Introduction
As one of the most common forms of cancer,
colorectal cancer is the third most frequent cause of
cancer-related mortality worldwide, accounting for ~1.36 million
new cases and 0.69 million estimated mortalities each year
(1). Colorectal adenoma is a major
precursor to colorectal cancer, with an estimated 5% of adenomas
developing into carcinomas (2,3).
MicroRNAs (miRNAs/miRs) are small, non-coding RNAs that
post-transcriptionally modulate the stability of messenger RNAs to
regulate gene expression and serve crucial roles in colorectal
tumorigenesis (4). Therefore,
clarifying the alteration of miRNA expression profiles in the
progression from normal colorectal tissue to adenoma, and finally
to carcinoma, is essential for an improved understanding of the
biological mechanisms of colorectal tumorigenesis.
Global expression analysis based on high-throughput
data has become a powerful technique in miRNA analysis (5). In the present study, using
miRNA-sequencing, the miRNA expression profiles in matching normal
colorectal tissue, adenoma and carcinoma samples from patients were
detected. Critical miRNAs that may be involved in cancer
progression were investigated, and their biological functions were
assessed through comprehensive bioinformatic analysis. The present
results may facilitate a better understanding of the roles of
miRNAs in colorectal tumorigenesis and promote novel strategies for
the prevention and treatment of colorectal carcinoma.
Materials and methods
Ethics statements and patient
selection
The present study was approved by The Ethics
Committee of Zhongshan Hospital, Fudan University (Shanghai,
China). Written informed consent was obtained from the 6 male
patients with colorectal adenoma and carcinoma and three adjacent
samples were collected using colonoscopy from each patient with
normal mucosa, adenoma and carcinoma between January 2014 and July
2014. The age of the patients ranged between 49–64 years (mean age
± standard deviation, 57.67±6.83 years).
RNA isolation
Total RNA was extracted from tissue samples using
the RNAprep Pure kit (For Tissue) (Tiangen Biotech Co., Ltd.)
following the manufacturer's protocol. The concentration and purity
of RNA samples were determined by absorbance detection and
electrophoresis.
miRNA-sequencing
The sequencing libraries were constructed according
to the protocol for the Illumina small RNA sample preparation kit
(6). Sequencing was performed on the
Illumina HiSeq 2000 sequencer (Illumina, Inc.). Library
construction and sequencing were performed at the Genergy Biotech
Co., Ltd. miRNA expression was analyzed by miRdeep version 2.0.0.7
(6).
Screening for differentially expressed
miRNAs
ANOVA and least significant difference post hoc test
were performed in GraphPad Prism version 5 (GraphPad Software,
Inc.) to compare the mean differences of the expression among the
three groups in normal colorectal tissues, colorectal adenomas and
tumor tissues. miRNAs with P<0.05 and fold-change >2 were
considered significantly altered. miRNAs among all 6 patients that
were consistently and significantly increased or decreased in
adenoma and cancer were identified using an FDR cutoff value of
0.05 and absolute log2-fold change greater than 1, and selected for
the next stage of analysis. Hierarchical clustering was performed
to further sort the selected differentially expressed miRNAs with
similar expression patterns, using Gene Cluster 3.0 with the
following settings: Distance metric, Pearson correlation; linkage
rule, average linkage (bonsai.hgc.jp/~mdehoon/software/cluster/software.htm).
Prediction of miRNA target genes
The target genes of these miRNAs were predicted
using TargetScan (www.targetscan.org) (7)
and miRanda (www.microrna.org/microrna/home.do) (8) databases. The overlapping predictions
obtained from the two databases were considered as potential target
genes.
Gene ontology (GO) and Kyoto
encyclopedia of genes and genomes (KEGG) analyses
GO and KEGG analyses were applied to analyze the
main functions of the target genes and identify the significant
pathways. The GO (geneontology.org) (9)
and KEGG (www.genome.jp/kegg) (10) databases were used as previously
reported (5,11). Enrichment analysis provided a measure
of the significance of the GO and KEGG pathways. A threshold of
P<0.05 indicated statistical significance.
