Introduction
Burkitt lymphoma (BL) is an aggressive form of
B-cell lymphoma that mostly affects children and adolescents. In
BL, dysregulation of the oncogenic transcription factor Myc is
considered to be the major driving force of lymphoma development
(1,2). However, the molecular mechanisms of the
pathogenesis of BL have not been fully elucidated.
Long non-coding RNAs (lncRNAs) are defined as
cellular RNA molecules, >200 base pairs in length, without
protein-coding capacity, which act as key regulators at
transcriptional and post-transcriptional levels (3,4).
Increasing evidence indicates that lncRNAs are involved in various
biological processes, including DNA replication, stem cell
pluripotency, proliferation and apoptosis (4,5). LncRNAs
serve a critical role in the development of various types of human
cancer, including haematopoietic malignancies, and may be potential
targets for tumour treatment (5).
Human plasmacytoma variant translocation 1
(PVT1), also known as the Pvt1 oncogene, is a lncRNA that is
homologous to the mouse plasmacytoma variant translocation gene.
PVT1 is located on chromosome 8, ~55 kb distal to the MYC
proto-oncogene bHLH transcription factor (c-Myc) gene, and
is frequently involved in translocations that occur in variant BL
(6,7). The overexpression of PVT1 is one
of the most frequent events in a variety of malignant diseases,
including melanoma (8),
hepatocellular carcinoma (9,10), thyroid cancer and colorectal cancer
(11,12). A number of studies have demonstrated
that lncRNA PVT1 interacts with the proliferation-associated
nucleolar proteins NOP2 or c-Myc, stabilizes these proteins against
degradation, and negatively modulates microRNA (miRNA) as a
competing endogenous RNA or a molecular sponge, in order to exert a
tumour-promoting effect (8,10,13,14). A
large genome-wide association study identified one high-risk single
nucleotide polymorphism (SNP; rs2608053) for classic Hodgkin
lymphoma at 8q24 near the Myc/PVT1 locus, which is
associated with patient outcome (15). In a meta-analysis, two independent
SNPs, rs13255292 and rs4733601, at 8q24.21 were identified for
diffuse large B cell lymphoma (16).
However, the functional role and molecular mechanism of PVT1
in BL remain unclear.
In the present study, knockdown of PVT1 was
able to inhibit Raji cell growth by regulating cell cycle
progression. Furthermore, it was revealed that PVT1 may
serve an important role in G0/G1 arrest,
which may be associated with the expression of c-Myc and
cell cycle-associated genes. Together, these results indicated that
lncRNA PVT1 may serve a critical role in Raji cell
proliferation, and may be considered a candidate target for novel
treatment of human BL.
Materials and methods
Cell culture and transfection
The Raji cell line was purchased from the Cell Bank
of Type Culture Collection of Chinese Academy of Sciences
(Shanghai, China. http://www.cellbank.org.cn/index.asp). Cells were
cultured in RPMI-1640 medium (Gibco; Thermo Fisher Scientific,
Inc., Waltham, MA, USA), supplemented with 10% heat-inactivated
foetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.) at
37°C in a humidified incubator with 5% CO2. Four small
interfering RNA (siRNA) sequences targeting PVT1 (siRNA54,
siRNA176, siRNA845, siRNA1055) and a scrambled control (SC) siRNA
were designed and synthesized by Shanghai GenePharma Co., Ltd.
(Shanghai, China). The sequences of the siRNA are as follows:
PVT1-siRNA54: 5′-CCUGAUGGAUUUACAGUGATT-3′,
PVT1-siRNA176: 5′-GCUGAAUGCCUCAUGGAUUTT-3′,
PVT1-siRNA845: 5′-CCUGUUACACCUGGGAUUUTT-3′,
PVT1-siRNA1055: 5′-GCUUCUCCUGUUGCUGCUATT-3′, SC-siRNA:
5′-GCUACGAUCUGCCUAAGAUTT-3′. Raji cells (3–4×105
cells/ml) in the exponential growth phase were grown for 24 h, then
PVT1-siRNA54, -siRNA176, -siRNA845 and-siRNA1055 were
transfected into Raji cells using HiPerfect (Qiagen, Inc.,
Valencia, CA, USA) according to the manufacturer's protocol. In
addition to non-silencing SC siRNA control, cells in the mock
transfection group were treated with HiPerfect agents only. The
total concentration of siRNA applied in eachcase was 100 nM. At 24
and 48 h post-transfection, silencing of PVT1 RNA was
examined.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA from the Raji cells of post-transfection
was isolated using a TRIzol® total RNA isolation system
(Invitrogen; Thermo Fisher Scientific, Inc.). RNA purity and
concentration were measured using a spectrophotometer, and RNA was
reverse transcribed into first-strand cDNA using random hexamer
primers and the reverse transcriptase Superscript II kit (Toyobo
Life Science, Osaka, Japan), according to the manufacturer's
protocol. The 2−ΔCt method (17) was used to analyse the relative
changes in gene expression in RT-qPCR experiments with SYBR Green
(Toyobo Life Science, Japan). The primers were designed and
synthesized by Shanghai GeneChem Co., Ltd. (Shanghai, China). The
primer sequences are listed in Table
I. GAPDH was used as a reference gene. The total PCR
reaction volume was 20 µl and reaction conditions were as follows:
Enzyme activation at 95°C for 10 min, followed by 40 cycles at 95°C
for 15 sec, 60°C for 15 sec and 72°C for 32 sec. At the end of each
run a melting curve was performed, starting at 65°C and reaching
95°C with an increase of 1°C/2 sec, to verify primer specificities,
specificity of amplification and absence of primer dimers. RT-qPCR
was repeated in at least three separate experiments.
 | Table I.Sequences of primers used for reverse
transcription-quantitative polymerase chain reaction. |
Table I.
Sequences of primers used for reverse
transcription-quantitative polymerase chain reaction.
| Gene | Forward primer
(5′-3′) | Reverse primer
(5′-3′) |
|---|
| PVT1 |
GTCTTGGTGCTCTGTGTTC |
CCCGTTATTCTGTCCTTCT |
| CCNG2 |
GGTTTCACCTTCATAAGAGCC |
GCTGAGTTTGATTGAGGCTAC |
| CDKN1A |
AGCGACCTTCCTCATCCACC |
AAGACAACTACTCCCAGCCCCATA |
| HUS1 |
ATGGGTCACAATGCGGCTACT |
GCTAACATCGGAAAACTTATCTCG |
| CDKN1B |
GGGCAAGTACGAGTGGCAAGAG |
CAAATGCGTGTCCTCAGAGTTAGC |
| CDKN3 |
AGTCCCAAACCTTCTGGATCTCTAC |
CTCCCAAGTCCTCCATAGCAGTG |
| RBL2 |
TTCTGGTAGTGCTGGCTGGTG |
GGGTGACTGAAGTTCGTGCTG |
| CCNE1 |
AAAGGTTTCAGGGTATCAGTGGTG |
TCTCTGTGGGTCTGTATGTTGTGTG |
| CCND1 |
CCCTCGGTGTCCTACTTCAAATGT |
GGAAGCGGTCCAGGTAGTTCAT |
| CDC20 |
TCACCAGAGCTTGCACTCCAC |
ACCTGCCGTTACATTCCTTCC |
| GAPDH |
GGACCTGACCTGCCGTCTAG |
GTAGCCCAGGATGCCCTTGA |
Western blot analysis
Cells were washed with PBS (10 mM, pH 7.4) and
incubated in 200 µl cell lysis buffer (Beyotime Institute of
Biotechnology) on ice for 30 min, and centrifuged at 13,000 × g for
15 min at 4°C. The protein content of the cell lysate was
determined by Bio-Rad protein assay (Bio-Rad Laboratories, Inc.,
Hercules, CA, USA), according to the manufacturer's protocol. Whole
cell extracts equivalent to 100 µg total protein were separated by
8% SDS-PAGE and transferred to nitrocellulose membranes (Gibco;
Thermo Fisher Scientific, Inc.) at 18 V for 10–15 min. The blots
were immersed in blocking buffer (10% non-fat dry milk, 1%
Tween-20, 20 mM Tris-buffered saline, pH 7.5) for 1 h at room
temperature, and were then incubated with appropriate anti-human
primary antibodies [rabbit immunoglobulin G (IgG) anti-c-Myc
(1:1,000 dilution; cat. no., sc-40, Santa Cruz Biotechnology, Inc.,
Dallas, TX, USA), anti-P21 (1:1,000 dilution; cat. no., ab109520,
Abcam, Cambridge, MA, USA) or anti-cyclin E1 (CCNE1) (1:1,000
dilution; cat. no., ab33911, Abcam, Cambridge, MA, USA USA), or
mouse anti-GAPDH/IgG (1:1,000 dilution; cat. no., ab8245, Abcam,
Cambridge, MA, USA) USA] in blocking buffer overnight at 4°C. Blots
were then incubated with anti-rabbit (cat. no., sc-2357) or
anti-mouse (cat. no., sc-516102) horseradish peroxidase-conjugated
secondary antibodies (1:2,000 dilution; Santa Cruz Biotechnology,
Inc.) for 1 h at room temperature and bands were detected by
chemiluminescence using Enhanced Chemiluminescence Hyperfilm (EMD,
Millpore, USA).
