Introduction
Liver cancer is one of the most prevalent types of
malignancy worldwide as the annual estimated rate reached 782,500
novel cases and ~745,500 liver cancer-associated mortalities in
2012 (1). In China, patients account
for ~50% of all these cases and mortalities (1). Hepatocellular carcinoma (HCC) is the
major histologic liver cancer subtype, representing 80% of all
liver malignancies (2). Although
surgical resection and liver transplantation are used in the
treatment of early-stage HCC, the overall prognosis remains poor,
due to high recurrence rates (3). A
number of studies have reported that gene expression signatures are
associated with the prognosis of HCC (4–6).
Therefore, identifying novel biomarkers that predict prognosis and
may guide individualized treatment for HCC would greatly benefit
patients.
ATPase family AAA domain-containing protein 3
(ATAD3) is a mitochondrial membrane-bound ATPase that was
first identified as a component of the mouse liver inner
mitochondrial membrane using a proteomic approach (7) and was subsequently discovered to be
overexpressed in head and neck carcinomas (8). Subsequent studies have reported that
ATAD3 serves important roles in Caenorhabditis elegans and
Drosophila melanogaster development, indicating that
ATAD3 is associated with proliferation and differentiation
(9,10). In primates, the ATAD3 gene
cluster contains ATAD3A, ATAD3B and ATAD3C. ATAD3A is
the ancestral form of ATAD3, while ATAD3B and
ATAD3C are similar, however, they contain important mutated
residues (11). These three genes
are located side-by-side at the end of chromosome 1 (locus
1p36.33).
ATAD3 is a member of the family of
AAA-ATPases, which are involved in a number of cellular processes,
including transcription, replication, translation, proteolysis, and
vesicular transport (12). In HeLa
cells, ATAD3 was reported in a large multi-molecular complex
associated with mitochondrial DNA (mtDNA) that serves a role in
mtDNA replication and transcription (13). ATAD3 protein has displacement
loop binding activity, which allows it to form or segregate
mitochondrial nucleoids (14).
However, Bogenhagen et al (15) have reported that ATAD3A and
ATAD3B indirectly interact with mtDNA, mediated by topology
rather than the C-terminal AAA domain. Therefore, they do not have
the opportunity to bind to mtDNA D-loops. Subsequent results have
demonstrated that ATAD3A controls mitochondrial dynamics between
the outer and inner membranes, and that the N-terminal region of
ATAD3A is outside the inner membrane, while the C-terminal region
is within the matrix (16,17). ATAD3 deficiency is associated
with aberrant mtDNA organization and cholesterol metabolism in the
central nervous system (18).
ATAD3 expression was originally reported to
produce autoimmune responses in patients with lung adenocarcinoma
or uterine cervical cancer (19,20) and
to be associated with tumorigenesis (11). Studies have reported that
ATAD3 expression is linked with the progression of head and
neck cancers (8), non-Hodgkin's
lymphoma (21), lung cancer
(22), uterine cervical cancer
(23), prostate cancer (24), and glioma (25). However, to the best of our knowledge,
there have been no studies investigating associations between
ATAD3 expression and HCC. In the present study, the
prognostic value of ATAD3 gene cluster expression was
investigated in HCC to determine it potential as a biomarker for
this disease.
Materials and methods
ATAD3 gene cluster expression in HCC
and normal liver tissues
Box plots comparing expression levels of the
ATAD3 gene cluster in HCC (n=369) vs. normal liver tissues
(n=50) were downloaded from the online tool Gene Expression
Profiling Interactive Analysis (http://gepia.cancer-pku.cn/), which uses data derived
from The Cancer Genome Atlas (TCGA; http://tcga-data.nci.nih.gov/tcga). Significance
cut-off level was set at P=0.05.
Patient information
Clinical data and ATAD3A, ATAD3B and
ATAD3C mRNA levels of the 360 patients were obtained from
the online websites OncoLnc (http://www.oncolnc.org/) and TCGA. The present study's
results are partially based on data generated by TCGA Research
(http://cancergenome.nih.gov/). The
included clinical data were race, sex, age, body mass index (BMI),
tumor node metastasis (TNM) stage (the seventh AJCC staging system)
(26), survival time (days), and
survival status.
Survival analysis
ATAD3A, ATAD3B and ATAD3C mRNA
expression levels from TCGA were individually divided into two
groups by their 50% cut-off values, resulting into the
high-expression (n=180) and low-expression groups (n=180). Overall
survival (OS) was analyzed by the Cox proportional hazards
regression model adjusted by sex, age, and tumor stage.
