Introduction
Osteosarcoma (OS) is the most common primary
malignant bone tumor frequently occurring in children and
adolescents (1). Originating from
mesenchymal cells, OS often results in pulmonary metastasis and has
poor overall prognosis (2,3). Amputation and chemotherapy are the most
common treatment options for OS; however, ~40% of treated patients
experience tumor metastasis, which ultimately leads to an adverse
clinical outcome (4,5). Therefore, it is imperative to
understand the mechanisms triggering OS metastasis for the
management of this disease.
Long non-coding RNAs (lncRNAs) are a group of
evolutionarily conserved non-coding RNAs >200 nucleotides long
with no or limited protein coding capacity (6). Based on the location and sequence,
lncRNAs can be classified into five types: Sense, antisense,
bidirectional, intronic and intergenic (7,8). A
number of previous studies have indicated that lncRNAs are involved
in almost every aspect of cell biology and contribute to tumor
development by various mechanisms (9–11).
Although multiple lncRNAs are abnormally expressed in cancer
tissues and function as potent oncogenes or tumor suppressors
(12–14), only a small number of lncRNAs have
had clear underlying mechanisms identified. lncRNAs are crucial for
OS initiation and progression (15,16). For
instance, nuclear paraspeckle assembly transcript 1, small
nucleolar RNA host gene 4 and tumor protein p73 antisense RNA 1
have been identified as oncogenic lncRNAs associated with poor
prognosis of patients with OS (17–19).
Homeobox A transcript at the distal tip (HOTTIP) is
an lncRNA transcribed from the 5′tip of homeobox A (HOXA) locus
that controls HOXA gene expression (20). HOTTIP is significantly upregulated in
human cancers; it is functionally linked with carcinogenesis and
represents a potential prognostic biomarker in cancer (21). The majority of studies on HOTTIP
support its oncogenic roles, including promoting cell
proliferation, inhibiting apoptosis and facilitating cell migration
(22). Li et al (23), reported that HOTTIP was upregulated
in OS, promoted cell proliferation, migration and invasion in
vitro, and higher HOTTIP expression levels were associated with
poor survival. HOTTIP also increased chemoresistance of OS cells
(24). However, its roles and
mechanisms in OS migration, invasion and epithelial-mesenchymal
transition (EMT) remain unclear. Notably, HOTTIP could activate the
Wnt/β-catenin signaling pathway in OS cells (24). Since c-Myc is a vital target gene and
effector of the Wnt/β-catenin pathway, which is associated with the
malignant phenotypes of OS, including migration, invasion and EMT
(25), this study hypothesized that
HOTTIP could also induce c-Myc expression in OS cells. Considering
the previously described attributes of HOTTIP activity, it was
hypothesized that HOTTIP may be involved in OS migration, invasion
and EMT.
Materials and methods
Patients and tissues
Twenty-five pairs of human OS tissues and adjacent
non-tumoral tissues were obtained from patients (range, 14–28 years
old; 10 male and 15 female patients) with OS who received surgical
treatment at the Department of Orthopedics, Changhai Hospital,
Second Military Medical University (Shanghai, China), between
January 2016 and June 2017. All samples were obtained with informed
consent and approved by the Ethics Committee of Changhai Hospital,
Second Military Medical University (Shanghai, China). The tissues
were frozen in liquid nitrogen immediately and stored at −80°C.
Cell culture
Human normal osteoblastic cell line hFOB1.19 and OS
cell lines SaoS2, HOS, U2OS and MG63 were purchased from The Cell
Bank of Type Tissue Collection of the Chinese Academy of Science or
the American Type Culture Collection. HOS cells were cultured in
RPMI-1640 (Gibco; Thermo Fisher Scientific, Inc.), while other cell
lines were cultured in DMEM (Gibco; Thermo Fisher Scientific,
Inc.). All media were supplemented with 10% fetal bovine serum
(FBS; Gibco; Thermo Fisher Scientific, Inc.) and cells were
cultured at 37°C in a humidified atmosphere with 5%
CO2.
Reverse transcription-quantitative PCR
(RT-qPCR)
RT-qPCR was used to determine the expression levels
of HOTTIP, β-catenin and c-Myc in tissues and cells (including
hFOB1.19, SaoS2, HOS, U2OS and MG63 cell lines). Total RNA was
extracted from tissues (≥100 mg) and cells (≥1×105)
using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.)
following the manufacturer's protocol. cDNA was synthesized using
GoScript Reverse Transcription Mix, Random Primers (Promega
Corporation). qPCR was performed in an ABI7500 instrument using
Toyobo SYBR Green Realtime PCR Master Mix (cat. no., QPK-201;
Toyobo Life Sciences). The following thermocycling conditions were
used: Initial denaturation at 95°C for 3 min; followed by 40 cycles
of 95°C for 15 sec and 60°C for 1 min; finally dissociation at 60°C
for 10 min. β-actin was used as an internal control. Fold changes
were calculated using the relative quantification
(2−ΔΔCq) method (26).
The primers used were as follows: HOTTIP, forward
5′-CCTAAAGCCACGCTTCTTTG-3′, reverse 5′-TGCAGGCTGGAGATCCTACT-3′;
β-actin, forward 5′-TCTTCGCCTTAATACTTGT-3′, reverse
5′-AAGCCTTCATACATCTCAA-3′; β-catenin, forward
5′-CCTTTGTCCCGCAAATCATG-3′, reverse 5′-CGTACGGCGCTGGGTATC-3′;
c-Myc, forward 5′-TACATCCTGTCGGTCCAA-3′, reverse
5′-AACTGTTCTCGCCTCTTC-3′.
Small interfering RNAs (siRNAs),
overexpression plasmids and transfections
HOTTIP siRNAs and negative control (si-NC) were
purchased from Shanghai GenePharma Co, Ltd. The sequences were as
follows: NC siRNA, 5′-GGUGGAACAAUUGCUUUUA-3′; HOTTIP siRNA 1,
5′-AAAUUGCUCACUAACAGUGUG-3′; HOTTIP siRNA 2,
5′-UUUUCUUGUCCCAAAAUAGAG-3′. The plasmids containing the coding
sequences of the two isoforms of c-Myc gene were constructed by PCR
using KOD-plus-Ver.2 kit (KOD-211; Toyobo Life Science) and cloned
into pcDNA3.1 with BamHI and HindIII as restriction enzymes.