Results
Identification of differentially
expressed miRNAs
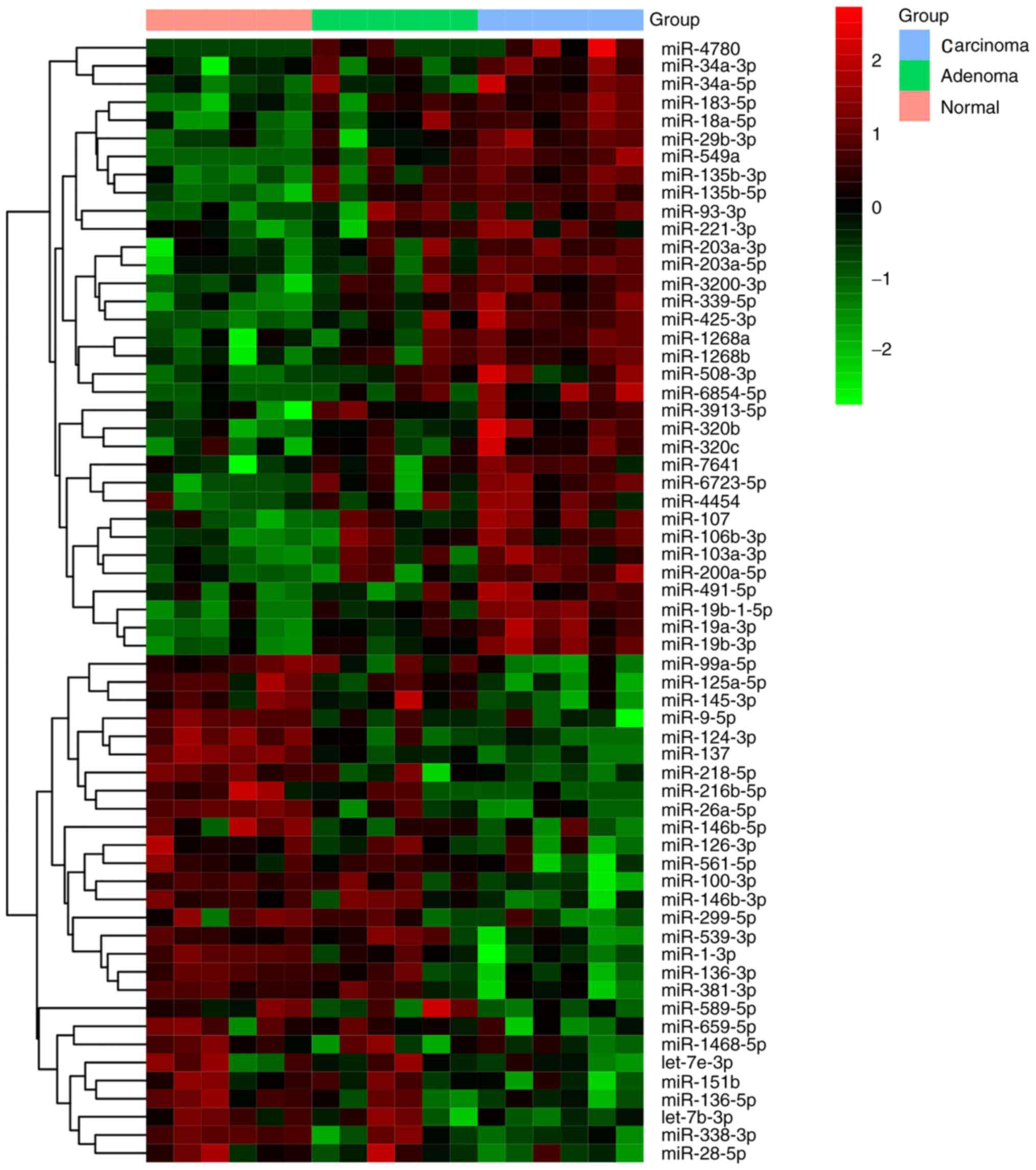
Microarray analysis revealed 334 differentially
expressed miRNAs in one normal colorectal, adenoma and carcinoma
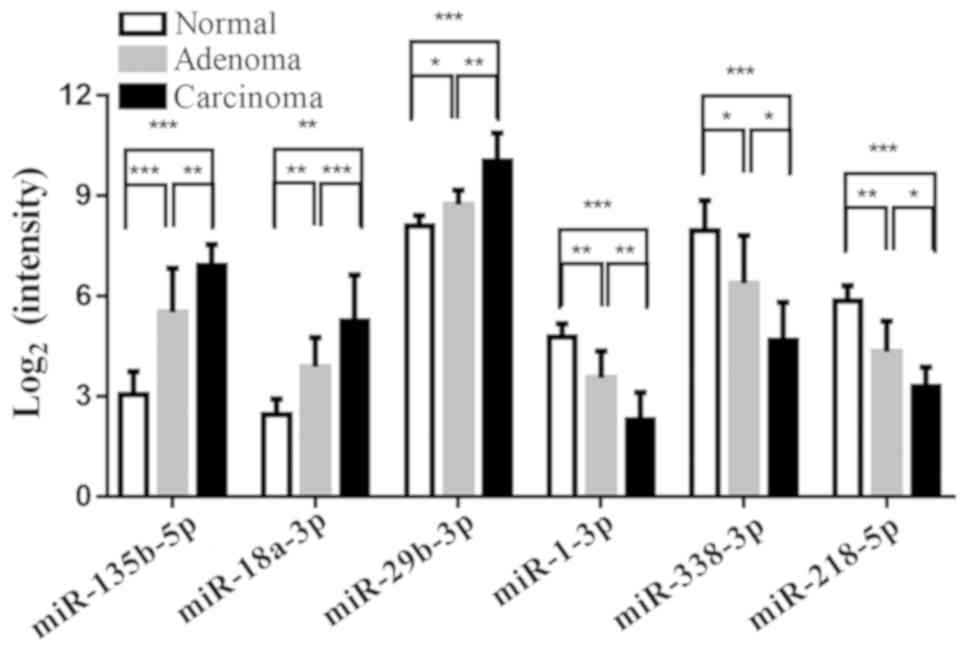
group compared with the other two groups. Among them, 34 miRNAs
were consistently upregulated in tissue undergoing colorectal
adenoma-carcinoma transition, including miR-135b-5p, miR-18a-5p and
miRNA-29b-3p (the three with the largest fold changes), while 28
miRNAs were consistently downregulated, including miR-1-3p,
miR-338-3p and miR-218-5p (the three with the largest fold changes)
(Fig. 1). The results of
hierarchical cluster analysis of the miRNAs with the largest up and