PVT1 short hairpinRNA
(shRNA)-expressing plasmid construction and cloning screen
To stably knockdown the expression of PVT1, a
shRNA sequence targeting PVT1 (siRNA1055) was cloned into
the pGV248-lentivirus vector (Shanghai GenePharma Co., Ltd.).
Subsequently, PVT1 knockdown vectors were reconstructed and
sequenced. pGV248 vector containing the negative control (NC) shRNA
was used as a control. Subsequently, 293T cells from the Cell Bank
of Type Culture Collection of Chinese Academy of Sciences
(Shanghai, China) were cultured in Dulbecco's modified Eagle's
medium (Gibco; ThermoFisher Scientific, Inc., Waltham, MA, USA)
containing 10% FBS, maintained at 37°C in a humidified incubator
with 5% CO2 and transfected with pGV248-shRNAs, Helper
1.0 and Helper 2.0 (Shanghai GenePharma Co., Ltd.) using
Lipofectamine® 2000 reagent (Invitrogen; Thermo Fisher
Scientific, Inc.). The media were replaced with 10 ml fresh medium
after incubation overnight. The virus-containing supernatants
(LV-PVT1-shRNA and LV-NC-shRNA) were collected after 48 h.
Raji cells were infected, respectively, with LV-PVT1-shRNA
and LV-NC-shRNA (multiplicity of infection=250) at 37°C for 72 h,
and were then selected using 4 µg/ml puromycin to screen single
cell clones for 2 weeks and expand the culture for 4 weeks. Green
fluorescence of cells was observed by inverted fluorescence
microscopy. The knockdown efficiency was measured using RT-qPCR.
Cell proliferation and cell cycle distribution were analysed in
positive clone Raji cells. In subsequent assays, Raji cells were
divided into three groups: Blank control group (cells without
infection), NC group (cells with LV-NC-shRNA) and PVT1
knockdown group (cells with LV-PVT1-shRNA).
Cell proliferation assay
For the quantitative determination of cellular
proliferation, a Cell Counting kit-8 (CCK8; Dojindo Molecular
Technologies, Inc., Kumamoto, Japan) assay was performed. The assay
was performed for Raji cells carrying LV-PVT1-shRNA and
LV-NC-shRNA. Cells were seeded at a density of 5×104
cells/well in 96-well plates and cultured at 37°C with 5%
CO2 in a humidified incubator. According to the
manufacturer's protoocols 10 µl CCK8 solution was added 4 h prior
to the end of incubation at 37°C at 24, 48, 72 and 96 h. Cell
proliferation was measured using a spectrophotometer (Bio-Rad
Laboratories, Inc.) at an absorbance wavelength of 450 nm. All
experiments were repeated three times.
Cell cycle assay
Raji cells containing LV-PVT1-shRNA or
LV-NC-shRNA were harvested. The cells were fixed with 70% ethanol
at −20°C overnight and stained with propidium iodide (5 µg/ml;
Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) in the presence of
ribonuclease A (1 mg/ml; Sigma-Aldrich; Merck KGaA) for 30 min at
room temperature. The cell cycle distribution was analysed via flow
cytometry (BD Biosciences, San Jose, CA, USA). All experiments were
repeated three times.
RT-PCR-based array analysis
Human Cell Cycle RT2 RNA QC PCR arrays
(Qiagen, Inc.) were used to screen a panel of 84 representative
cell cycle-associated genes in Raji cells infected with
LV-PVT1-shRNA. Total RNA was isolated from the
LV-PVT1-shRNA and LV-NC-shRNA cells using the Qiagen RNeasy
Mini kit (Qiagen, Inc.) according to the manufacturer's protocol.
RNA was quantified by the NanoDrop® ND-1000 (NanoDrop;
Thermo Scientific, Inc., Wilmington, DE, USA), and quality was
assessed by visualizing 18S and 28S ribosomal RNA bands separated
by 1% agarose gel electrophoresis with ethidium bromide staining
(Sigma-Aldrich; Merck KGaA). According to manufacturer's protocol,
the first-strand cDNA was obtained using an RT2 First
Strand kit (Qiagen, Inc.). qPCR was conducted using 2X
RT2 SYBR Green qPCR Master Mix (Qiagen, Inc.) on a
Bio-Rad Real-Time PCR system (Bio-Rad Laboratories, Inc.) according
to the RT2 Profiler PCR array protocols under the
following conditions: 95°C for 10 min, then 40 cycles at 95°C for
15 sec and 60°C for 1 min. At the end of each run a melting curve
was performed, extending at 60°C for 1 min. Microarray data were
normalized for housekeeping genes (ACTB, B2M, GAPDH, HPRT1,
RPLP0) by calculating the ΔCq and 2−ΔΔCq (17) for each gene of interest in the plate.
Fold changes of each gene between the LV-PVT1-shRNA and
LV-NC-shRNA groups were calculated as 2−ΔΔCt, and
scatter plots were analysed.
Statistical analysis
All data are expressed as the means ± standard
deviation. Differences among three or more groups were compared
using one-way analysis of variance (ANOVA), followed by SNK post
hoc test. All data were analysed using SPSS v13.0 software (SPSS,
Inc., Chicago, IL, USA). P<0.05 was considered to indicate a
statistically significant difference.
Results
PVT1 siRNAs or shRNA suppress PVT1 RNA
and c-Myc protein expression in Raji cells
To determine the efficiency of PVT1
inhibition following siRNA (siRNA54, siRNA176, siRNA845, siRNA1055)
transfection, PVT1 RNA expression was analysed. RT-qPCR data
were obtained from at least three independent experiments. The
relative qPCR formula: 2−ΔCt ×100% was used. As shown in
Fig. 1A, at 24 and 48 h after
transient transfection, the relative expression levels of
PVT1 RNA in Raji cells transfected with PVT1 siRNA
(siRNA1055)were significantly lower than in cells transfected with
the control SC siRNA (P<0.05). Conversely, siRNA54, siRNA176,
siRNA845 had no significant effect on the expression of PVT1
RNA when compared respectively with the control SC siRNA. A
specific siRNA (siRNA1055) against PVT1 was used to stably
knockdown PVT1 expression. Subsequently, lentiviral vectors
carrying LV-PVT1-shRNA or LV-NC-shRNA were successfully
constructed. Raji cells stably carrying PVT1-shRNA or
NC-shRNA were screened. Raji cells expressing LV-PVT1-shRNA
were observed to exhibit green fluorescence under an inverted
fluorescence microscope (data not shown). As shown in Fig. 1B, following stable infection with the
LV-PVT1-shRNA expression vector, the expression levels of
PVT1 RNA were significantly reduced compared with in the
LV-NC-shRNA and blank control cell groups (P<0.05).