Transfections were performed using Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc.), according to the
manufacturer's instructions. Briefly, cells (1×105
cells/well; U-2OS and MG63 cell lines were used since HOTTIP was
highly expressed in the two cell lines) were seeded into a 6-well
plate and incubated at 37°C overnight. Prior to the transfection,
the siRNA/plasmid-Lipofectamine solution was prepared by mixing 200
µl MEM separately with Lipofectamine (5 µl) or siRNA/plasmid (2 µg)
and then mixing the solutions together. Finally, the
siRNA/plasmid-Lipofectamine solution was added into each well, and
the cells were incubated at 37°C for 24 h (for plasmid) or 48 h
(for siRNA) for subsequent experiments.
Cell Counting Kit-8 (CCK-8) assay and
trypan blue staining
For CCK-8 assay, U-2OS and MG63 cells were seeded in
96-well plates (5×104 cells/well) and incubated at 37°C
in a humidified atmosphere with 5% CO2 for 12 h.
Subsequently, fresh medium containing 10% FBS was added to the
culture plate and 10 µl CCK-8 (Dojindo Molecular Technologies)
reagent was added to each well at 0, 24, 48 and 72 h and incubated
at 37°C for another 2 h. The absorbance was measured at 450 nm
using a microplate reader (Molecular Devices). For Trypan blue
staining, triplicates of U-2OS and MG63 cells (2×104
cells/well) were seeded in 96-well plates initially, then
trypsinized at different time points (0, 24, 36 and 48 h) and
stained with trypan blue at room temperature for 5 min; stained
cells were counted using a hemocytometer and cell viability
([(total cells)-(blue cells)]x100%) curves were plotted.
Wound healing assay
Cell migration ability was determined by wound
healing assay. Transfected MG63 cells were seeded in 6-well plate
(3×105 cells/well) for 24 h until confluence was
reached. A scratch was made using a sterile 200 µl pipette tip in
the central axis of the wells. The cells were washed with PBS and
cultured with serum-free medium for 24 h. At 48 or 72 h, the
migration of the cells into the scratch was observed under an
inverted microscope (magnification, ×200) and the distance between
the edges of the wound was calculated.
Matrigel assay
Cell invasive ability was determined by Matrigel
assay. Transfected U-2OS cells were seeded (3×104
cells/well) with medium containing 0.1% FBS in the upper chamber of
an insert coated with Matrigel and allowed to migrate into the
lower chamber supplemented with medium containing 10% FBS for 24 h
(37°C; 5% CO2). Cells that invaded through the membrane
were fixed by 4% paraformaldehyde for 20 min and stained with 0.5%
crystal violet solution for 30 min at room temperature. Three
replicates were obtained.
Western blotting
Cellular proteins (≥1×105 cells/well;
U-2OS and MG63 cell lines) were extracted by using RIPA buffer
(Beyotime Institute of Biotechnology) containing
phenylmethylsulphonyl fluoride (Beyotime Institute of
Biotechnology). Proteins (~40 µg), quantified by bicinchoninic acid
protein assay kit (Beyotime Institute of Biotechnology), were
separated by 8% SDS-PAGE, transferred to a nitrocellulose membrane
(EMD Millipore) and incubated with 5% skimmed milk for 1 h at room
temperature. The membranes were subsequently incubated with the
following primary antibodies at a dilution of 1:1,000 at 4°C
overnight: E-cadherin (cat. no., 3195), vimentin (cat. no., 5741),
zinc-finger E-box-binding homeobox 1 (ZEB1; cat. no., 3396), Snail
(cat. no., 3879), Slug (cat. no., 9585), β-catenin (cat. no.,
8480), c-Myc (cat. no., 5605) and β-actin (cat. no., 4970). β-actin
was used as an internal control. HRP-linked anti-rabbit IgG (cat.
no., 7074; 1:1,000 dilution) was used as the secondary antibody.
All antibodies were purchased from Cell Signaling Technology, Inc.
Western blot bands were visualized using ECL Western Blotting
Detection System (EMD Millipore).
Statistical analysis
Statistical analyses were performed using GraphPad
Prism 6.0 software (GraphPad Software, Inc.). All data are
presented as the mean ± standard deviation. Experimental results
were assessed using paired Student's t-test, unpaired Student's
t-test or one-way analysis of variance with Dunnett's post hoc
test. P<0.05 was considered to indicate a statistically
significant difference.