downregulated changes are shown in Fig.
2.
The target genes of these 62 miRNAs were predicted
using the miRNA target site databases TargetScan and miRanda, and
the overlapping results are listed in Table SI, including 3,715 genes.
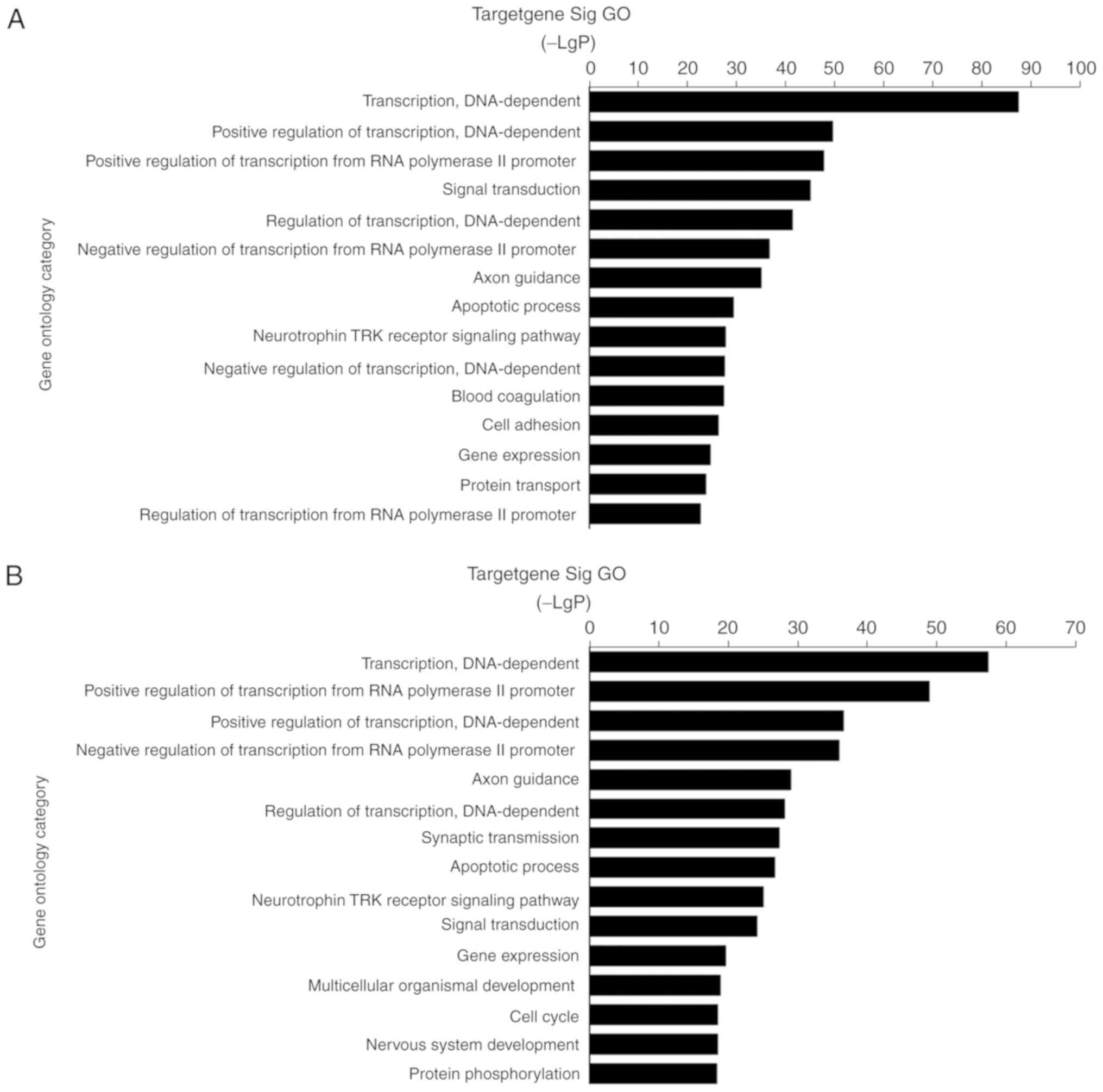
Enriched GO terms
Using GO analysis, it was found that the potential
target genes of the consistently upregulated miRNAs were
significantly enriched in 561 GO terms, while which of consistently
downregulated miRNAs were significantly enriched in 475 GO terms.
The potential targets of differentially expressed miRNAs were
mainly enriched in transcription regulation, signaling and
metabolism of biological process, extracellular matrix (ECM), and
signal transducer activity of molecular function during colorectal
tumorigenesis, such as ‘transcription, DNA-dependent (GO:0006351)’,
‘signal transduction (GO:0007165)’, ‘small molecule metabolic
process (GO:0044281)’, ‘apoptotic process (GO:0006915)’, ‘cell
adhesion (GO:0007155)’ and ‘protein transport (GO:0015031)’,
consistent with the proliferative and invasive capacity of
colorectal cancer cells (Fig. 3). In
addition, other GO functions associated with target genes included
‘activation of MAPK activity (GO:0000187)’, ‘cell cycle
(GO:0007049)’, ‘cell-cell signaling (GO:0007267)’,‘cell
differentiation (GO:0030154)’ and ‘protein ubiquitination
(GO:0016567)’.