Additionally, no significant differences between the LV-NC-shRNA
group and the blank control cell group were identified
(P>0.05).
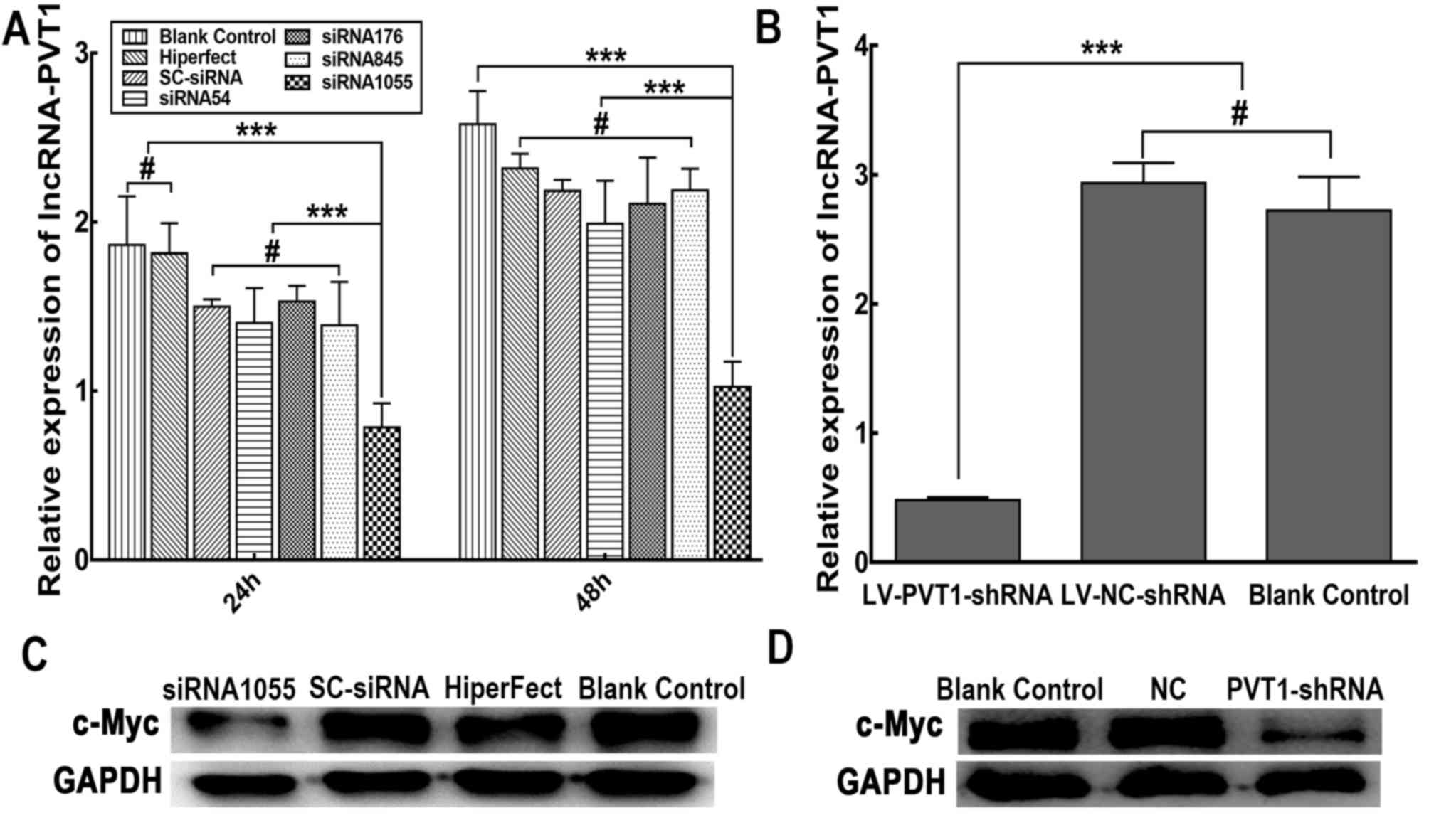 | Figure 1.Effect of PVT1 siRNAs or shRNA
on PVT1 RNA and c-Myc protein expression in Raji cells. (A)
Suppression of PVT1 RNA expression was measured by reverse
transcription-quantitative polymerase chain reaction at 24 and 48 h
after transient transfection with PVT1 siRNAs (siRNA54,
siRNA176, siRNA845 and siRNA1055). Non-silencing SC
siRNA-transfected, mock-transfected (HiPerfect reagents only) and
non-treated (blank control) cells were used as controls.
GAPDH was used as the reference gene. (B) Expression levels
of PVT1 RNA in Raji cells stably infected with
LV-PVT1-shRNA and LV-NC-shRNA. (C) Expression of c-Myc
protein in Raji cells at 48 h after PVT1-siRNA transfection,
assessed by western blotting. (D) Protein expression levels of
c-Myc in Raji cells stably infected with LV-PVT1-shRNA.
Representative images are shown. The results are expressed as the
mean values of three independent experiments ± standard deviation.
***P<0.01, #P>0.05. c-Myc, MYC proto-oncogene bHLH
transcription factor; lncRNA, long non-coding RNA; LV, lentiviral
vector; NC, negative control; PVT1, plasmacytoma variant
translocation 1; SC, scrambled; shRNA, short hairpin RNA; siRNA,
small interfering RNA. |
The protein expression levels of c-Myc were assessed
by western blotting. As shown in Fig.
1C, western blot analysis confirmed that c-Myc protein
expression was decreased in the Raji cells transfected with
PVT1 siRNA1055 after 48 h compared with in the SC siRNA and
blank control cell groups. There were no clear differences in c-Myc
expression among the SC siRNA group, mock-transfected cells and
blank control cells. Similarly, c-Myc protein expression was
decreased in Raji cells stably carrying LV-PVT1-shRNA
compared with either the LV-NC-shRNA group or blank control group
(Fig. 1D). There was no difference
identified between c-Myc expression in the LV-NC-shRNA group and
blank control group. These findings indicated that PVT1
siRNA and shRNA were effective at reducing c-Myc protein
expression.
