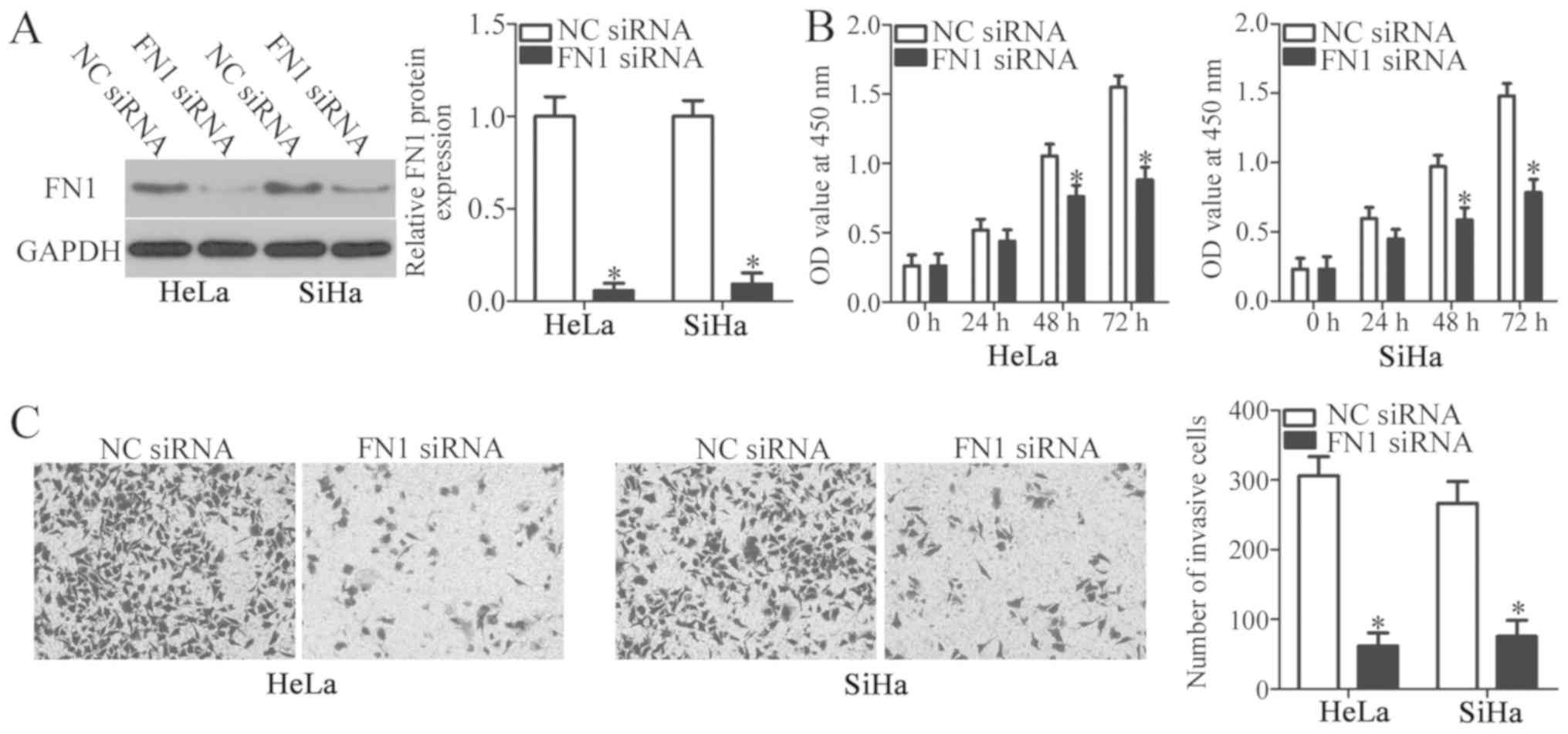
cancer, FN1 siRNA was introduced into HeLa and SiHa cells to
decrease FN1 expression. The protein level of FN1 was efficiently
knocked down in HeLa and SiHa cells following FN1 siRNA
transfection (P<0.05; Fig. 4A).