Joint-effects analysis
ATAD3A and ATAD3B expression indicated
statistically significant associations with OS in the patient
cohort with HCC. Therefore, a joint-effects analysis of the
combination of ATAD3A and ATAD3B with group I (low
ATAD3A and ATAD3B expression), group II (low
ATAD3A and high ATAD3B expression), group III (high
ATAD3A and low ATAD3B expression), and group IV (high
ATAD3A and ATAD3B expression) was performed. Sex,
age, and tumor stage were adjusted in the Cox proportional hazards
regression model.
Gene co-expression network
analysis
In order to predict gene function and to construct a
pathway for the ATAD3 genes, the Database for Annotation,
Visualization and Integrated Discovery (DAVID; http://david.ncifcrf.gov/content.jsp?file=citation.htm)
was used to carry out the enriched Gene Ontology (GO) terms and
Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways (27). Cytoscape 3.6.0 software (https://cytoscape.org/) was used to construct
biological networks (28).
Statistical analysis
Median survival time (MST) and OS were calculated
using the Kaplan-Meier method with log-rank tests. The Cox
proportional hazards regression model was used to perform
univariate and multivariate survival analyses. Hazard ratios (HR)
and 95% confidence intervals (CI) were calculated subsequent to
adjusting for sex, age, and tumor stage. All statistical analyses
were performed with SPSS version 22.0 (IBM Corp., Armonk, NY, USA),
with P-values <0.05 considered to indicate a statistically
significant difference.
Results
Analysis of ATAD3 gene cluster
expression in HCC and normal liver tissues
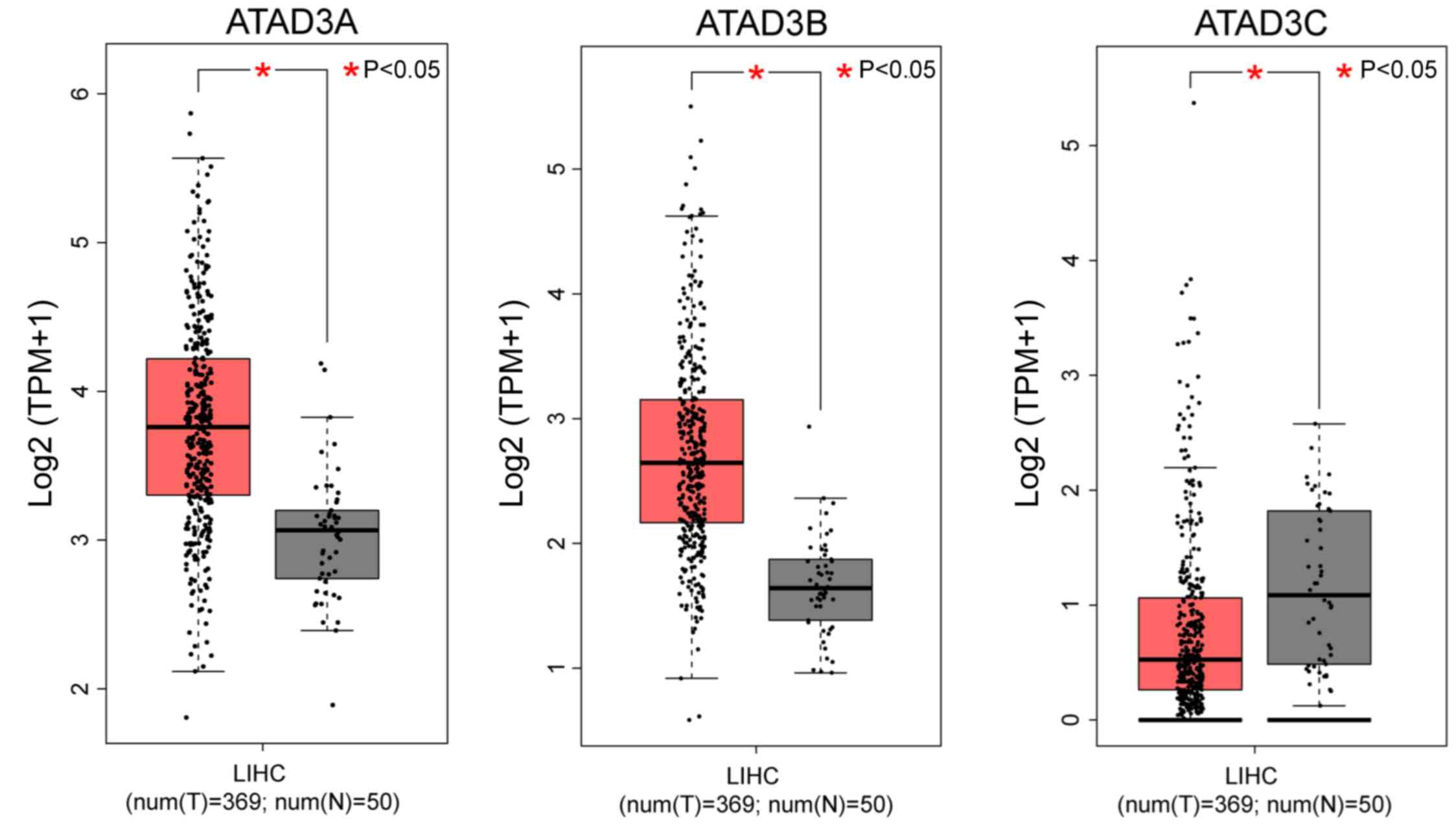
Expression data from 369 HCC and 50 normal liver
samples were analyzed by box plots, and the results indicated that
ATAD3A and ATAD3B were significantly overexpressed in
HCC tissues compared with normal liver tissues (P<0.05; Fig. 1). However, the expression level of
ATAD3C was significantly reduced in HCC tissues compared
with normal liver tissues (P<0.05; Fig. 1).