Results
HOTTIP overexpression in OS tissues
and cell lines
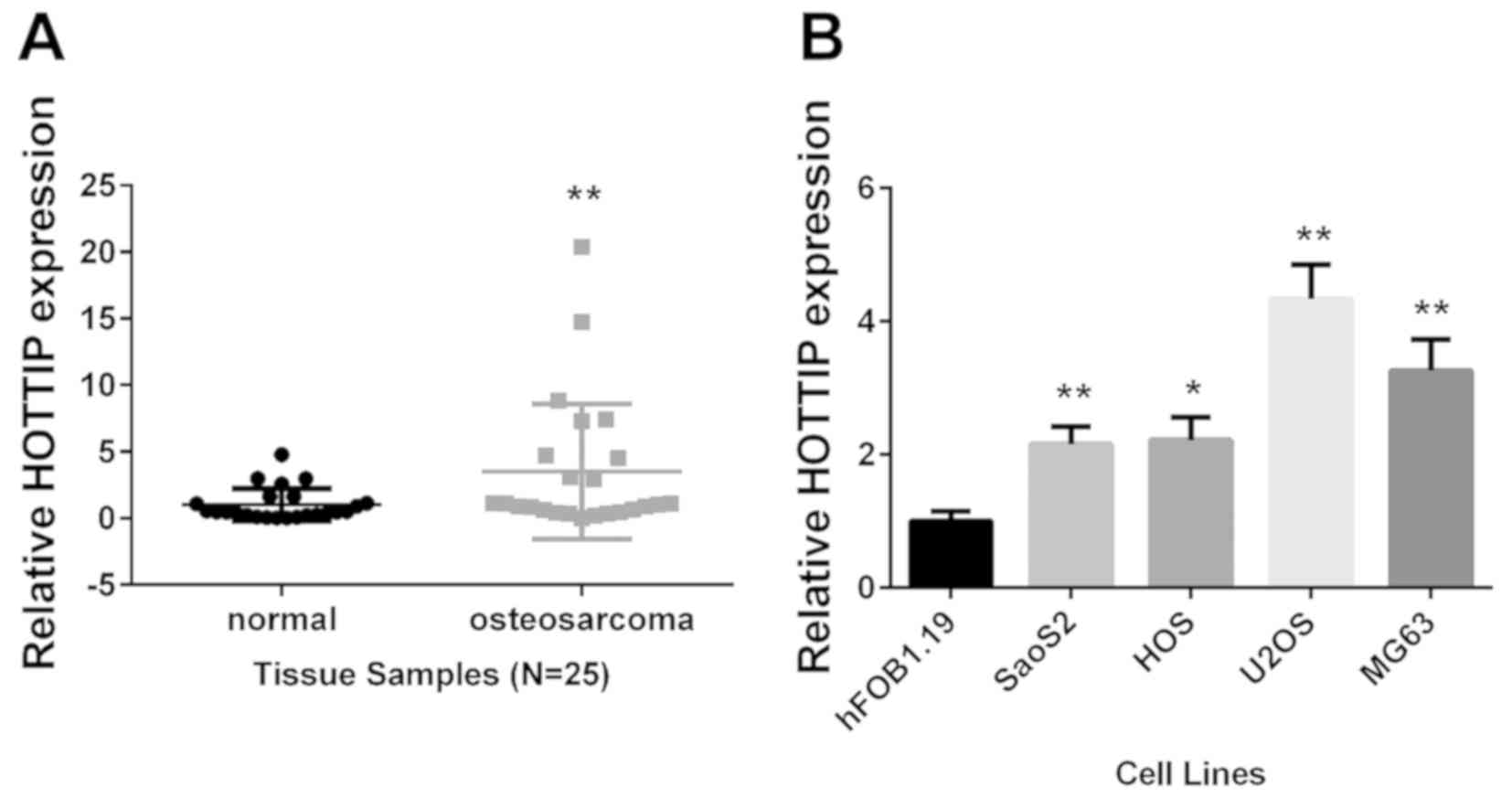
RT-qPCR was used to detect the expression levels of
lncRNA HOTTIP in 25 pairs of OS tissues and adjacent non-tumor
tissues. The result revealed a significant overexpression of HOTTIP
in OS tissues (P<0.01; Fig. 1A).
This was in agreement with the previous reports (23,24).
HOTTIP expression was also analyzed in vitro; compared with
the non-tumor human osteoblastic cell line hFOB1.19, HOTTIP was
significantly upregulated in the four OS cell lines (Fig. 1B). The overexpression of HOTTIP
suggested it may serve tumor-promoting roles in OS development.
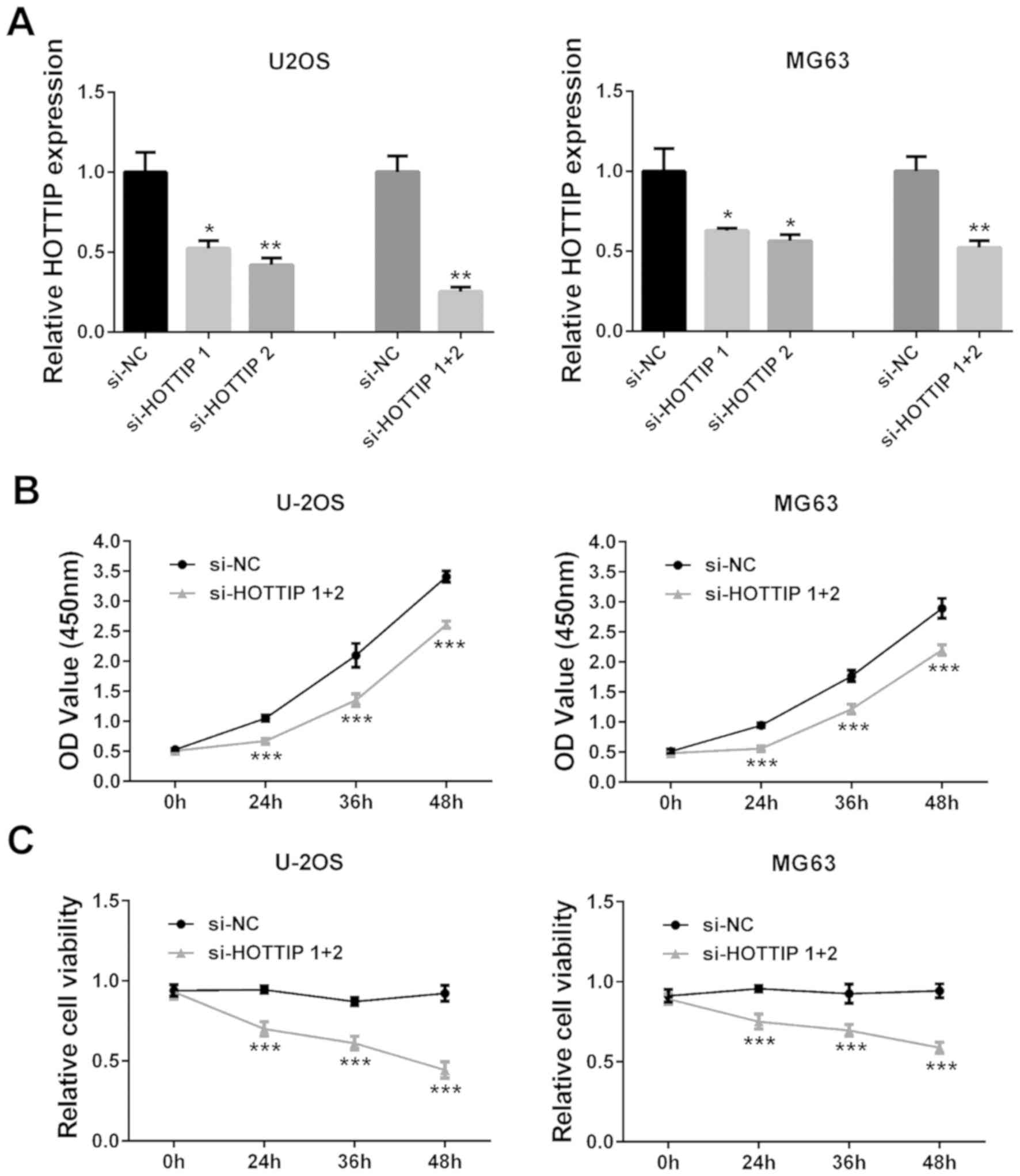
Knockdown of HOTTIP inhibits OS cell
viability, invasion, migration and EMT
siRNA-mediated silencing was used to knock down
HOTTIP expression levels in two OS cell lines, U2OS and MG63, as
HOTTIP was indicated to be highly expressed in these cells.