CCK-8 and Transwell Matrigel invasion assays were performed to
identify the effects of FN1 inhibition on the proliferation and
invasion of cervical cancer cells, respectively. FN1 silencing
significantly decreased proliferation at 48 and 72 h (P<0.05;
Fig. 4B) and invasion (P<0.05;
Fig. 4C) of HeLa and SiHa cells
compared with the NC siRNA cells. These results indicated that the
functional roles of FN1 inhibition were similar to those induced by
miR-432 upregulation in cervical cancer cells, further identifying
FN1 as a downstream target of miR-432.
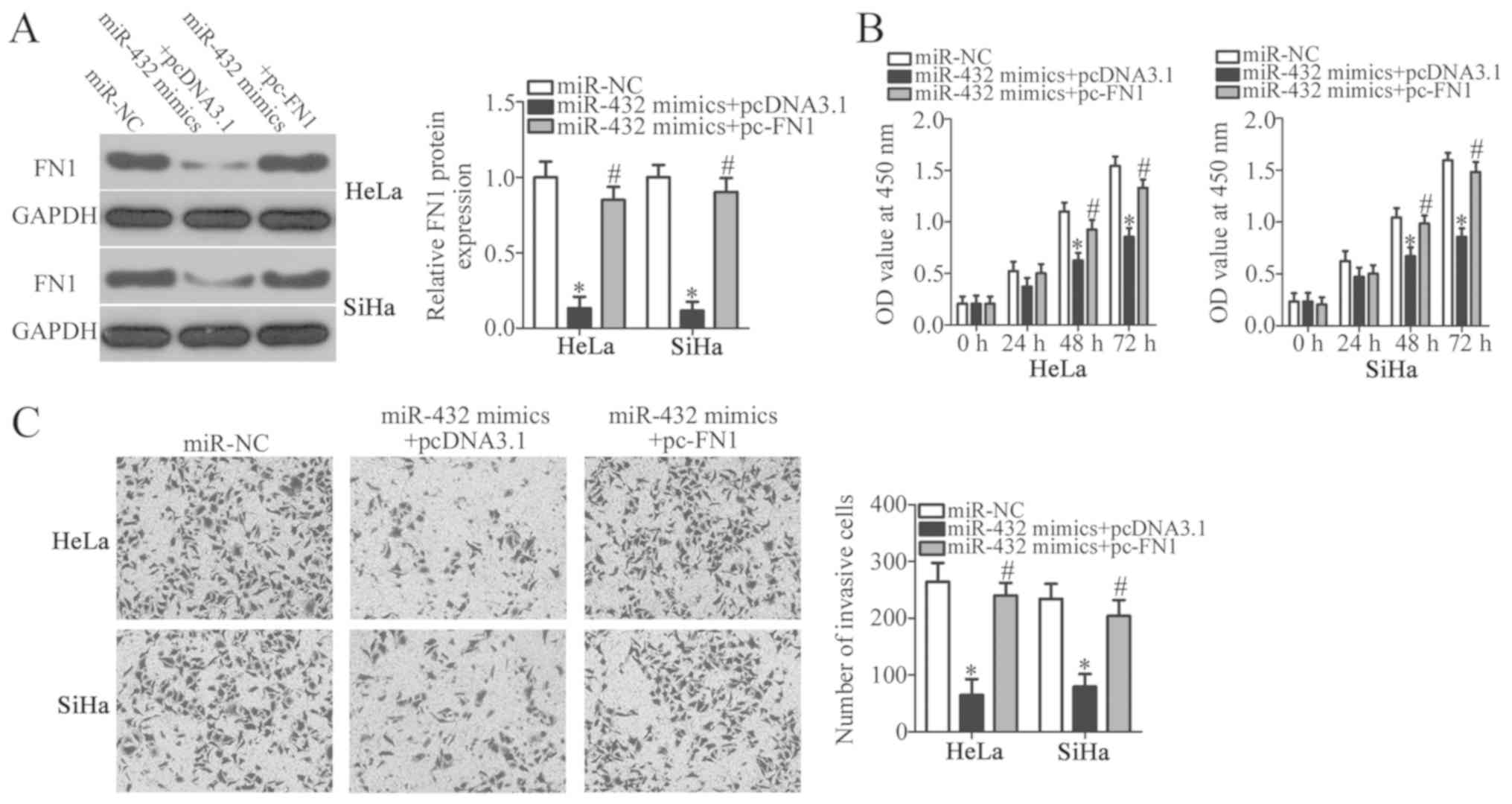
FN1 restoration rescues the
suppressive effects of miR-432 on the malignant phenotypes of
cervical cancer cells
miR-432 overexpression suppressed the proliferation
and invasion of cervical cancer cells, and FN1 was identified as a
direct target gene of miR-432, therefore it was hypothesized that
FN1 downregulation was essential for the miR-432-mediated malignant
phenotypes of cervical cancer cells. To confirm this hypothesis,
miR-432 mimics along with pcDNA3.1-FN1 (pc-FN1) or blank pcDNA3.1
plasmid were co-transfected into HeLa and SiHa cells. Following
transfection, western blot analysis revealed that the decreased
level of FN1 protein in HeLa and SiHa cells caused by miR-432
overexpression was almost recovered by co-transfection with pc-FN1
(P<0.05; Fig. 5A). CCK-8 and
Transwell Matrigel invasion assays were further performed. The
proliferation and invasion of HeLa and SiHa cells were
significantly suppressed upon miR-432 overexpression; however, the
inhibition of proliferation (P<0.05; Fig. 5B) and invasion (P<0.05; Fig. 5C) associated with miR-432
overexpression were reversed by FN1 restoration. This suggests that
the tumour suppressive roles of miR-432 in cervical cancer cells
may be, at least in part, attributable to the downregulation of
FN1.
Discussion
Numerous studies have demonstrated the dysregulation
of miRNAs in cervical cancer (28–30).
miRNAs serve as oncogenes or tumour suppressors and are involved in