TCGA database patient
characteristics
Clinical characteristics of the 360 patients from
the TCGA database are presented in Table
I. The cohort included 244 male and 116 female patients, and
the median age was 61 years. The analysis indicated that TNM stage
was significantly associated with OS (P<0.001; HR=2.50; 95%
CI=1.72–3.63), whereas neither race, sex, age nor BMI were
associated with OS.
 | Table I.Demography and clinical
characteristics of 360 patients with hepatocellular carcinoma in
The Cancer Genome Atlas database. |
Table I.
Demography and clinical
characteristics of 360 patients with hepatocellular carcinoma in
The Cancer Genome Atlas database.
|
|
|
| Overall
survival |
|---|
|
|
|
|
|
|---|
| Variables | Patients
(n=360) | MST (days) | HR (95% CI) | Log-rank
P-value |
|---|
| Race |
|
Asian | 155 | NA | 1.29
(0.89–1.87) | 0.188 |
|
White+other | 196 | 1,397 |
|
|
|
Missing | 9 |
|
|
|
| Sex |
|
Male | 244 | 2,486 | 1.21
(0.84–1.73) | 0.311 |
|
Female | 116 | 1,560 |
|
|
| Age (years) |
|
<61 | 186 | 2,116 | 1.09
(0.77–1.54) | 0.622 |
|
≥61 | 171 | 1,622 |
|
|
|
Missing | 3 |
|
|
|
| BMI |
|
≤25 | 193 | 2,456 | 0.87
(0.60–1.27) | 0.473 |
|
>25 | 137 | 2,116 |
|
|
|
Missing | 30 |
|
|
|
| TNM stage |
|
I+II | 252 | 2,532 | 2.50
(1.72–3.63) | <0.001 |
|
III+IV | 87 | 770 |
|
|
|
Missing | 21 |
|
|
|
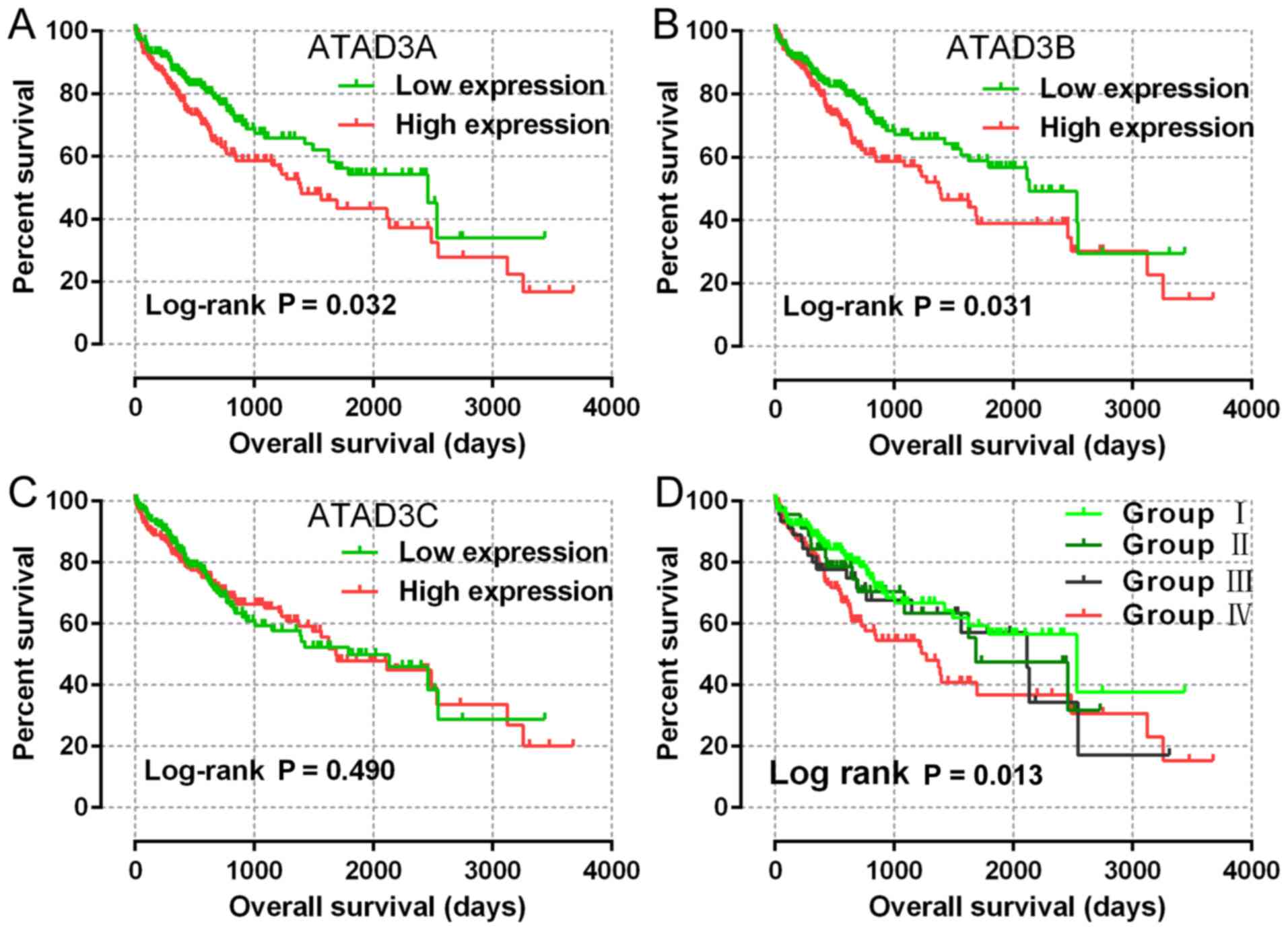
Survival analysis of ATAD3 mRNA levels
with OS
ATAD3A, ATAD3B and ATAD3C mRNA
expression data were available for all patients from the TCGA
database. The patients were divided into two groups based on the
50% cut-off level for each mRNA. The correlations between each gene
and OS were analyzed. The results indicated that the expression