Individual and simultaneous transfections of two siRNAs against
HOTTIP successfully decreased HOTTIP expression levels in the two
OS cell lines (P<0.05; Fig. 2A);
co-transfection with si-HOTTIP 1 and si-HOTTIP 2 was used in all
subsequent experiments. CCK-8 assay revealed that HOTTIP knockdown
notably inhibited cell viability compared with the si-NC group
(P<0.001; Fig. 2B). In addition,
trypan blue staining also demonstrated that HOTTIP knockdown
significantly reduced OS cell viability (P<0.001; Fig. 2C).
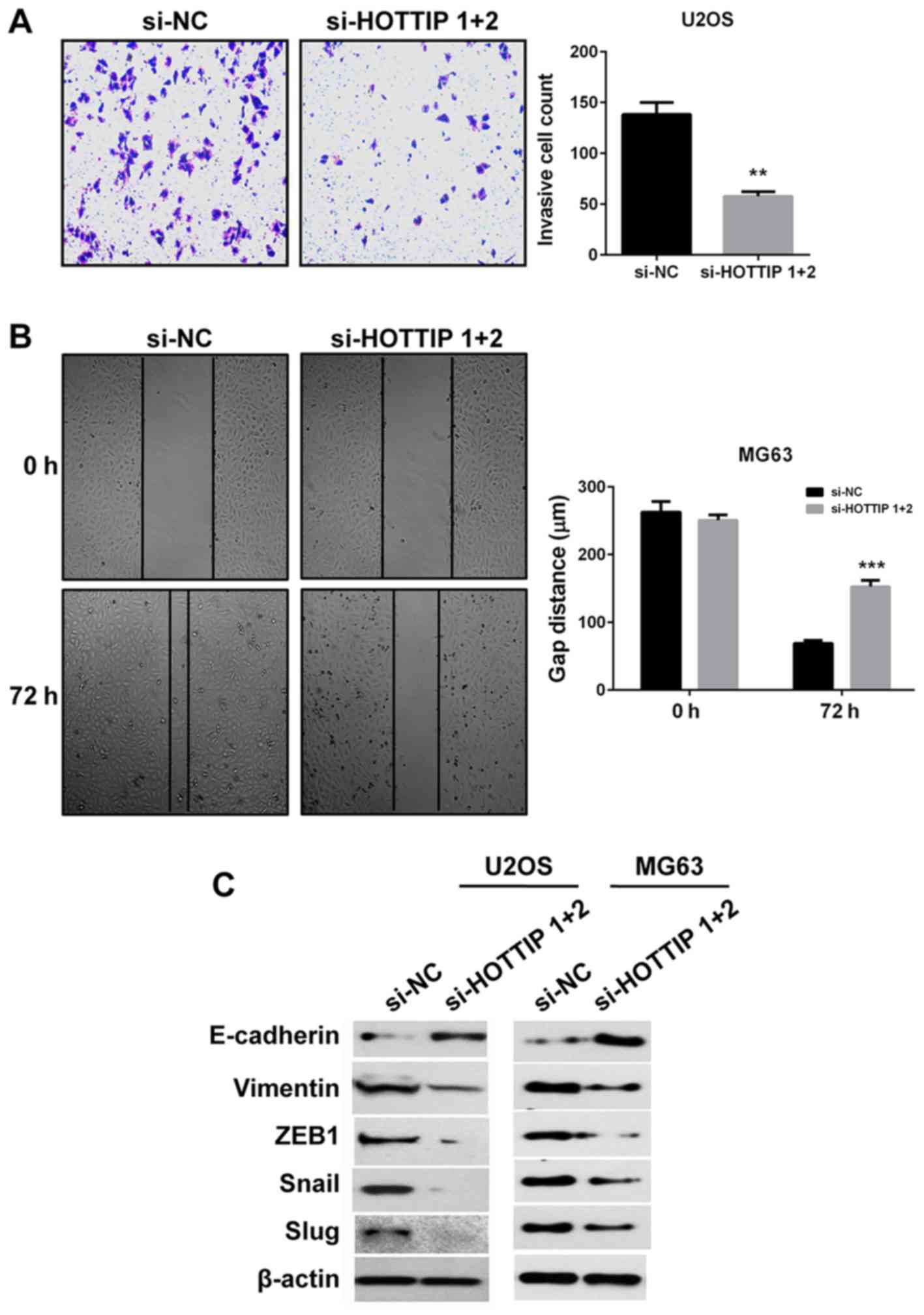
To determine the role of HOTTIP in OS pro-metastatic
processes in vitro, Matrigel and wound healing assays were
performed to evaluate the invasive and migratory capacities of the
cells, respectively. Due to the large size of MG63 cells and
therefore their inability to pass through the holes of the
Transwell, Matrigel assay was only performed in U2OS cells. Whereas
for wound healing assay, MG63 cells were used since they exhibited
higher migration capacity. The results demonstrated that HOTTIP
knockdown inhibited cell invasion and migration compared with si-NC
transfected cells (Fig. 3A and B).
Since EMT is also a key pro-metastatic process, the expression of
EMT markers was detected by western blotting. Following HOTTIP
knockdown, the protein expression levels of the epithelial marker
E-cadherin were upregulated, whereas the protein expression levels
of the mesenchymal markers vimentin, ZEB1, Snail and Slug were
downregulated in two OS cell lines (Fig.