Effect of PVT1 knockdown on the
proliferation and cell cycle distribution of Raji cells
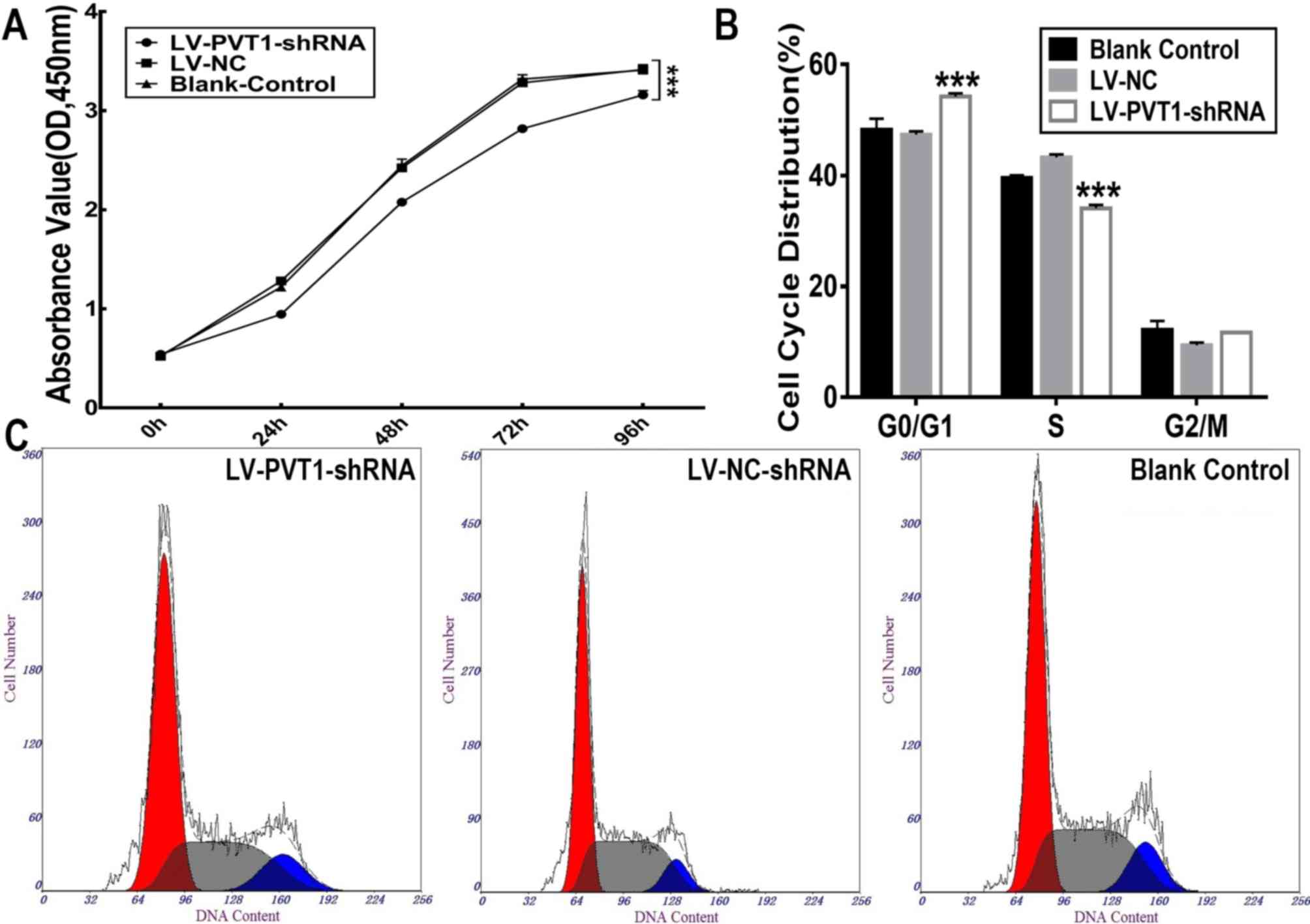
A CCK8 assay was performed to quantify cell
proliferation. As shown in Fig. 2A,
the proliferative activity of Raji cells carrying the
LV-PVT1-shRNA was significantly inhibited (P<0.01)
compared with Raji cells carrying LV-NC-shRNA and blank control
cells (P<0.01). No significant difference in proliferation of
LV-NC-shRNA group and blank control cells was identified.
PVT1 siRNA had a similar effect on the proliferative
activity of Raji cells (data not shown).
Proliferation of Raji cells was inhibited following
PVT1 knockdown. Additionally, the effect of decreased
PVT1 expression on the cell cycle was examined. According to
flow cytometric analysis, the percentages of cells in
G0/G1, S and G2/M phases were 54.
20±0.61, 34.07±0.64 and 11.70±0.00% in Raji cells carrying
LV-PVT1-shRNA, respectively. In Raji cells carrying
LV-NC-shRNA, the percentages of cells in
G0/G1, S and G2/M phases were
47.37±0.60, 43.27±0.55 and 9.37±0.51%, respectively. There was a
significant decrease in the percentage of cells in the S phase and
a marked accumulation of cells in the G0/G1
phase in Raji cells carrying LV-PVT1-shRNA compared with
those carrying LV-NC-shRNA and blank control cells (Fig. 2B and C). There was no significant
difference identified between the Raji cells carrying LV-NC-shRNA
and blank control cells. These results indicated that knockdown of
PVT1 expression in Raji cells induced
G0/G1 phase arrest.
Alteration in expression pattern of
cell cycle-associated genes in Raji cells as a result of PVT1
knockdown
Differential mRNA expression levels of 84 genes
involved in the cell cycle were assessed. The results of the PCR
array revealed that 54 genes were upregulated and 26 genes were
downregulated in the LV-PVT1-shRNA group compared with the
LV-NC-shRNA group. The other four genes exhibited too low an
expression level. There were more upregulated genes than
downregulated genes in Raji cells carrying LV-PVT1-shRNA
(Fig. 3A; Table II). The present study demonstrated
that 16/84 examined genes were upregulated at least two-fold in the
LV-PVT1-shRNA group.
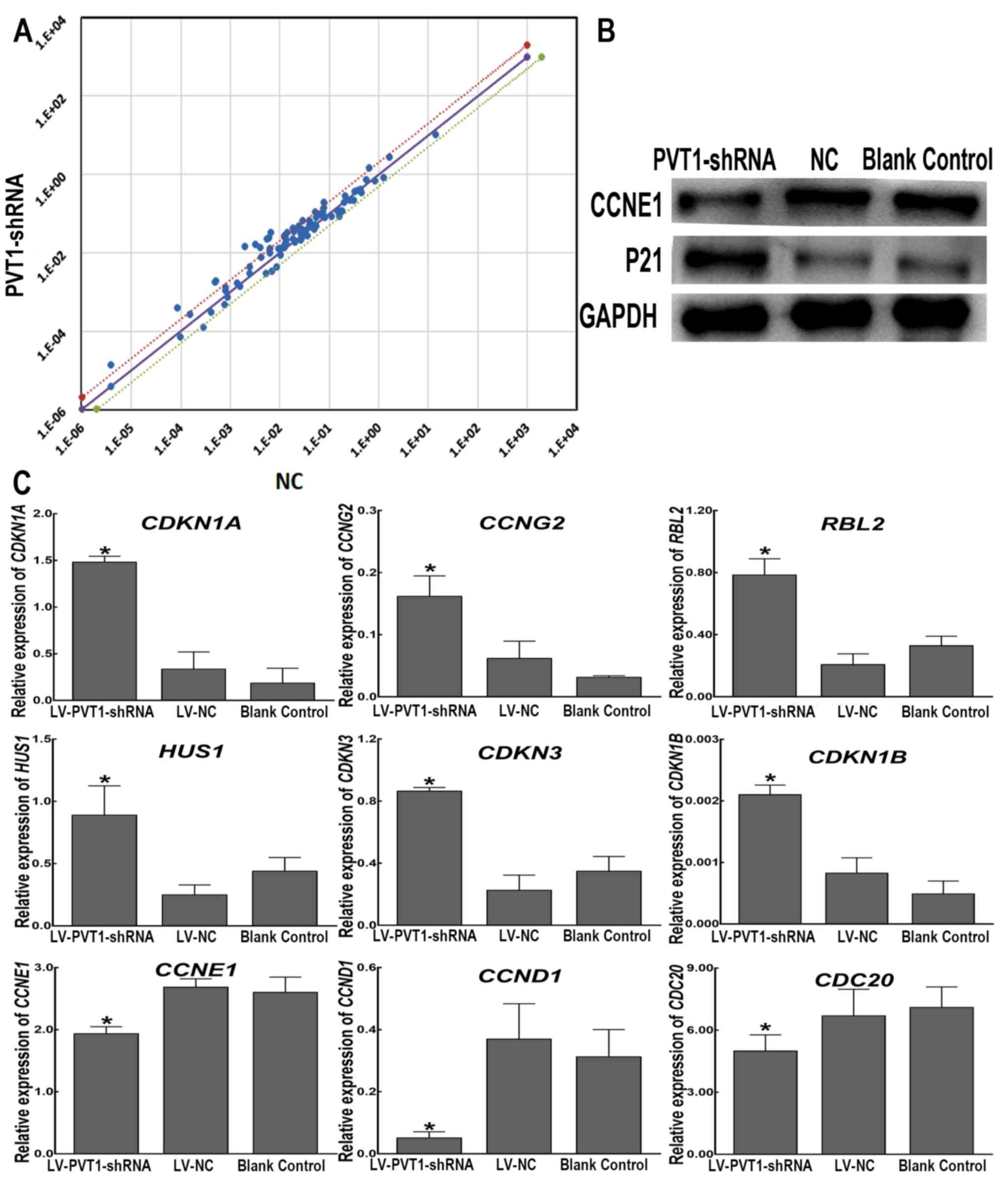 | Figure 3.Differential expression of cell
cycle-associated genes in Raji cells with PVT1 knockdown.