regulating a number of tumour-associated biological behaviours,
including the cell cycle, cell proliferation, apoptosis and
metastasis (2). Therefore,
investigating the roles of cervical cancer-associated miRNAs and
the underlying molecular mechanisms is essential for the
development of novel diagnostic biomarkers and effective
therapeutic targets. In the present study, the expression levels
and clinical value of miR-432 in cervical cancer were investigated.
Furthermore, the detailed roles of miR-432 in the development of
cervical cancer were determined. Notably, the underlying molecular
mechanisms by which miR-432 may affect the development of cervical
cancer were investigated.
miR-432 is deregulated in several types of human
cancer. For instance, miR-432 was downregulated in lung
adenocarcinoma tissues and cell lines. Downregulation of miR-432
was significantly associated with the clinical stage of patients
with lung adenocarcinoma. Patients with lung adenocarcinoma that
have low miR-432 levels presented with poorer prognoses than those
patients with high miR-432 levels (18). miR-432 was also expressed at low
levels in neuroblastoma (19),
hepatocellular carcinoma (20) and
prostate cancer (21). However, the
expression status of miR-432 in cervical cancer remains unclear. In
the present study, RT-qPCR analysis was used to detect miR-432
expression in cervical cancer tissues and cell lines. The results
of the present study revealed that miR-432 was expressed at low
levels in cervical cancer tissues and cell lines. In addition,
decreased miR-432 expression was associated with FIGO stage,
myometrium invasion and lymph node metastasis in patients with
cervical cancer. These results suggest that miR-432 may be a
potential biomarker for the diagnosis of patients with these
specific types of cancer.
miR-432 deregulation contributes to the
carcinogenesis and progression of several types of human
malignancy. For example, miR-432 re-expression repressed cell
proliferation and induced cell cycle arrest in lung adenocarcinoma.