3C). These results indicated that HOTTIP may have a role in the
promotion of OS invasion, migration and EMT.
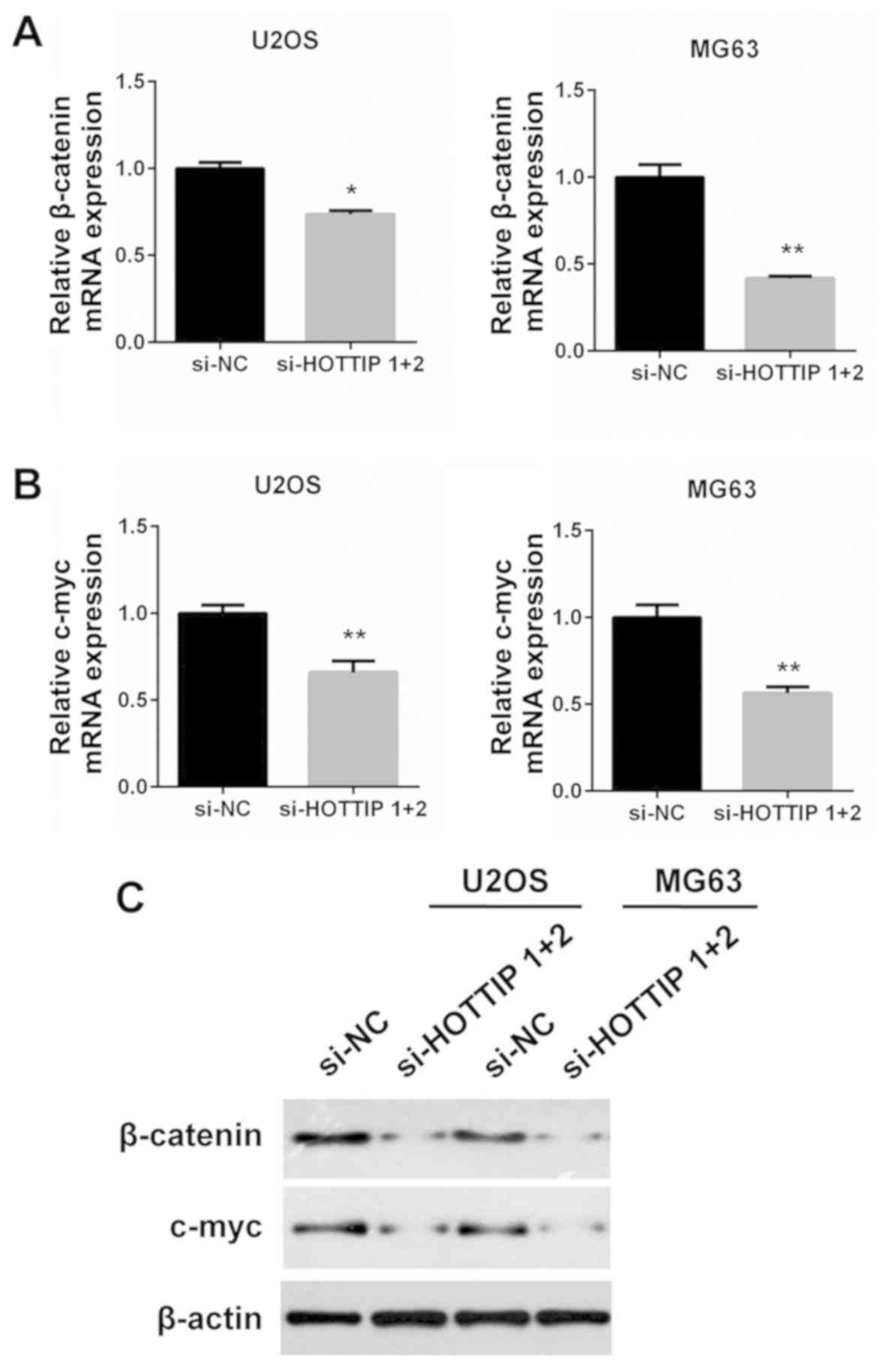
Reciprocal regulation between HOTTIP
and c-Myc
Based on a previous report by Li et al
(24), HOTTIP activates the
Wnt/β-catenin pathway in OS cells. Since c-Myc is an important
effector of the Wnt/β-catenin pathway, RT-qPCR and western blotting
were used to examine whether HOTTIP regulated the expression of
c-Myc in OS cells. Knockdown of HOTTIP led to a decrease of
β-catenin and c-Myc mRNA expression (P<0.05; Fig. 4A and B). The decreased expression was
also observed in their protein levels (Fig. 4C). Therefore, it was concluded that
HOTTIP may promote c-Myc expression in OS cells. Conversely, HOTTIP
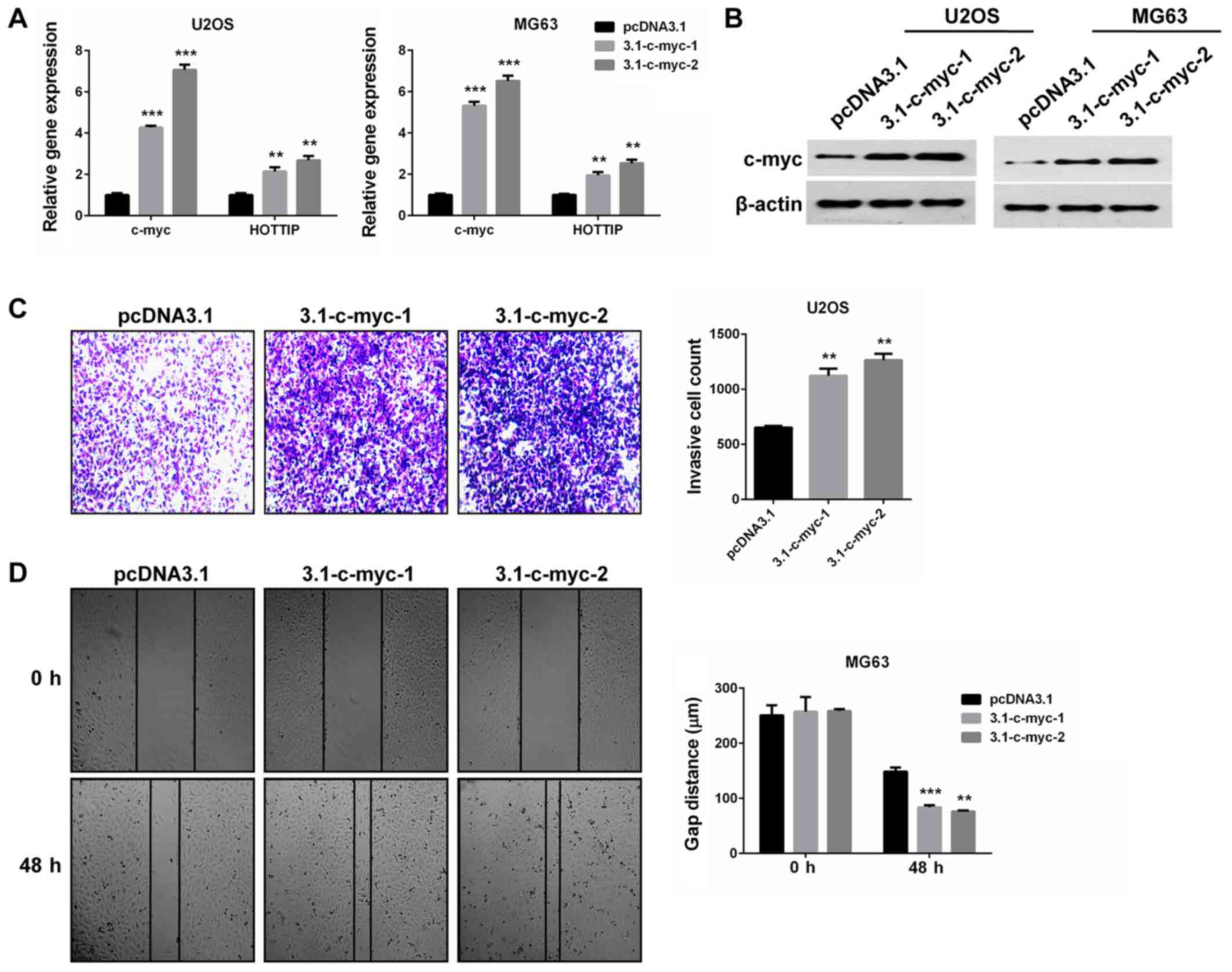
was upregulated by c-Myc overexpression in the two OS cell lines.