(A) There are three straight lines in the coordinate system, and
the black line represents the fold changes (2−ΔΔCt) of
1. Dots above the black line represent genes that are upregulated,
and dots below the black line indicate genes that are downregulated
in the LV-PVT1-shRNA group compared with the LV-NC-shRNA
group. A point within the range of the two dotted lines signifies a
differential expression of >1-fold but <2-fold in Raji cells
infected with LV-PVT1-shRNA compared with cells infected
with LV-NC-shRNA. Points outside the two dotted lines are
>2-fold differentially expressed. (B) Protein expression levels
of P21 and CCNE1 in Raji cells were determined using western blot
analysis following PVT1 knockdown. GAPDH served as a loading
control. (C) Reverse transcription-quantitative polymerase chain
reaction analysis of nine genes in Raji cells stably infected with
LV-PVT1-shRNA compared with the genes in the negative
control cells infected with LV-NC-shRNA. GAPDH was used as a
reference gene. The graphs depict the mean mRNA expression changes
± standard deviation of three independent experiments. *P<0.05
vs. LV-NC-shRNA. CCNE1, cyclin E1; CCND1, cyclin D1;
CCNG2, cyclin G2; CDC20, cell division cycle 20
homolog (S. cerevisiae); CDKN1A, cyclin-dependent
kinase inhibitor 1A (P21, Cip1); CDKN1B, cyclin-dependent
kinase inhibitor 1B (P27, Kip1); CDKN3, cyclin-dependent
kinase inhibitor 3; HUS1, HUS1 checkpoint homolog (S.
pombe); LV, lentiviral vector; NC, negative control;
PVT1, plasmacytoma variant translocation 1; RBL2,
Retinoblastoma-like 2 (p130); shRNA, short hairpin RNA. |
 | Table II.Differentially expressed genes
associated with the cell cycle in Raji cells followingPVT1
knockdown. |
Table II.
Differentially expressed genes
associated with the cell cycle in Raji cells followingPVT1
knockdown.
| Gene symbol | Accession no. | Gene
description | Fold change |
|---|
| CCNG2 | NM_004354 | Cyclin G2 | 7.03 |
| RBL2 | NM_005611 | Retinoblastoma-like
2 (p130) | 5.26 |
| CDKN1A | NM_000389 | Cyclin-dependent
kinase inhibitor 1A (P21, Cip1) | 4.78 |
| CCNT1 | NM_001240 | Cyclin T1 | 4.17 |
| CASP3 | NM_004346 | Caspase 3,
apoptosis-related cysteine peptidase | 3.96 |
| RBL1 | NM_002895 | Retinoblastoma-like
1 (p107) | 3.70 |
| RB1 | NM_000321 | Retinoblastoma
1 | 3.68 |
| HUS1 | NM_004507 | HUS1 checkpoint
homolog (S. pombe) | 3.38 |
| CCND2 | NM_001759 | Cyclin D2 | 2.51 |
| CDC6 | NM_001254 | Cell division cycle
6 homolog (S. cerevisiae) | 2.34 |
| CDKN1B | NM_004064 | Cyclin-dependent
kinase inhibitor 1B (p27, Kip1) | 2.31 |
| TFDP2 | NM_006286 | Transcription
factor Dp-2 (E2F dimerization partner 2) | 2.23 |
| CDC16 | NM_003903 | Cell division cycle
16 homolog (S. cerevisiae) | 2.21 |
| GADD45A | NM_001924 | Growth arrest and
DNA-damage-inducible, alpha | 2.08 |
| CDKN3 | NM_005192 | Cyclin-dependent
kinase inhibitor 3 | 2.07 |
| ABL1 | NM_005157 | C-abl oncogene 1,
non-receptor tyrosine kinase | 2.04 |
| BRCA2 | NM_000059 | Breast cancer 2,
early onset | −2.22 |
| CCNE1 | NM_001238 | Cyclin E1 | −2.05 |
| CCND1 | NM_053056 | Cyclin D1 | −1.98 |
| CDK6 | NM_001259 | Cyclin-dependent
kinase 6 | −1.85 |
| CDC20 | NM_001255 | Cell division cycle
20 homolog (S. cerevisiae) | −1.81 |
| ATM | NM_000051 | Ataxia
telangiectasia mutated | −1.67 |
| CKS2 | NM_001827 | CDC28 protein
kinase regulatory subunit 2 | −1.56 |
| CDK4 | NM_000075 | Cyclin-dependent
kinase 4 | −1.52 |
Candidate gene selected from PCR microarray analyses
was validated. As shown in Fig. 3B,
western blotting revealed that P21, encoded by cyclin-dependent
kinase inhibitor 1A (CDKN1A) was increased and CCNE1 was
decreased in the LV-PVT1-shRNA group compared within the
LV-NC-shRNA group and in blank control cells. As shown in Fig. 3C, the RT-qPCR assay demonstrated that
the expression levels of cyclin G2 (CCNG2),
Retinoblastoma-like 2 (RBL2, p130), CDKN1A, HUS1
checkpoint homolog (HUS1), cyclin dependent kinase inhibitor
3 (CDKN3) and cyclin dependent kinase inhibitor 1B
(CDKN1B)were upregulated in Raji cells infected with
LV-PVT1-shRNA compared with the LV-NC-shRNA group and blank
control cells. Although the expression of CDKN1B was very
low in the three groups, there was statistically significant
difference between the LV-PVT1-shRNA group and the
LV-NC-shRNA group (P<0.05). In addition, CCNE1, CCND1 and
cell division cycle 20 (CDC20) were significantly
downregulated in the LV-PVT1-shRNA group compared with in
the LV-NC-shRNA group and blank control cells (P<0.05). The
RT-qPCR assay results of the 9 selected differentially expressed
genes did not change notably compared with the PCR array
results.
Discussion
PVT1 is aberrantly expressed in a variety of
tumour types and acts as a potential oncogene that promotes cancer
cell proliferation (8–12). Gain of PVT1 lncRNA expression
is required for high MYC protein levels in 8q24-amplified human
cancer cells (13). PVT1 RNA
and MYC protein expression are correlated in primary human tumours,
and copy number of PVT1 co-increases in >98% of
MYC-copy-increase types of cancer (13). Although it was reported in 1990 that
the PVT1 locus may be a site of variant translocations,
including in human BL (6), to the
best of our knowledge, there are no reports about the role of
PVT1 in BL. To investigate the role of PVT1 in BL
cell proliferation, RNA interference was used to inhibit its
expression in the human BL-derived Raji cell line. The results of
the present study revealed that the expression levels of
PVT1 RNA and c-Myc protein decreased following transfection
of Raji cells with PVT1 siRNA targeting the 1,055-1,074 nt
region of the PVT1 sequence. No significant effect of the
other three siRNAs targeting PVT1 (siRNA54, siRNA176 and
siRNA845) on the expression levels of PVT1 RNA was
identified. The specific siRNA against PVT1, siRNA1055, was
successfully incorporated into a lentiviral vector. Raji cells
carrying the PVT1-shRNA lentiviral vector were screened.
Once Raji cells stably expressed PVT1-shRNA, the levels of
PVT1 RNA and c-Myc protein were markedly reduced, which is
consistent with the effect of PVT1 siRNA1055.