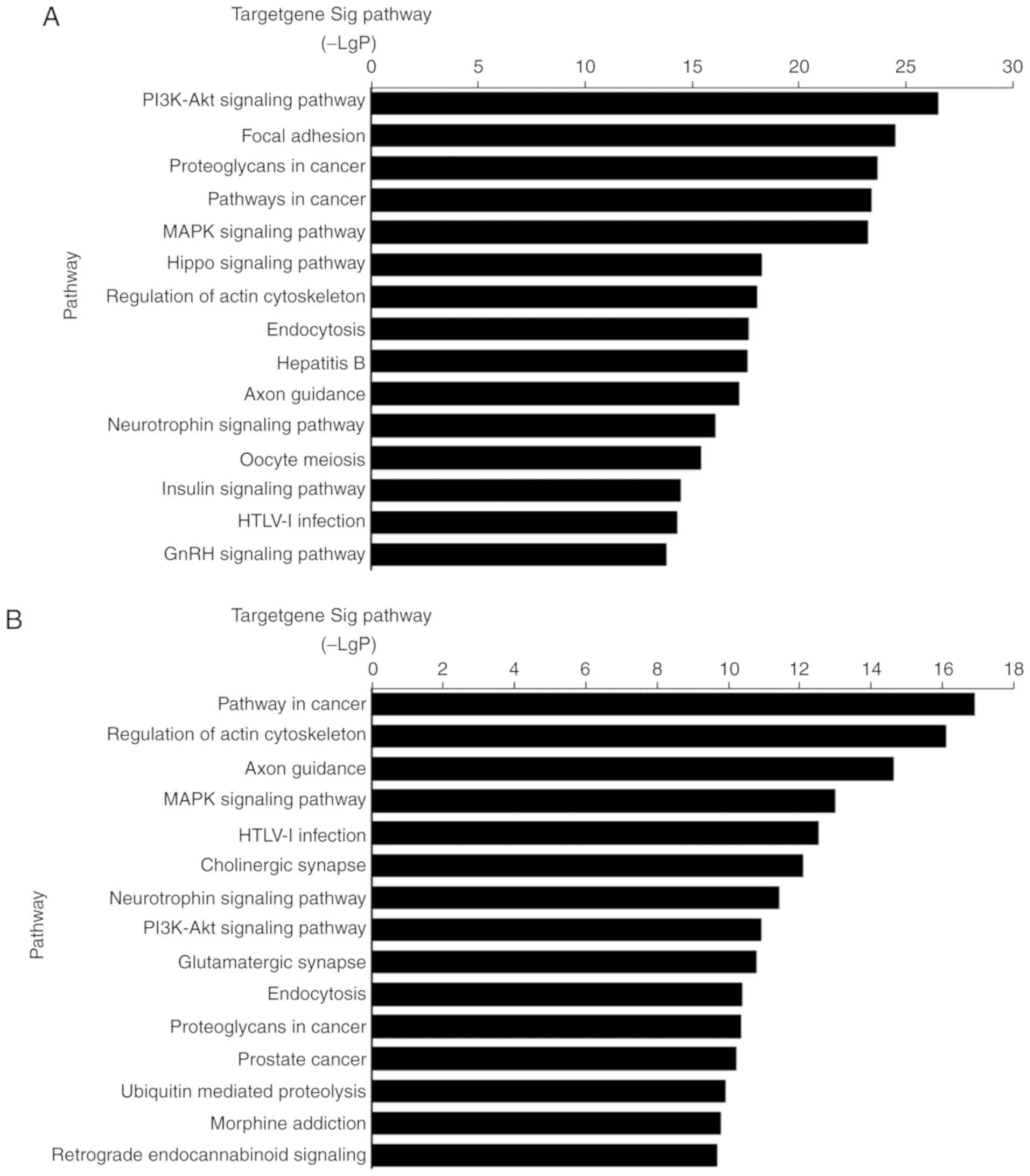
Significant KEGG pathways
Using KEGG analysis, it was found that the potential
target genes of upregulated miRNAs were significantly enriched in
135 signaling pathways, while those of downregulated miRNAs were
significantly enriched in 106 signaling pathways. The majority of
these signaling pathways overlapped. The results revealed that
potential target genes of the upregulated and downregulated miRNAs
are involved in the ‘MAPK signaling pathway’, which is consistent
with the GO analysis (Fig. 4).