The two isoforms of c-Myc were confirmed to be overexpressed
(P<0.001; Fig. 5A and B) and
increased HOTTIP expression levels (P<0.01; Fig. 5A), compared with cells transfected
with an empty vector. Furthermore, c-Myc overexpression promoted OS
cell invasion and migration in vitro (P<0.01; Fig. 5C and D). These findings indicate the
oncogenic role of c-Myc in OS cells, and provide a reciprocal
linkage of HOTTIP and c-Myc.
Restoration of c-Myc in
HOTTIP-silenced OS cells rescued OS cell invasion and
migration
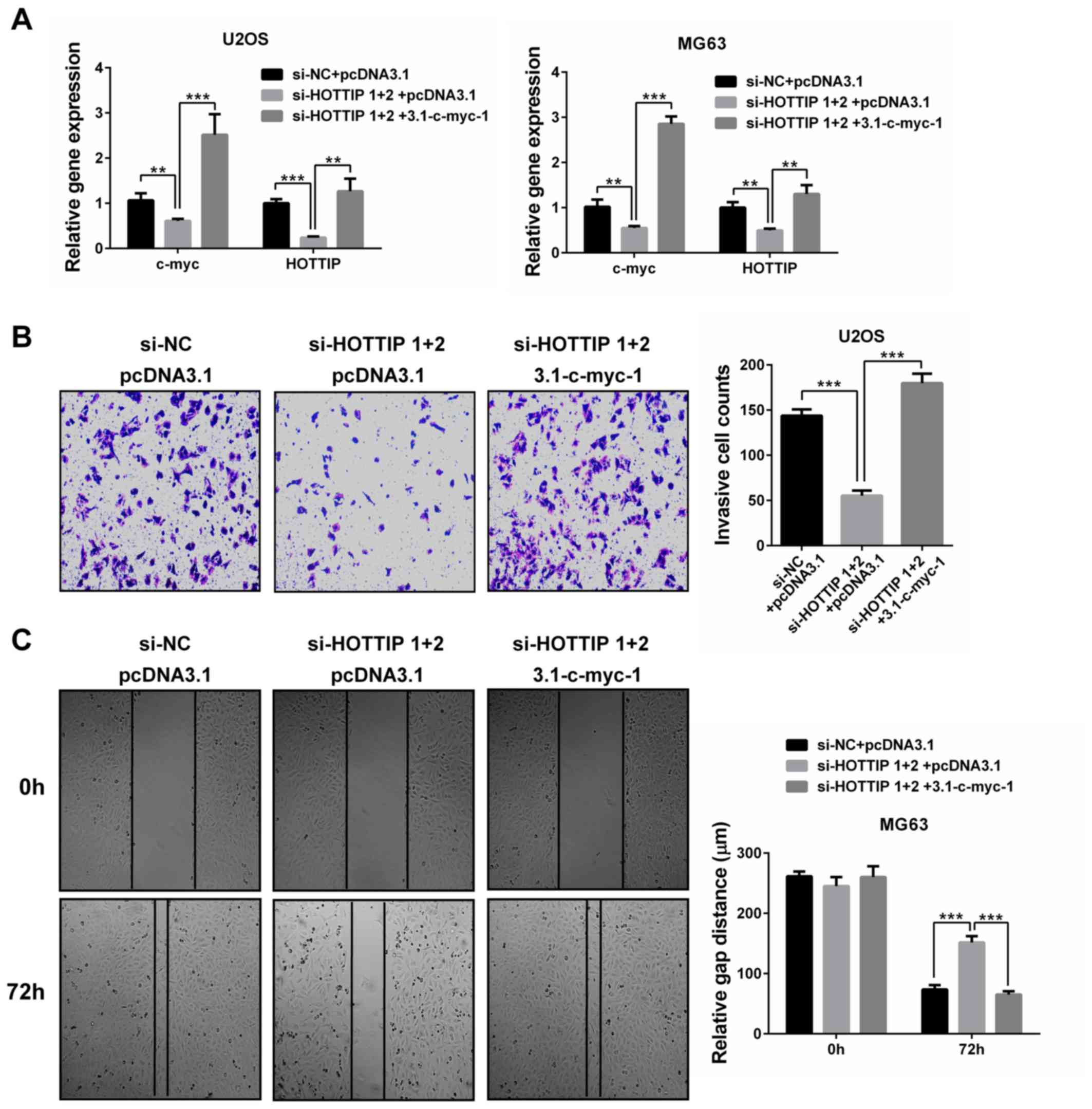
To test whether HOTTIP promoted OS cell invasion and
migration by upregulating c-Myc, c-Myc was overexpressed in
HOTTIP-silenced OS cells. RT-qPCR validated the overexpression of
c-Myc as well as the induction of HOTTIP expression in the
restoration group (P<0.01; Fig.