In the present study, the proliferation of Raji
cells carrying PVT1-shRNA was significantly decreased
compared with in control cells. However, downregulation of
PVT1 expression by shRNA could not induce apoptosis of Raji
cells (data not shown). Cell cycle distribution analysis indicated
that knockdown of PVT1 in Raji cells resulted in notable
G0/G1 phase arrest. Previous studies have
reported that PVT1 can promote the proliferation of cells,
including hepatocellular carcinoma (10) and thyroid cancer cells (11). The results of the present study were
consistent with those of previous studies. Taken together, the
results suggested that the PVT1 shRNA-mediated suppression
of Raji cell proliferation may occur via cell cycle arrest.
PVT1 may be associated with a series of
signalling pathways and genes in tumour development and progression
(18–20). According to topological measures,
Paci et al (20) revealed
that the lncRNA PVT1 is connected to 753 different mRNAs
(~50% of total mRNAs in the network), and the miR-200 family
members mediate >80% of these interactions. However, the
mechanism by which knockdown of PVT1 causes cell cycle
alteration in Raji cells remains largely unexplained. It has been
confirmed that PVT1 is located adjacent to the
proto-oncogene c-Myc, and the expression of PVT1-Myc
in the majority of tumours is positively correlated with the copy
number of 8q24 (13,15,21).
Tseng et al (13)
demonstrated that c-Myc protein expression is dependent on the
expression levels of PVT1; therefore, this lncRNA may
promote cell proliferation and tumourigenesis by regulating the
expression of c-Myc protein. Following transfection of Raji cells
with PVT1-siRNA or shRNA, the expression of c-Myc protein
decreased, which was consistent with previous reports (13). It has been demonstrated that c-Myc is
involved in regulation of the cell cycle and proliferation
(22). Yang et al (23) revealed that c-Myc regulates the
cyclin-dependent kinase1 (CDK1)/cyclin B1-dependent G2/M
cell cycle progression by controlling histone H4 acetylation.
Additionally, a study has reported that loss of c-Myc function
impedes G1-phase progression (22). The CDK inhibitors P27 and P21 appear
to be critical targets of c-Myc, which cooperates with the zinc
finger MIZ-type containing 1 to repress the transcription of
cell-cycle inhibitors, including P15, P21 and P27 (24). Yang et al (23) also reported that suppression of c-Myc
induces the upregulation of P21 and the downregulation of P27 in
Raji cells. Recently, Wang et al (25) reported that the epigenetically
induced lncRNA EPIC1 promotes cell cycle progression by
interacting with c-Myc via the 129–283 nt region of EPIC1.
EPIC1 knockdown reduces the affinity of c-Myc to its target
genes, including CDKN1A, CCNA2, CDC20 and CDC45
(25). The results of the present
study suggested that the regulation of G0/G1
cell cycle progression by PVT1 may be associated with c-Myc
expression or c-Myc regulation of cell cycle-associated genes.
To further evaluate the alterations in cell
cycle-associated gene expression in Raji cells with PVT1
knockdown, a cell cycle PCR microarray was used to detect
alterations in cell cycle-associated genes in Raji cells stably
expressing PVT1-shRNA. The results revealed that cell
cycle-associated genes, including CCNG2, RBL2, CDKN1A, HUS1,
CDKN3 and CDKN1B, were upregulated in Raji cells with
PVT1 knockdown. The majority of the aforementioned genes
inhibit cell cycle progression. CDKN1A encodes a cell
cycle-dependent kinase inhibitor, which binds and inhibits the
activity of cell cycle-associated genes with cyclin and CDK
complexes, including CCND1-CDK4 and CCNE-CDK2. P21 is
a member of the cell cycle regulator CIP/KIP family, which is
involved in the regulation of numerous important biological
behaviours, the most important of which is the regulation of cell
cycle progression, particularly cell cycle arrest at the
G1 phase (26,27). Therefore, P21 serves a key role in
cell quiescence, senescence and differentiation (28). In addition, a recent study identified
a positive correlation between PVT1 and miR-1207-5P, and a
negative correlation between miR-1207-5P and signal transducer and
activator of transcription 6 (STAT6) in breast cancer
(29). miR-1207-5P, produced by
PVT1 transcription and targeting STAT6, promotes the
proliferation of breast cancer cells by regulating P21 and CDKN1B
(29). In nasopharyngeal cancer
cells, PVT1 promotes cancer stem cell-like properties by
inhibiting miR-1207 and activating the phosphoinositide
3-kinase/protein kinase B signalling pathway (30). In the present study, following the
downregulation of PVT1 expression in Raji cells, the RNA
expression levels of CDKN1A and P21 protein expression were
increased, and the cell cycle was blocked in
G0/G1 phase, suggesting that PVT1 may
promote cell cycle progression by inhibiting the expression of P21.
In pancreatic cancer cell lines, PVT1 can promote
epithelial-to-mesenchymal transition by downregulating the
expression of P21, thus promoting cell proliferation and migration
(31). In addition, a study by Cui
et al (32) suggested that
PVT1 promotes cell proliferation and migration by
downregulating P21 in pancreatic cancer cells. Therefore, the
results of the present study were consistent with the
literature.
CCNG2, which has been shown to be associated
with various types of tumour, can cause cell cycle arrest in the
G1 phase (33–37). A study by Cui et al (37) indicated that CCNG2 expression
is decreased in prostate cancer and that PC-3 cells transfected
with CCNG2 exhibit a lower survival rate, a higher
percentage of cells in the G0/G1 phases and
lower CDK2 protein expression, suggesting that CCNG2 may
serve important roles as a negative regulator of prostate cancer
cells. The results of the present study demonstrated that the
downregulation of PVT1 in Raji cells increased the
expression of CCNG2, which suggested that PVT1 may
also promote cell cycle progression by inhibiting the expression of
CCNG2. The RBL2 gene, encoding the proline rich
protein BstNI subfamily 2 (pRb2) protein, is most abundant in the
G0 phase. RBL2 maintains G0 arrest in
quiescent or differentiated cells, controls the transition from
G1 to S phase, and is a key regulator of growth arrest
in cellular senescence (38). AKT
inhibition reduces cell viability, induces cell accumulation in
G0/G1 phase and triggers apoptosis, which
proves to be largely dependent on RBL2/pRb2 itself, as shown by
RBL2/pRb2 silencing (39). The HUS1
protein, encoded by the HUS1 gene, is a component of an
evolutionarily conserved and genotoxin-activated checkpoint complex
that is involved in cell cycle arrest in response to DNA damage. In
the present study, the expression levels of the cell cycle
progression regulators RBL2 and HUS1 were increased
in Raji cells with PVT1 knockdown, suggesting that the
involvement of PVT1 in cell cycle progression may be
associated with the expression of RBL2 and HUS1.
CDKN3 (also called CDI1 or KAP), encoded by the CDKN3 gene,
is a member of the dual specificity protein phosphatase family,
which is essential for mitosis and G1/S phase
transition, due to its role in regulating the CDK1 (also called
CDC2) signalling axis (40), and
CDKN3 serves an important role in regulating cell division.
CDKN3 may also function as an oncogene by inducing
G0/G1 phase progression, apoptosis and
metastasis in ovarian cancer cell lines (41). In the present study, the expression
levels of CDKN3 were increased in Raji cells with
PVT1 knockdown, suggesting that CDKN3 may be involved
in the cell cycle arrest by the induction of PVT1
downregulation.
In the present study, CCNE1, CCND1 and
CDC20 were significantly downregulated in Raji cells
following knockdown of PVT1. CCNE1 is a positive
regulator of the cell cycle that controls the transition of cells
from the G1 to the S phase, and serves an important role
in cell proliferation and tumourigenesis. Overexpression of
CCNE1 can increase tumour incidence and susceptibility to
multiple tumourigenesis in mice (42–44).