Target genes were also significantly enriched in the ‘PI3K/AKT
signaling pathway’, ‘focal adhesion’ and the ‘Hippo signaling
pathway.’
Discussion
In the present study, high throughput screening of
miRNA expression profiles was performed by sequencing miRNA from
adjacent non-tumor, colorectal adenoma and carcinoma tissue samples
from the same patient. Careful screening revealed 334 miRNAs as
significantly differentially expressed in the three tissues types.
These differentially expressed miRNAs may be promising potential
diagnostic markers for the progression of colorectal cancer. In
support of these findings, a study by Slattery et al
(12) also identified ~600
differentially expressed miRNAs in colorectal carcinoma progression
using an Agilent microarray platform (12).
In the present study, the potential function of the
62 miRNAs which were consistently upregulated or downregulated
during the progression from normal to adenoma, and subsequently to
carcinoma, was investigated. It was revealed that miR-135b-5p,
miR-18a-5p and miR-29b-3p were consistently upregulated during this
transition. Several studies have confirmed the overexpression of
miR-135b-5p in colorectal tumors (13–15).
Valeri et al (13) reported
that miR-135b-5p overexpression was triggered in mice and humans by
loss of adenomatous polyposis coli, PTEN/PI3K pathway deregulation
and overexpression of proto-oncogene tyrosine-protein kinase Src,
and it was also demonstrated that miR-135b-5p promoted tumor
transformation and progression. Zhang et al (14) and Liu et al (16) reported that miR-135b-5p acted as an
upstream factor of the PI3K/AKT pathway, while the results
presented in this study demonstrated that the PI3K/AKT pathway was
enriched for targets of differentially expressed miRNAs. Yau et
al (17) reported that
miR-18a-5p expression was significantly increased in tumor tissues
and stool samples of patients with colorectal carcinoma, and that
it may serve as a potential biomarker for a non-invasive diagnosis
of this cancer. In the present study, miR-29b-3p was significantly
upregulated in colorectal tumors. However, Inoue et al
(18) reported that miR-29b-3p
expression was significantly downregulated in tumor tissue compared
with normal mucosa, and that it was an independent positive
prognostic factor of disease-free survival. Wang et al
(19) reported that miR-29b-3p
suppressed tumor growth and metastasis in colorectal cancer by
downregulating T-cell lymphoma invasion and metastasis 1 expression
and inhibiting epithelial to mesenchymal transition. Therefore,
further study is required to clarify the impact of altered
miR-29b-3p expression in colorectal tumors.
The present study also revealed that miR-1-3p,
miR-338-3p and miR-218-5p were significantly downregulated
throughout the transition from normal tissue to adenoma and then
carcinoma. A recent meta-analysis of The Cancer Genome Atlas and
Gene Expression Omnibus data performed by Wang et al
(20) demonstrated significant
miR-1-3p downregulation in colorectal cancer. Specifically,
miR-1-3p was found to suppress tumor proliferation via the mothers
against decapentaplegic homolog 3, hypoxia-inducible factor 1, and
MAPK and PI3K pathways (21,22). Sun et al (23) reported that miR-338-3p expression was
reduced in colorectal cancer and was negatively correlated with
advanced TNM stage and local invasion. It was also reported that
miRNA-338-3p suppressed cell proliferation of human colorectal
carcinoma by targeting the smoothened gene (24). Furthermore, several studies reported
miR-218-5p downregulation in colorectal cancer, which exerted its
tumor suppressor effects via the PI3K/AKT pathway, diphthamide
biosynthesis 1 and metastasis-associated in colon cancer 1, among
others (25–28).
Based on the present results, the predicted target
genes of differentially expressed miRNAs were enriched in cell
cycle, cell adhesion and the PI3K and MAPK signaling pathways. The
majority of these GO terms and KEGG pathways have been demonstrated
to serve significant roles in colorectal tumorigenesis. Sun et
al (29) found that the MAPK
pathway was associated with cell proliferation, differentiation,
migration, senescence and apoptosis in colorectal carcinoma.