In addition, miR-432 overexpression increased the chemosensitivity
of lung adenocarcinoma cells to cisplatin (18). In neuroblastoma, enforced miR-432
expression decreased cell proliferation, promoted G0-G1 cell cycle
arrest and induced neurite projections (19). In hepatocellular carcinoma,
restoration of miR-432 expression inhibited cell proliferation and
colony formation, increased G0-G1 cell cycle arrest in vitro
and decreased tumour growth in vivo through deactivation of
the Wnt/β-catenin signalling pathway (20). In prostate cancer, ectopic miR-432
expression inhibited the activation of the Wnt/β-catenin signalling
pathway and participated in the regulation of cell proliferation
and apoptosis. Nevertheless, to the best of our knowledge, the
functional roles of miR-432 in the progression and development of
cervical cancer have not been investigated. In the present study,
functional experiments demonstrated that resumption of miR-432
expression restrained the proliferation and invasion of cervical
cancer cells. These results suggest that miR-432 may represent a
useful therapeutic target for managing patients with these types of
malignant tumour.
Several genes have been identified as direct targets
of miR-432, including E2F transcription factor 3 and anexelekto in
lung adenocarcinoma (18), nestin
and REST corepressor 1 in neuroblastoma (19) and tripartite motif-containing protein
29 and Pygopus homolog 2 in prostate cancer (21). In the present study, the underlying
molecular mechanism by which miR-432 regulates the development of
cervical cancer was also investigated. FN1, a member of the FN
family (24), was identified as a
direct target of miR-432 in cervical cancer. The expression of FN1
was previously reported to be upregulated in numerous types of
human malignant tumours, including breast cancer (31), gastric cancer (32), ovarian cancer (33) and thyroid cancer (34). A number of studies have demonstrated
the key role that FN1 serves in tumourigenesis and tumour
progression; FN1 serves oncogenic roles and regulates a variety of
cellular processes (24–27). In the present study, it was revealed
that FN1 was markedly expressed in cervical cancer, and that
inhibition of FN1 could inhibit the proliferation and invasion of
cervical cancer cells. These data suggest that targeting FN1 may be
a promising approach for treating patients with cervical
cancer.
In conclusion, miR-432 was downregulated in cervical
cancer, which was correlated with FIGO stage, myometrium invasion
and lymph node metastasis. Enforced miR-432 expression suppressed
the proliferation and invasion of cervical cancer cells by directly
targeting FN1. The results of the present study provide a
theoretical basis for the application of the miR-432/FN1 pathway in
the treatment of patients with cervical cancer. However, there were
two limitations to the present study: First, the influence of
miR-432 on the cell cycle of cervical cancer was not investigated.
Secondly, the sample size of the present study was small. Future
studies will aim to resolve these two limitations.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analysed during the present
study are available from the corresponding author upon reasonable
request.
Authors' contributions
WW and SW designed this study. SW and BG performed
RT-qPCR, CCK-8 assay and luciferase reporter assay. Transwell
Matrigel invasion assay was carried out by HY. XL and XW conducted
western blot analysis. All authors have made a significant
contribution to the findings and methods, and have read and
approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of The Third People's Hospital of Linyi (Linyi, China).
All patients provided written informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Banno K, Iida M, Yanokura M, Kisu I, Iwata
T, Tominaga E, Tanaka K and Aoki D: MicroRNA in cervical cancer:
OncomiRs and tumor suppressor miRs in diagnosis and treatment.
ScientificWorldJournal 2014. 1780752014.
|
|
3
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2012. CA Cancer J Clin. 62:10–29. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Handler AS, Henderson VA, Rosenfeld A,
Rankin K, Jones B and Issel LM: Illinois breast and cervical cancer
program: Implementing effective public-private partnerships to
assure population health. J Public Health Manag Pract. 21:459–466.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kogo R, How C, Chaudary N, Bruce J, Shi W,
Hill RP, Zahedi P, Yip KW and Liu FF: The microRNA-218~Survivin
axis regulates migration, invasion, and lymph node metastasis in
cervical cancer. Oncotarget. 6:1090–1100. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Griffiths-Jones S, Grocock RJ, van Dongen
S, Bateman A and Enright AJ: miRBase: microRNA sequences, targets
and gene nomenclature. Nucleic Acids Res. 34:D140–D144. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Shu L, Zhang Z and Cai Y: MicroRNA-204
inhibits cell migration and invasion in human cervical cancer by
regulating transcription factor 12. Oncol Lett. 15:161–166.