CCND1, which belongs to the G1 cyclins, serves an
important role in cell cycle regulation and promotes cell cycle
progression from the G1 phase to S phase through its
interaction with CDKs. CCND1 is overexpressed and/or
amplified in numerous human types of cancer (45,46). A
previous study reported that CCND1 allows for the progression from
the G1 to the S phase by binding and sequestering P21
(47). A study by Li et al
(48) suggested that PVT1
promotes cell cycle progression by increasing the expression of
CCND1. The mechanism involved indicated that PVT1
promotes the progression of clear cell renal cell carcinoma partly
via activation of the epidermal growth factor receptor pathway
(48). In melanoma cells, silencing
of PVT1 significantly inhibits cell proliferation, migration
and invasion, and arrests the cell cycle at
G0/G1 stage by significantly decreasing
CCND1 expression (49).
CDC20, which has important functions in chromosome segregation and
mitotic exit, is abnormally expressed in a wide range of tumours,
including human bladder carcinoma, pancreatic, colorectal breast
and lung cell cancer (50). It has
been reported that CDC20 were significantly downregulated by lncRNA
EPIC1 knockdown in both tumor samples and cell lines (25). Therefore, the results of the present
study suggested that the role of PVT1 in regulating
G0/G1 cell cycle progression was likely to be
associated with the expression levels of CCNE1, CCND1 and
CDC20.
In conclusion, the results of the present study
revealed that knockdown of PVT1 may inhibit the
proliferation of Raji cells by arresting cells in the
G0/G1 phase. The role of PVT1 in
regulating cell cycle progression may be associated with the
expression of c-Myc and cell cycle-associated genes, including
CCNG2, RBL2, CDKN1A, HUS1, CDKN3, CDKN1B, CCNE1, CCND1 and
CDC20. However, it remains unclear how PVT1
influences the expression levels of cell cycle-associated genes.
Furthermore, it is not clear which signalling pathways could be
directly associated with PVT1. Therefore, further studies
are required to understand the potential molecular mechanisms that
underlie the regulation of cell cycle-associated genes by
PVT1 in Raji cells. Certainly, the expression pattern of
PVT1 in BL should be investigated following the collection
of clinical samples. It will be useful to develop a comprehensive
understanding of the role of PVT1 and its potential as an
oncotarget in BL.
Acknowledgements
The authors would like to thank associate Professor
Gexiu Liu and senior experimentalist Mrs Shaohua Chen, School of
Medicine, Jinan University, Guangzhou, for providing valuable
technical support and advice.
Funding
The project was supported by grants from the
Guangdong Science and Technology Project (grant no. 2015A050502029)
and the Overseas Chinese Affairs Office of the State Council Key
Discipline Construction Fund (grant no. 51205002).
Availability of data and materials
The datasets used and/or analysed in the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
CZ and YX performed the experiments and interpreted
results. DH and YL developed the original concept, designed the
study and contributed to the writing of the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Molyneux EM, Rochford R, Griffin B, Newton
R, Jackson G, Menon G, Harrison CJ, Israels T and Bailey S:
Burkitt's lymphoma. Lancet. 379:1234–1244. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Boxer LM and Dang CV: Translocations
involving c-myc and c-myc function. Oncogene. 20:5595–5610. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Fang Y and Fullwood MJ: Roles, functions,
and mechanisms of long non-coding RNAs in cancer. Genomics
Proteomics Bioinformatics. 14:42–54. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Dykes IM and Emanueli C: Transcriptional
and post-transcriptional gene regulation by long non-coding RNA.
Genomics Proteomics Bioinformatics. 15:177–186. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Rodríguez-Malavé NI and Rao DS: Long
noncoding RNAs in hematopoietic malignancies. Brief Funct Genomics.
15:227–238. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Shtivelman E and Bishop JM: Effects of
translocations on transcription from PVT. Mol Cell Biol.
10:1835–1839. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Huppi K and Siwarski D: Chimeric
transcripts with an open reading frame are generated as a result of
translocation to the Pvt-1 region in mouse B-cell tumors. Int J
Cancer. 59:848–851. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wang BJ, Ding HW and Ma GA: Long noncoding
RNA PVT1 promotes melanoma progression via endogenous sponging
miR-26b. Oncol Res. 26:675–681. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ding C, Yang Z, Lv Z, Du C, Xiao H, Peng
C, Cheng S, Xie H, Zhou L, Wu J and Zheng S: Long non-coding RNA
PVT1 is associated with tumor progression and predicts recurrence
in hepatocellular carcinoma patients. Oncol Lett. 9:955–963. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang F, Yuan JH, Wang SB, Yang F, Yuan SX,
Ye C, Yang N, Zhou WP, Li WL, Li W and Sun SH: Oncofetal long
noncoding RNA PVT1 promotes proliferation and stem cell-like
property of hepatocellular carcinoma cells by stabilizing NOP2.
Hepatology. 60:1278–1290. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhou Q, Chen J, Feng J and Wang J: Long
noncoding RNA PVT1 modulates thyroid cancer cell proliferation by
recruiting EZH2 and regulating thyroid-stimulating hormone receptor
(TSHR). Tumour Biol. 37:3105–3113. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Takahashi Y, Sawada G, Kurashige J, Uchi
R, Matsumura T, Ueo H, Takano Y, Eguchi H, Sudo T, Sugimachi K, et
al: Amplification of PVT-1 is involved in poor prognosis via
apoptosis inhibition in colorectal cancers. Brit J Cancer.
110:164–171. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tseng YY, Moriarity BS, Gong W, Akiyama R,
Tiwari A, Kawakami H, Ronning P, Reuland B, Guenther K, Beadnell
TC, et al: PVT1 dependence in cancer with MYC copy-number increase.
Nature. 512:82–86. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Huang F, Chen W, Peng J, Li Y, Zhuang Y,
Zhu Z, Shao C, Yang W, Yao H and Zhang S: LncRNA PVT1 triggers
Cyto-protective autophagy and promotes pancreatic ductal
adenocarcinoma development via the miR-20a-5p/ULK1 Axis. Mol
Cancer. 17:982018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ghesquières H, Larrabee BR, Casasnovas O,
Maurer MJ, McKay JD, Ansell SM, Montgomery D, Asmann YW, Farrell K,
Verney A, et al: A susceptibility locus for classical Hodgkin
lymphoma at 8q24 near MYC/PVT1 predicts patient outcome in two
independent cohorts. Brit J Haematol. 180:286–290. 2018. View Article : Google Scholar
|
|
16
|
Cerhan JR, Berndt SI, Vijai J, Ghesquières
H, McKay J, Wang SS, Wang Z, Yeager M, Conde L, de Bakker PI, et
al: Genome-wide association study identifies multiple
susceptibility loci for diffuse large B cell lymphoma. Nat Genet.
46:1233–1238. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2 (-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang Y, Mo WJ, Wang X, Zhang TT, Qin Y,
Wang HL, Chen G, Wei DM and Dang YW: Microarray-based
bioinformatics analysis of the prospective target gene network of
key miRNAs influenced by long non-coding RNA PVT1 in HCC. Oncol
Rep. 40:226–240. 2018.PubMed/NCBI
|
|
19
|
Zhang Y, Dang YW, Wang X, Yang X, Zhang R,
Lv ZL and Chen G: Comprehensive analysis of long non-coding RNA
PVT1 gene interaction regulatory network inhepatocellular carcinoma
using gene microarray and bioinformatics. Am J Transl Res.