level of ATAD3A (P=0.017, HR=1.54, 95% CI=1.08–2.20;
adjusted P=0.032; adjusted HR=1.52; 95% CI=1.04–2.22) and
ATAD3B (P=0.026, HR=1.49, 95% CI=1.05–2.13; adjusted
P=0.031, adjusted HR=1.52, 95% CI=1.04–2.21) were significantly
correlated with OS (Table II;
Fig. 2A and B). ATAD3C
expression level was not significantly associated with OS (Table II; Fig.
2C). Furthermore, a joint-effects analysis of ATAD3A and
ATAD3B with OS was performed, which demonstrated that
patients with high expression levels of both ATAD3A and
ATAD3B had a worse OS compared with those with low
expression levels of ATAD3A and ATAD3B (P=0.007,
HR=1.77, 95% CI=1.16–2.69; adjusted P=0.013, adjusted HR=1.76, 95%
CI=1.13–2.75) (Table III; Fig. 2D).
 | Table II.Prognostic survival analysis of
ATAD3 gene expression in The Cancer Genome Atlas
database. |
Table II.
Prognostic survival analysis of
ATAD3 gene expression in The Cancer Genome Atlas
database.
|
|
|
|
|
|
| Overall
survival |
|---|
|
|
|
|
|
|
|
|
|---|
| Gene | Patients
(n=360) | No. of events
(%) | MST (days) | HR (95% CI) | P-value | Adjusted HR (95%
CI)a | Adjusted
P-valuea |
|---|
| ATAD3A |
|
Low | 180 | 52 (28.9) | 2,456 | 1.54
(1.08–2.20) | 0.017 | 1.52
(1.04–2.22) | 0.032 |
|
High | 180 | 74 (41.1) | 1,386 |
|
|
|
|
| ATAD3B |
|
Low | 180 | 55 (30.6) | 2,131 | 1.49
(1.05–2.13) | 0.026 | 1.52
(1.04–2.21) | 0.031 |
|
High | 180 | 71 (39.4) | 1,386 |
|
|
|
|
| ATAD3C |
|
Low | 180 | 61 (33.9) | 1,791 | 1.00
(0.70–1.42) | 0.993 | 0.88
(0.61–1.27) | 0.490 |
|
High | 180 | 65 (36.1) | 1,685 |
|
|
|
|
 | Table III.Joint-effects analysis of the
combination of ATAD3A and ATAD3B expression in The
Cancer Genome Atlas database. |
Table III.