6A). Furthermore, c-Myc restoration rescued OS cell invasion
and migration (P<0.001; Fig. 6B and
C). These results suggested that c-Myc may be the mediator of
the oncogenic role of HOTTIP in OS cells.
Discussion
Mainly arising from the mesenchymal cells of the
long bones, OS is the most common bone malignancy in the world
(1). Despite the progress on its
treatment options, including surgery and chemotherapy, most
patients with OS still experience recurrence and have a short
overall survival time (3).
Metastasis is the major cause of the unsatisfactory clinical
outcome. A recent study has suggested that dysregulated expression
of several lncRNAs is implicated in OS progression (16). HOTTIP is among these lncRNAs, and two
reports have identified its expression and tumor-promoting
activities in OS (23,24). However, the role and mechanism of
HOTTIP in OS cell migration, invasion and EMT remain unclear.
Overexpression of HOTTIP has been reported in a
number of human cancers; with HOTTIP associated with multiple
cancer-associated malignancy phenotypes and signaling pathways, in
addition to contributing to cancer development (21). In the present study, higher
expression of HOTTIP was confirmed in OS tissues and all cell lines
compared to their matched controls. Moreover, knockdown of HOTTIP
by siRNAs reduced the viability of two OS cell lines. These
findings were in agreement with the previous studies on the role of
HOTTIP in OS (23,24).
Furthermore, the present study suggested that HOTTIP
may also contribute to the aggressive phenotypes of OS. The in
vitro experiments consistently demonstrated that knockdown of
HOTTIP inhibited OS cell migration, invasion and EMT, which
indicated the pro-metastatic activity of HOTTIP in OS. This was in
agreement with previous studies, which have identified the
promotion of cancer invasion and EMT by HOTTIP in gastric cancer,
esophageal squamous cell carcinoma and glioma (27–29). In
the first report of HOTTIP in OS by Li et al, the analysis
of the association between HOTTIP expression levels and several
clinicopathological features of patients with OS revealed that high
HOTTIP expression was associated with distant metastasis (23). In vitro experiments from the
same study demonstrated that knockdown of HOTTIP inhibited OS cell
migration and invasion (23).
Therefore, the results of the present study validated their
findings and verified the promotion of EMT by HOTTIP in OS
cells.
The relationship between HOTTIP and the
Wnt/β-catenin signaling pathway in OS cells has been described by
Li et al (24). HOTTIP
overexpression leads to increased expression of cyclin D1,
cyclin-dependent kinase 4 and β-catenin proteins, which are the
effectors of the Wnt/β-catenin signaling pathway (24). The activation of Wnt/β-catenin
signaling by HOTTIP is associated with the pro-growth and
chemoresistance roles of HOTTIP in OS cells (24). Recently, a study from colorectal
cancer revealed that knockdown of HOTTIP inhibited cell
proliferation and migration, and significantly suppressed the
expression of glycogen synthase kinase 3β, β-catenin and c-Myc
(30). The results of the present
study demonstrated that knockdown of HOTTIP decreased the
expression of β-catenin and c-Myc at both mRNA and protein levels.
In addition, c-Myc overexpression increased HOTTIP expression.
Thus, HOTTIP and c-Myc may form a positive feedback loop. The
reciprocal regulation between HOTTIP and c-Myc may explain the
upregulation of HOTTIP in OS, as elevated c-Myc expression may
determine the upregulation of HOTTIP. Conversely, c-Myc
overexpression may promote OS cell migration and invasion in
vitro, which may be related to the pro-metastatic role of
HOTTIP in OS cells.
In conclusion, the results of the present study
demonstrated that lncRNA HOTTIP was overexpressed in OS and may
facilitate migration, invasion and EMT in vitro by forming a
positive feedback loop with c-Myc. However, there are still several
limitations. Firstly, the oncogenic roles of HOTTIP in OS cells
need to be confirmed by in vivo data in the near future.
Secondly, the molecular mechanisms governing the reciprocal
regulation of HOTTIP and c-Myc remain largely unclear. Finally,
HOTTIP may have an impact on OS progression via other pathways. The
findings of the present study support that HOTTIP may serve as a
promising therapeutic target in the treatment of OS.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
YT and FJ designed the study. YT performed the
experiments and analyzed the data. FJ wrote the manuscript. Both
authors have read and approved the final version of this
manuscript.
Ethics approval and consent to
participate
Ethical approval for this study was obtained from
the Ethics Committee of Changhai Hospital, Second Military Medical
University (Shanghai, China), and written informed consent was
obtained from all patients at the initial stage of this study.
Patient consent for publication
All patients provided written informed consent for
the publication of their data.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Yang Y, Han L, He Z, Li X, Yang S, Yang J,
Zhang Y, Li D, Yang Y and Yang Z: Advances in limb salvage
treatment of osteosarcoma. J Bone Oncol. 10:36–40. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Abarrategi A, Tornin J, Martinez-Cruzado
L, Hamilton A, Martinez-Campos E, Rodrigo JP, González MV, Baldini
N, Garcia-Castro J and Rodriguez R: Osteosarcoma: Cells-of-Origin,
cancer stem cells and targeted therapies. Stem Cells Int.