2018.PubMed/NCBI
|
|
8
|
Srivastava SK, Ahmad A, Zubair H, Miree O,
Singh S, Rocconi RP, Scalici J and Singh AP: MicroRNAs in
gynecological cancers: Small molecules with big implications.
Cancer Lett. 407:123–138. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhao Y, Liu X and Lu YX: MicroRNA-143
regulates the proliferation and apoptosis of cervical cancer cells
by targeting HIF-1alpha. Eur Rev Med Pharmacol Sci. 21:5580–5586.
2017.PubMed/NCBI
|
|
10
|
Zhang H, Lu Y, Wang S, Sheng X and Zhang
S: MicroRNA-152 acts as a tumor suppressor microRNA by inhibiting
Kruppel-like factor 5 in human cervical cancer. Oncol Res.
27:335–340. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Gong M, Chen C, Zhao H, Sun M and Song M:
miR-506 suppresses cervical cancer cell proliferation both in vitro
and in vivo. Neoplasma. 65:331–338. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang Z, Wang J, Wang X, Song W, Shi Y and
Zhang L: MicroRNA-21 promotes proliferation, migration, and
invasion of cervical cancer through targeting TIMP3. Arch Gynecol
Obstet. 297:433–442. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yu LM, Wang WW, Qi R, Leng TG and Zhang
XL: MicroRNA-224 inhibition prevents progression of cervical
carcinoma by targeting PTX3. J Cell Biochem. 119:10278–10290. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chen Z, Zhang M, Qiao Y, Yang J and Yin Q:
MicroRNA-1297 contributes to the progression of human cervical
carcinoma through PTEN. Artif Cells Nanomed Biotechnol. 46 (Suppl
2):S1120–S1126. 2018. View Article : Google Scholar
|
|
15
|
Kanekura K, Nishi H, Isaka K and Kuroda M:
MicroRNA and gynecologic cancers. J Obstet Gynaecol Res.
42:612–617. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Gonzalez-Quintana V, Palma-Berré L,
Campos-Parra AD, López-Urrutia E, Peralta-Zaragoza O, Vazquez-Romo
R and Pérez-Plasencia C: MicroRNAs are involved in cervical cancer
development, progression, clinical outcome and improvement
treatment response (Review). Oncol Rep. 35:3–12. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang F, Li B and Xie X: The roles and
clinical significance of microRNAs in cervical cancer. Histol
Histopathol. 31:131–139. 2016.PubMed/NCBI
|
|
18
|
Chen L, Kong G, Zhang C, Dong H, Yang C,
Song G, Guo C, Wang L and Yu H: MicroRNA-432 functions as a tumor
suppressor gene through targeting E2F3 and AXL in lung
adenocarcinoma. Oncotarget. 7:20041–20053. 2016.PubMed/NCBI
|
|
19
|
Das E and Bhattacharyya NP: MicroRNA-432
contributes to dopamine cocktail and retinoic acid induced
differentiation of human neuroblastoma cells by targeting NESTIN
and RCOR1 genes. FEBS Lett. 588:1706–1714. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Jiang N, Chen WJ, Zhang JW, Xu C, Zeng XC,
Zhang T, Li Y and Wang GY: Downregulation of miR-432 activates
Wnt/beta-catenin signaling and promotes human hepatocellular
carcinoma proliferation. Oncotarget. 6:7866–7879. 2015.PubMed/NCBI
|
|
21
|
Li JB, Liu F, Zhang BP, Bai WK, Cheng W,
Zhang YH and Yu LJ: LncRNA625 modulates prostate cancer cells
proliferation and apoptosis through regulating the Wnt/beta-catenin
pathway by targeting miR-432. Eur Rev Med Pharmacol Sci.