Joint-effects analysis of the
combination of ATAD3A and ATAD3B expression in The
Cancer Genome Atlas database.
|
|
|
|
|
|
|
| Overall
survival |
|---|
|
|
|
|
|
|
|
|
|
|---|
| Group | ATAD3A | ATAD3B | Patients
(n=360) | MST (days) | HR (95% CI) | P-value | Adjusted HR (95%
CI)a | Adjusted
P-valuea |
|---|
| I | Low | Low | 133 | 2,532 | N/A | 0.060 | N/A | 0.102 |
| II | Low | High | 47 | 1,685 | 1.24
(0.68–2.26) | 0.482 | 1.50
(0.80–2.84) | 0.211 |
| III | High | Low | 47 | 2,116 | 1.32
(0.75–2.32) | 0.333 | 1.49
(0.80–2.78) | 0.210 |
| IV | High | High | 133 | 1,271 | 1.77
(1.16–2.69) | 0.007 | 1.76
(1.13–2.75) | 0.013 |
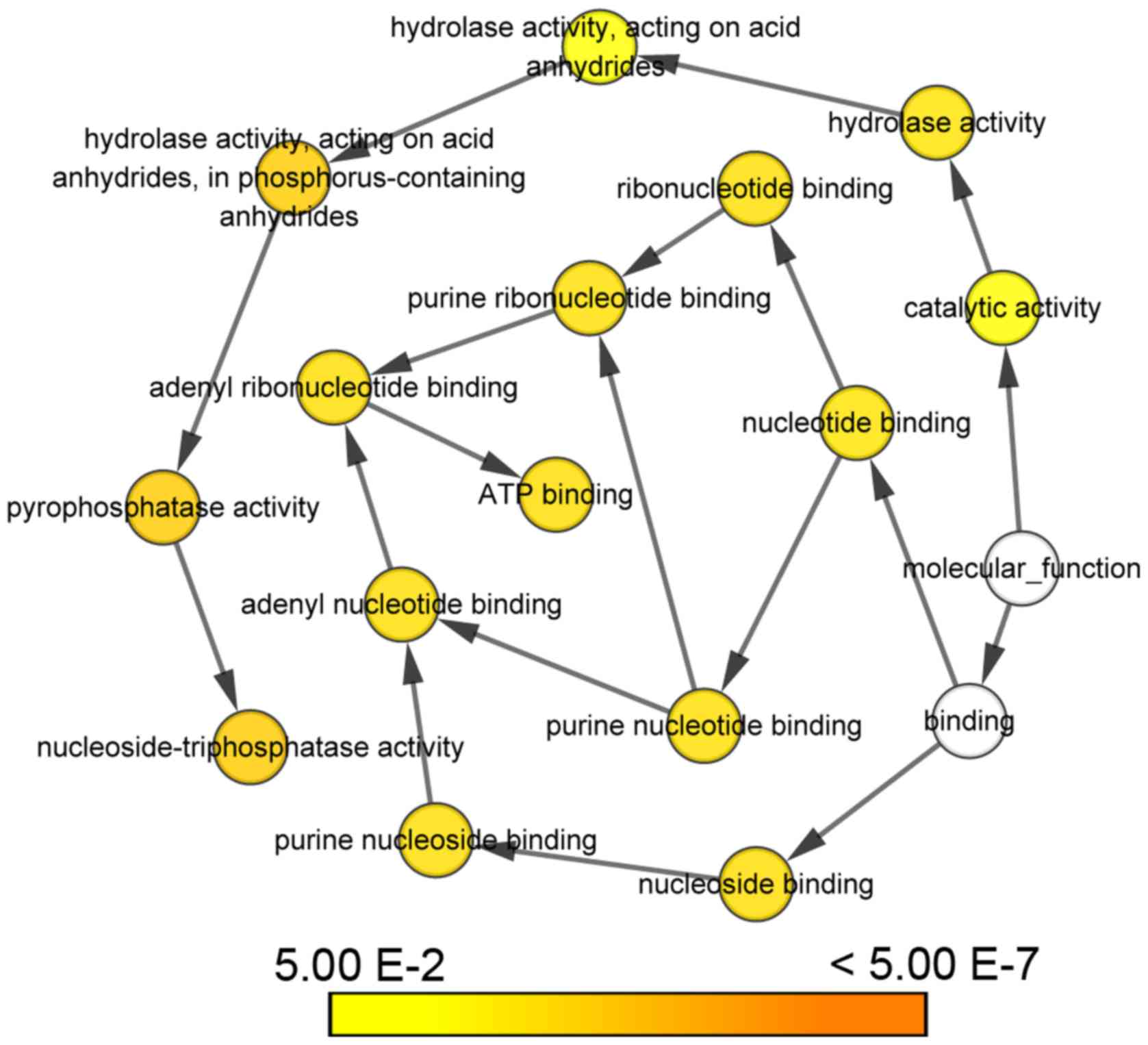
GO functional analysis of ATAD3
genes
KEGG pathway analysis revealed that the ATAD3
gene cluster was associated with ATP binding, cell growth and cell
division. Particularly, ATAD3A was a possible negative regulator of
apoptosis (Table IV). Biological
networks constructed by Cytoscape indicated that ATAD3 genes
serve important roles in ATP binding, nucleoside binding,
nucleotide binding, purine nucleotide and ribonucleotide binding,
adenyl nucleotide and ribonucleotide binding, catalytic activity,
hydrolase activity, pyrophosphatase activity and
nucleoside-triphosphatase activity (Fig.