2016:36317642016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Meazza C and Scanagatta P: Metastatic
osteosarcoma: A challenging multidisciplinary treatment. Expert Rev
Anticancer Ther. 16:543–556. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Osasan S, Zhang M, Shen F, Paul PJ, Persad
S and Sergi C: Osteogenic sarcoma: A 21st century review.
Anticancer Res. 36:4391–4398. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Vos HI, Coenen MJ, Guchelaar HJ and Te Loo
DM: The role of pharmacogenetics in the treatment of osteosarcoma.
Drug Discov Today. 21:1775–1786. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bonasio R and Shiekhattar R: Regulation of
transcription by long noncoding RNAs. Annu Rev Genet. 48:433–455.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Mercer TR, Dinger ME and Mattick JS: Long
non-coding RNAs: Insights into functions. Nat Rev Genet.
10:155–159. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ponting CP, Oliver PL and Reik W:
Evolution and functions of long noncoding RNAs. Cell. 136:629–641.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Li X and Li N: LncRNAs on guard. Int
Immunopharmacol. 65:60–63. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zampetaki A, Albrecht A and Steinhofel K:
Long non-coding RNA structure and function: Is there a link? Front
Physiol. 9:12012018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Balas MM and Johnson AM: Exploring the
mechanisms behind long noncoding RNAs and cancer. Noncoding RNA
Res. 3:108–117. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Pop-Bica C, Gulei D, Cojocneanu-Petric R,
Braicu C, Petrut B and Berindan-Neagoe I: Understanding the role of
non-coding RNAs in bladder cancer: From dark matter to valuable
therapeutic targets. Int J Mol Sci. 18(pii): E15142017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Khorshidi A, Dhaliwal P and Yang BB:
Noncoding RNAs in tumor angiogenesis. Adv Exp Med Biol.
927:217–241. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang G, Liu C, Deng S, Zhao Q, Li T, Qiao
S, Shen L, Zhang Y, Lü J, Meng L, et al: Long noncoding RNAs in
regulation of human breast cancer. Brief Funct Genomics.
15:222–226. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li Z, Dou P, Liu T and He S: Application
of long noncoding RNAs in osteosarcoma: Biomarkers and therapeutic
targets. Cell Physiol Biochem. 42:1407–1419. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chen R, Wang G, Zheng Y, Hua Y and Cai Z:
Long non-coding RNAs in osteosarcoma. Oncotarget. 8:20462–20475.
2017.PubMed/NCBI
|
|
17
|
Li P, Huang R, Huang T, Cheng S, Chen Y
and Wang Z: Long non-coding RNA NEAT1 promotes proliferation,
migration and invasion of human osteosarcoma cells. Int J Med Sci.
15:1227–1234. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Xu R, Feng F, Yu X, Liu Z and Lao L:
LncRNA SNHG4 promotes tumour growth by sponging miR-224-3p and
predicts poor survival and recurrence in human osteosarcoma. Cell
Prolif. 51:e125152018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chen X, Zhou Y, Liu S, Zhang D, Yang X,
Zhou Q, Song Y and Liu Y: LncRNA TP73-AS1 predicts poor prognosis
and functions as oncogenic lncRNA in osteosarcoma. J Cell Biochem.
Sep 14–2018.doi: 10.1002/jcb.27556 (Epub ahead of print).
|
|
20
|
Wang KC, Yang YW, Liu B, Sanyal A,
Corces-Zimmerman R, Chen Y, Lajoie BR, Protacio A, Flynn RA, Gupta
RA, et al: A long noncoding RNA maintains active chromatin to
coordinate homeotic gene expression. Nature. 472:120–124. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Fan Y, Yan T, Chai Y, Jiang Y and Zhu X:
Long noncoding RNA HOTTIP as an independent prognostic marker in
cancer. Clin Chim Acta. 482:224–230. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lian Y, Cai Z, Gong H, Xue S, Wu D and
Wang K: HOTTIP: A critical oncogenic long non-coding RNA in human
cancers. Mol Biosyst. 12:3247–3253. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Li F, Cao L, Hang D, Wang F and Wang Q:
Long non-coding RNA HOTTIP is up-regulated and associated with poor
prognosis in patients with osteosarcoma. Int J Clin Exp Pathol.
8:11414–11420. 2015.PubMed/NCBI
|
|
24
|
Li Z, Zhao L and Wang Q: Overexpression of
long non-coding RNA HOTTIP increases chemoresistance of
osteosarcoma cell by activating the Wnt/β-catenin pathway. Am J
Transl Res. 8:2385–2393. 2016.PubMed/NCBI
|
|
25
|
Zhang M, Wang D, Zhu T and Yin R: RASSF4
overexpression inhibits the proliferation, invasion, EMT and Wnt
signaling pathway in osteosarcoma cells. Oncol Res. 25:83–91. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ye H, Liu K and Qian K: Overexpression of
long noncoding RNA HOTTIP promotes tumor invasion and predicts poor
prognosis in gastric cancer. Onco Targets Ther. 9:2081–2088.
2016.PubMed/NCBI
|
|
28
|
Chen X, Han H, Li Y, Zhang Q, Mo K and
Chen S: Upregulation of long noncoding RNA HOTTIP promotes
metastasis of esophageal squamous cell carcinoma via induction of
EMT. Oncotarget. 7:84480–84485. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhang S, Wang W, Liu G, Xie S, Li Q, Li Y
and Lin Z: Long non-coding RNA HOTTIP promotes hypoxia-induced
epithelial-mesenchymal transition of malignant glioma by regulating
the miR-101/ZEB1 axis. Biomed Pharmacother. 95:711–720. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Liu T, Yu T, Hu H and He K: Knockdown of
the long non-coding RNA HOTTIP inhibits colorectal cancer cell
proliferation and migration and induces apoptosis by targeting
SGK1. Biomed Pharmacother. 98:286–296. 2018. View Article : Google Scholar : PubMed/NCBI
|