21:2586–2595. 2017.PubMed/NCBI
|
|
22
|
Guo L, Liu X, Wang L, Sun H, Huang K, Li
X, Tang F, Li S, Yuan X and Wang C: Outcome of international
Federation of gynecology and obstetrics stage IIb cervical cancer
from 2003 to 2012: An evaluation of treatments and prognosis: A
retrospective study. Int J Gynecol Cancer. 25:910–918. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Cai X, Liu C, Zhang TN, Zhu YW, Dong X and
Xue P: Down-regulation of FN1 inhibits colorectal carcinogenesis by
suppressing proliferation, migration, and invasion. J Cell Biochem.
119:4717–4728. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ye Y, Zhuang J, Wang G, He S, Ni J and Xia
W: MicroRNA-139 targets fibronectin 1 to inhibit papillary thyroid
carcinoma progression. Oncol Lett. 14:7799–7806. 2017.PubMed/NCBI
|
|
26
|
Wang J, Deng L, Huang J, Cai R, Zhu X, Liu
F, Wang Q, Zhang J and Zheng Y: High expression of Fibronectin 1
suppresses apoptosis through the NF-κB pathway and is associated
with migration in nasopharyngeal carcinoma. Am J Transl Res.
9:4502–4511. 2017.PubMed/NCBI
|
|
27
|
Xu X, Liu Z, Zhou L, Xie H, Cheng J, Ling
Q, Wang J, Guo H, Wei X and Zheng S: Characterization of
genome-wide TFCP2 targets in hepatocellular carcinoma: Implication
of targets FN1 and TJP1 in metastasis. J Exp Clin Cancer Res.
34:62015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Diaz-Gonzalez Sdel M, Deas J,
Benitez-Boijseauneau O, Gómez-Cerón C, Bermúdez-Morales VH,
Rodríguez-Dorantes M, Pérez-Plasencia C and Peralta-Zaragoza O:
Utility of microRNAs and siRNAs in cervical carcinogenesis. Biomed
Res Int 2015. 3749242015.
|
|
29
|
Yang F, Guo L, Cao Y, Li S, Li J and Liu
M: MicroRNA-7-5p promotes cisplatin resistance of cervical cancer
cells and modulation of cellular energy homeostasis by regulating
the expression of the PARP-1 and BCL2 genes. Med Sci Monit.
24:6506–6516. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Shang A, Zhou C, Bian G, Chen W, Lu W,
Wang W and Li D: miR-381-3p restrains cervical cancer progression
by downregulating FGF7. J Cell Biochem. 120:778–789. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ruiz-Garcia E, Scott V, Machavoine C,
Bidart JM, Lacroix L, Delaloge S and Andre F: Gene expression
profiling identifies Fibronectin 1 and CXCL9 as candidate
biomarkers for breast cancer screening. Br J Cancer. 102:462–468.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhang H, Sun Z, Li Y, Fan D and Jiang H:
MicroRNA-200c binding to FN1 suppresses the proliferation,
migration and invasion of gastric cancer cells. Biomed
Pharmacother. 88:285–292. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Helleman J, Jansen MP, Span PN, van
Staveren IL, Massuger LF, Meijer-van Gelder ME, Sweep FC, Ewing PC,
van der Burg ME, Stoter G, et al: Molecular profiling of platinum
resistant ovarian cancer. Int J Cancer. 118:1963–1971. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Sponziello M, Rosignolo F, Celano M,
Maggisano V, Pecce V, De Rose RF, Lombardo GE, Durante C, Filetti
S, Damante G, et al: Fibronectin-1 expression is increased in
aggressive thyroid cancer and favors the migration and invasion of
cancer cells. Mol Cell Endocrinol. 431:123–132. 2016. View Article : Google Scholar : PubMed/NCBI
|