3).
 | Table IV.Gene ontology analysis of
ATAD3 genes. |
Table IV.
Gene ontology analysis of
ATAD3 genes.
| Gene | Category | Term | Description |
|---|
| ATAD3A | BP | 0016049 | Cell growth |
|
| BP | 0043066 | Negative regulation
of apoptotic process |
|
| CC | 0005739 | Mitochondrion |
|
| CC | 0005743 | Mitochondrial inner
membrane |
|
| CC | 0016021 | Integral component
of membrane |
|
| CC | 0042645 | Mitochondrial
nucleoid |
|
| MF | 0005524 | ATP binding |
| ATAD3B | BP | 0051301 | Cell division |
|
| CC | 0005743 | Mitochondrial inner
membrane |
|
| MF | 0005524 | ATP binding |
| ATAD3C | MF | 0005524 | ATP binding |
Discussion
ATAD3, a member of the ATPase family, is exclusively
present in multicellular eukaryotes at the interface between the
outer and inner mitochondrial membranes, where it controls
mitochondrial dynamics, mitochondrial fission, proliferation, and
cholesterol transport (16,29,30). In
particular, one cellular function of ATAD3 is protecting
mtDNA integrity in multicellular organisms (14,16). A
number of studies have demonstrated that ATAD3 is linked to
the progression of various malignancies, including non-Hodgkin's
lymphoma (21), lung adenocarcinoma
(22), uterine cervical cancer
(23) and prostate cancer (24). However, to the best of our knowledge,
there have been no previous reports that have identified the
association of ATAD3 with HCC. Therefore, this is the first
study to indicate an association between ATAD3 expression
and HCC outcomes.
In the present study, the expression of all
ATAD3 genes, including ATAD3A, ATAD3B and
ATAD3C, was analyzed with regard to the prognosis of
patients with HCC from TCGA database. The results indicated that
ATAD3A and ATAD3B expression were significantly
associated with OS in HCC. High ATAD3A expression or high
ATAD3B expression were associated with poor MST and OS in
patients with HCC. In addition, a joint-effects analysis
demonstrated that patients with high ATAD3A and
ATAD3B expression had reduced MST and OS rates. However, the
mechanism underlying the poor survival of patients with HCC with
high ATAD3A and ATAD3B expression requires further
investigation.
ATAD3A is the human homologue of murine TOB3
(20), which controls mitochondrial
dynamics at the interface of the inner and outer mitochondrial
membranes and regulates diverse cellular responses including
growth, cholesterol channeling and mitochondrial fission (16). ATAD3A has been reported to indirectly
interact with mtDNA, and silencing of ATAD3A increases the
condensation and decreases the multimerization of mtDNA (14). A report indicated that ATAD3A
was overexpressed in lung adenocarcinoma samples and associated
with significantly higher tumor recurrence and increased drug
resistance, and that silencing ATAD3A increased apoptosis in
lung adenocarcinoma cells (22).
Another study suggested that ATAD3A was highly expressed in
prostate cancer, and that downregulating ATAD3A expression
reduced prostate-specific antigen secretion and cisplatin
resistance (24). ATAD3A is also
associated with HPV infection, reduced autophagy and apoptosis, and
increased drug resistance in uterine cervical cancer (23) Additionally, ATAD3B, which is a
c-MYC and myogenin target gene, was reported to serve important
roles in tumor progression (8).
Regarding ATAD3B, a study demonstrated that
this family member was downregulated in radiation-treated Raji B
cells and was associated with proliferation and apoptosis
inhibition (21). Another study
reported that higher ATAD3B expression was associated with
poor survival in breast cancer and that ATAD3B was activated
through estrogen receptor-α-mediated, non-genomic, MAPK-regulated
transcription factors, including myogenin and c-Myc (31). Notably, one study suggested that
ATAD3B overexpression results in loss-of-function of
endogenous ATAD3A (32). This
was corroborated by a study that indicated ATAD3B, as a human
embryonic stem cell-specific mitochondrial protein, negatively
regulated ATAD3A and acted as an adaptor of mitochondrial
homeostasis and metabolism in human embryonic stem cells and lung
carcinoma cells (33). In the
present study, gene function network analysis also indicated that
ATAD3A negatively regulated apoptotic processes.