9:3904–3917. 2017.PubMed/NCBI
|
|
20
|
Paci P, Colombo T and Farina L:
Computational analysis identifies a sponge interaction network
between long non-coding RNAs and messenger RNAs in human breast
cancer. BMC Syst Biol. 8:832014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kong R, Zhang EB, Yin DD, You LH, Xu TP,
Chen WM, Xia R, Wan L, Sun M, Wang ZX, et al: Long noncoding RNA
PVT1 indicates a poor prognosis of gastric cancer and promotes cell
proliferation through epigenetically regulating p15 and p16. Mol
Cancer. 14:822015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bretones G, Delgado MD and León J: Myc and
cell cycle control. Biochim Biophys Acta. 1849:506–516. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yang Y Xue K, Li Z, Zheng W, Dong W, Song
J, Sun S, Ma T and Li W: c-Myc regulates the CDK1/cyclin B1
dependent-G2/M cell cycle progression by histone H4 acetylation in
Raji cells. Int J Mol Med. 41:3366–3378. 2018.PubMed/NCBI
|
|
24
|
Dang CV: MYC on the path to cancer. Cell.
149:22–35. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang Z, Yang B, Zhang M, Guo W, Wu Z, Wang
Y, Jia L, Li S; Cancer Genome Atlas Research Network, ; Xie W and
Yang D: lncRNA epigenetic landscape analysis identifies EPIC1 as an
oncogenic lncRNA that interacts with MYC and promotes cell-cycle
progression in cancer. Cancer Cell. 33:706–720.e9. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Dutto I, Tillhon M, Cazzalini O, Stivala
LA and Prosperi E: Biology of the cell cycle inhibitor p21
(CDKN1A): Molecular mechanisms and relevance in chemical
toxicology. Arch Toxicol. 89:155–178. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Cazzalini O, Scovassi AI, Savio M, Stivala
LA and Prosperi E: Multiple roles of the cell cycle inhibitor p21
(CDKN1A) in the DNA damage response. Mutat Res. 704:12–20. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Perucca P, Cazzalini O, Madine M, Savio M,
Laskey RA, Vannini V, Prosperi E and Stivala LA: Loss of p21 CDKN1A
impairs entry to quiescence and activates a DNA damage response in
normal fibroblasts induced to quiescence. Cell Cycle. 8:105–114.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yan C, Chen Y, Kong W, Fu L, Liu Y, Yao Q
and Yuan Y: PVT1-derived miR-1207-5p promotes breast cancer cell
growth by targeting STAT6. Cancer Sci. 108:868–876. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Cui M, Chang Y, Fang QG, Du W, Wu JF, Wang
JH, Liu ST and Luo SX: Non-coding RNA Pvt1 promotes cancer stem
cell-like traits in nasopharyngeal cancer via inhibiting miR-1207.
Pathol Oncol Res. 2018. View Article : Google Scholar
|
|
31
|
Wu BQ, Jiang Y, Zhu F, Sun DL and He XZ:
Long noncoding RNA PVT1 promotes EMT and cell proliferation and
migration through downregulating p21 in pancreatic cancer cells.
Technol Cancer Res Treat. 1:15330346177005592017.
|
|
32
|
Cui D, Yu CH, Liu M, Xia QQ, Zhang YF and
Jiang WL: Long non-coding RNA PVT1 as a novel biomarker for
diagnosis and prognosis of non-small cell lung cancer. Tumor Biol.
37:4127–4134. 2016. View Article : Google Scholar
|
|
33
|
Bennin DA, Don AS, Brake T, McKenzie JL,
Rosenbaum H, Ortiz L, DePaoli-Roach AA and Horne MC: Cyclin G2
associates with protein phosphatase 2A catalytic and regulatory B′
subunits in active complexes and induces nuclear aberrations and a
G1/S phase cell cycle arrest. J Biol Chem. 277:27449–27467. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Hasegawa S, Nagano H, Konno M, Eguchi H,
Tomokuni A, Tomimaru Y, Wada H, Hama N, Kawamoto K, Kobayashi S, et
al: Cyclin G2: A novel independent prognostic marker in pancreatic
cancer. Oncol Lett. 10:2986–2990. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yin G, Zhou H, Xue Y, Yao B and Zhao W:
MicroRNA-340 promotes the tumor growth of human gastric cancer by
inhibiting cyclin G2. Oncol Rep. 36:1111–1118. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zimmermann M, Arachchige-Don AP, Donaldson
MS, Patriarchi T and Horne MC: Cyclin G2 promotes cell cycle arrest
in breast cancer cells responding to fulvestrant and metformin and
correlates with patient survival. Cell Cycle. 15:3278–3295. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Cui DW, Cheng YJ, Jing SW and Sun GG:
Effect of cyclin G2 on proliferative ability of prostate cancer
PC-3 cell. Tumour Biol. 35:3017–3024. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Helmbold H, Galderisi U and Bohn W: The
switch from pRb/p105 to Rb2/p130 in DNA damage and cellular
senescence. J Cell Physiol. 227:508–513. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Pentimalli F, Forte IM, Esposito L,
Indovina P, Iannuzzi CA, Alfano L, Costa C, Barone D, Rocco G and
Giordano A: RBL2/p130 is a direct AKT target and is required to
induce apoptosis upon AKT inhibition in lung cancer and
mesothelioma cell lines. Oncogene. 37:3657–3671. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Nalepa G, Barnholtz-Sloan J, Enzor R, Dey
D, He Y, Gehlhausen JR, Lehmann AS, Park SJ, Yang Y, Yang X, et al:
The tumor suppressor CDKN3 controls mitosis. J Cell Biol.
201:997–1012. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Yu C, Cao H, He X, Sun P, Feng Y, Chen L
and Gong H: Cyclin-dependent kinase inhibitor 3 (CDKN3) plays a
critical role in prostate cancer via regulating cell cycle and DNA
replication signaling. Biomed Pharmacother. 96:1109–1118. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Bortner DM and Rosenberg MP: Induction of
mammary gland hyperplasia and carcinomas in transgenic mice
expressing human cyclin E. Mol Cell Biol. 17:453–459. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Ma Y, Fiering S, Black C, Liu X, Yuan Z,
Memoli VA, Robbins DJ, Bentley HA, Tsongalis GJ, Demidenko E, et
al: Transgenic cyclin E triggers dysplasia and multiple pulmonary
adenocarcinomas. Proc Natl Acad Sci USA. 104:4089–4094. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Loeb KR, Kostner H, Firpo E, Norwood T, D
Tsuchiya K, Clurman BE and Roberts JM: A mouse model for cyclin
E-dependent genetic instability and tumorigenesis. Cancer Cell.
8:35–47. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Santarius T, Shipley J, Brewer D, Stratton
MR and Cooper CS: A census of amplified and overexpressed human
cancer genes. Nat Rev Cancer. 10:59–64. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Cheng G, Zhang L, Lv W, Dong C, Wang Y and
Zhang J: Overexpression of cyclin D1 in meningioma is associated
with malignancy grade and causes abnormalities in apoptosis,
invasion and cell cycle progression. Med Oncol. 32:4392015.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Hall M, Bates S and Peters G: Evidence for
different modes of action of cyclin-dependent kinase inhibitors:
p15 and p16 bind to kinases, p21 and p27 bind to cyclins. Oncogene.
11:1581–1588. 1995.PubMed/NCBI
|
|
48
|
Li W, Zheng Z, Chen H, Cai Y and Xie W:
Knockdown of long non-coding RNA PVT1 induces apoptosis and cell
cycle arrest in clear cell renal cell carcinoma through the
epidermal growth factor receptor pathway. Oncol Lett. 15:7855–7863.
2018.PubMed/NCBI
|
|
49
|
Chen L, Ma D, Li Y, Li X, Zhao L, Zhang J
and Song Y: Effect of long non-coding RNA PVT1 on cell
proliferation and migration in melanoma. Int J Mol Med.
41:1275–1282. 2018.PubMed/NCBI
|
|
50
|
Kapanidou M, Curtis NL and Bolanos-Garcia
VM: Cdc20: At the crossroads between chromosome segregation and
mitotic exit. Trends Biochem Sci. 42:193–205. 2017. View Article : Google Scholar : PubMed/NCBI
|