ATAD3C has been reported to have 87% homology with
ATAD3A and is also associated with tumor progression
(11). However, to the best of our
knowledge, no studies have reported that ATAD3C expression
levels are associated with the prognosis of human patients with
cancer. In the current study, ATAD3C was not significantly
associated with patient survival with HCC.
In conclusion, previous studies have reported that
ATAD3A and ATAD3B are associated with tumor
progression, likely due to their roles in proliferation, apoptosis,
autophagy and increasing drug resistance (11,22,23). The
present study's gene network analysis also revealed that the ATAD3
protein family was associated with cell growth, cell division and
apoptosis. Results also demonstrated that ATAD3A and
ATAD3B expression levels were significantly associated with
the prognosis of patients with HCC. The present study revealed that
ATAD3A and ATAD3B may serve as potential biomarkers
for predicting the prognosis of patients with HCC. However,
experimental and multi-center studies of ATAD3 are required
to further confirm the present study's results.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors' contributions
BY, XL and GL designed the study. LA, QY and TY
analyzed the data and interpreted the results. XL and GL wrote the
manuscript. BY edited the manuscript. All authors discussed the
results and approved the final version of the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Aravalli RN, Cressman EN and Steer CJ:
Cellular and molecular mechanisms of hepatocellular carcinoma: An
update. Arch Toxicol. 87:227–247. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Llovet JM, Schwartz M and Mazzaferro V:
Resection and liver transplantation for hepatocellular carcinoma.
Semin Liver Dis. 25:181–200. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Xue C, Zhong Z, Ye S, Wang Y and Ye Q:
Association between the overexpression of PBOV1 and the prognosis
of patients with hepatocellular carcinoma. Oncol Lett.
16:3401–3407. 2018.PubMed/NCBI
|
|
5
|
Liu F, Pan Z, Zhang J, Ni J, Wang C, Wang
Z, Gu F, Dong W, Zhou W and Liu H: Overexpression of RHEB is
associated with metastasis and poor prognosis in hepatocellular
carcinoma. Oncol Lett. 15:3838–3845. 2018.PubMed/NCBI
|
|
6
|
Yu T, Wang X, Zhu G, Han C, Su H, Liao X,
Yang C, Qin W, Huang K and Peng T: The prognostic value of
differentially expressed CYP3A subfamily members for hepatocellular
carcinoma. Cancer Manag Res. 10:1713–1726. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Da Cruz S, Xenarios I, Langridge J,
Vilbois F, Parone PA and Martinou JC: Proteomic analysis of the
mouse liver mitochondrial inner membrane. J Biol Chem.
278:41566–41571. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Schaffrik M, Mack B, Matthias C, Rauch J
and Gires O: Molecular characterization of the tumor-associated
antigen AAA-TOB3. Cell Mol Life Sci. 63:2162–2174. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kamath RS, Fraser AG, Dong Y, Poulin G,
Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M, et
al: Systematic functional analysis of the Caenorhabditis elegans
genome using RNAi. Nature. 421:231–237. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hoffmann M, Bellance N, Rossignol R,
Koopman WJ, Willems PH, Mayatepek E, Bossinger O and Distelmaier F:
C. Elegans ATAD-3 is essential for mitochondrial activity and
development. PLoS One. 4:e76442009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Li S and Rousseau D: ATAD3, a vital
membrane bound mitochondrial ATPase involved in tumor progression.
J Bioenerg Biomembr. 44:189–197. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Frickey T and Lupas AN: Phylogenetic
analysis of AAA proteins. J Struct Biol. 146:2–10. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang Y and Bogenhagen DF: Human
mitochondrial DNA nucleoids are linked to protein folding machinery
and metabolic enzymes at the mitochondrial inner membrane. J Biol
Chem. 281:25791–25802. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
He J, Mao CC, Reyes A, Sembongi H, Di Re
M, Granycome C, Clippingdale AB, Fearnley IM, Harbour M, Robinson
AJ, et al: The AAA+ protein ATAD3 has displacement loop binding
properties and is involved in mitochondrial nucleoid organization.
J Cell Biol. 176:141–146. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bogenhagen DF, Rousseau D and Burke S: The
layered structure of human mitochondrial DNA nucleoids. J Biol
Chem. 283:3665–3675. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Gilquin B, Taillebourg E, Cherradi N,
Hubstenberger A, Gay O, Merle N, Assard N, Fauvarque MO, Tomohiro
S, Kuge O and Baudier J: The AAA+ ATPase ATAD3A controls
mitochondrial dynamics at the interface of the inner and outer
membranes. Mol Cell Biol. 30:1984–1996. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hubstenberger A, Merle N, Charton R,
Brandolin G and Rousseau D: Topological analysis of ATAD3A
insertion in purified human mitochondria. J Bioenerg Biomembr.
42:143–150. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Desai R, Frazier AE, Durigon R, Patel H,
Jones AW, Dalla Rosa I, Lake NJ, Compton AG, Mountford HS, Tucker
EJ, et al: ATAD3 gene cluster deletions cause cerebellar
dysfunction associated with altered mitochondrial DNA and
cholesterol metabolism. Brain. 140:1595–1610. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gires O, Münz M, Schaffrik M, Kieu C,
Rauch J, Ahlemann M, Eberle D, Mack B, Wollenberg B, Lang S, et al:
Profile identification of disease-associated humoral antigens using
AMIDA, a novel proteomics-based technology. Cell Mol Life Sci.
61:1198–1207. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Geuijen CA, Bijl N, Smit RC, Cox F,
Throsby M, Visser TJ, Jongeneelen MA, Bakker AB, Kruisbeek AM,
Goudsmit J and de Kruif J: A proteomic approach to tumour target
identification using phage display, affinity purification and mass
spectrometry. Eur J Cancer. 41:178–187. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Jiang Y, Liu X, Fang X and Wang X:
Proteomic analysis of mitochondria in Raji cells following exposure
to radiation: Implications for radiotherapy response. Protein Pept
Lett. 16:1350–1359. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Fang HY, Chang CL, Hsu SH, Huang CY,
Chiang SF, Chiou SH, Huang CH, Hsiao YT, Lin TY, Chiang IP, et al:
ATPase family AAA domain-containing 3A is a novel anti-apoptotic
factor in lung adenocarcinoma cells. J Cell Sci. 123:1171–1180.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chen TC, Hung YC, Lin TY, Chang HW, Chiang
IP, Chen YY and Chow KC: Human papillomavirus infection and
expression of ATPase family AAA domain containing 3A, a novel
anti-autophagy factor, in uterine cervical cancer. Int J Mol Med.
28:689–696. 2011.PubMed/NCBI
|
|
24
|
Huang KH, Chow KC, Chang HW, Lin TY and
Lee MC: ATPase family AAA domain containing 3A is an anti-apoptotic
factor and a secretion regulator of PSA in prostate cancer. Int J
Mol Med. 28:9–15. 2011.PubMed/NCBI
|
|
25
|
Hubstenberger A, Labourdette G, Baudier J
and Rousseau D: ATAD 3A and ATAD 3B are distal 1p-located genes
differentially expressed in human glioma cell lines and present in
vitro anti-oncogenic and chemoresistant properties. Exp Cell Res.
314:2870–2883. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Edge S: American joint committee on
cancer; ACS. AJCC cancer staging manual. 7th edition. New York.
Springer2009.
|
|
27
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Rone MB, Midzak AS, Issop L, Rammouz G,
Jagannathan S, Fan J, Ye X, Blonder J, Veenstra T and Papadopoulos
V: Identification of a dynamic mitochondrial protein complex
driving cholesterol import, trafficking, and metabolism to steroid
hormones. Mol Endocrinol. 26:1868–1882. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Li S, Lamarche F, Charton R, Delphin C,
Gires O, Hubstenberger A, Schlattner U and Rousseau D: Expression
analysis of ATAD3 isoforms in rodent and human cell lines and
tissues. Gene. 535:60–69. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ovaska K, Matarese F, Grote K, Charapitsa
I, Cervera A, Liu C, Reid G, Seifert M, Stunnenberg HG and
Hautaniemi S: Integrative analysis of deep sequencing data
identifies estrogen receptor early response genes and links ATAD3B
to poor survival in breast cancer. PLoS Comput Biol.
9:e10031002013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
He J, Cooper HM, Reyes A, Di Re M,
Sembongi H, Litwin TR, Gao J, Neuman KC, Fearnley IM, Spinazzola A,
et al: Mitochondrial nucleoid interacting proteins support
mitochondrial protein synthesis. Nucleic Acids Res. 40:6109–6121.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Merle N, Féraud O, Gilquin B,
Hubstenberger A, Kieffer-Jacquinot S, Assard N, Bennaceur-Griscelli
A, Honnorat J and Baudier J: ATAD3B is a human embryonic stem cell
specific mitochondrial protein, re-expressed in cancer cells, that
functions as dominant negative for the ubiquitous ATAD3A.
Mitochondrion. 12:441–448. 2012. View Article : Google Scholar : PubMed/NCBI
|