Bogaert and Prenen (30) concluded
the PI3K/AKT, WNT, p53, receptor tyrosine kinase/RAS GTPase and
transforming growth factor β pathways to be the five pathways
frequently altered in colorectal cancer. Notably, the predicted
target genes identified in the present study were also enriched in
the majority of these pathways.
It should be noted that there are several
limitations to the present study. First, only samples from 6
patients were used. Second, the expression of miRNAs was not
validated by other methods such as quantitative PCR. Third, the
majority of the results were based on bioinformatics analysis and
require further validation.
In the present study, a high-throughput screening of
miRNA expression profiles in colorectal cancer development was
performed by sequencing miRNA from adjacent normal, colorectal
adenoma and carcinoma tissues. The miRNAs that were consistently
and significantly upregulated or downregulated were selected, and
their biological targets were predicted using bioinformatics
analysis. These miRNAs may be promising diagnostic biomarkers and
potential treatment targets in colorectal carcinoma. These findings
should be further validated in future experiments.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by The Medical
Leading Project of Shanghai Municipal Science and Technology
Committee (grant nos. 16DZ2280900 and 19DZ2280100).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available on reasonable request from the corresponding
author on reasonable request.
Authors' contributions
LY, YZ and PZ designed the study. JL and YZ wrote
the manuscript. JL and SC conducted the experiments and analyzed
the data. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by The Ethics
Committee of Zhongshan Hospital, Fudan University (Shanghai, China)
and conducted according to their guidelines and regulations.
Patient consent for publication
All participants provided informed consent.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Muto T, Bussey HJ and Morson BC: The
evolution of cancer of the colon and rectum. Cancer. 36:2251–2270.
1975. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Shinya H and Wolff WI: Morphology,
anatomic distribution and cancer potential of colonic polyps. Ann
Surg. 190:679–683. 1979. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Masuda T, Hayashi N, Kuroda Y, Ito S,
Eguchi H and Mimori K: MicroRNAs as biomarkers in colorectal
cancer. Cancers (Basel). 9:1242017. View Article : Google Scholar
|
|
5
|
Zhan C, Yan L, Wang L, Jiang W, Zhang Y,
Xi J, Jin Y, Chen L, Shi Y, Lin Z and Wang Q: Landscape of
expression profiles in esophageal carcinoma by the cancer genome
Atlas data. Dis Esophagus. 29:920–928. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Friedlander MR, Chen W, Adamidi C,
Maaskola J, Einspanier R, Knespel S and Rajewsky N: Discovering
microRNAs from deep sequencing data using miRDeep. Nat Biotechnol.
26:407–415. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Agarwal V, Bell GW, Nam JW and Bartel DP:
Predicting effective microRNA target sites in mammalian mRNAs.
Elife. Aug 12–2015.doi: 10.7554/eLife.05005. View Article : Google Scholar
|
|
8
|
Betel D, Koppal A, Agius P, Sander C and
Leslie C: Comprehensive modeling of microRNA targets predicts
functional non-conserved and non-canonical sites. Genome Biol.
11:R902010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
The Gene Ontology Consortium: Expansion of
the Gene Ontology knowledgebase and resources. Nucleic Acids Res.
45:D331–D338. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kanehisa M, Furumichi M, Tanabe M, Sato Y
and Morishima K: KEGG: New perspectives on genomes, pathways,
diseases and drugs. Nucleic Acids Res. 45:D353–D361. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yan L, Zhan C, Wu J and Wang S: Expression
profile analysis of head and neck squamous cell carcinomas using
data from The Cancer Genome Atlas. Mol Med Rep. 13:4259–4265. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Slattery ML, Herrick JS, Pellatt DF,
Stevens JR, Mullany LE, Wolff E, Hoffman MD, Samowitz WS and Wolff
RK: MicroRNA profiles in colorectal carcinomas, adenomas and normal
colonic mucosa: Variations in miRNA expression and disease
progression. Carcinogenesis. 37:245–261. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Valeri N, Braconi C, Gasparini P, Murgia
C, Lampis A, Paulus-Hock V, Hart JR, Ueno L, Grivennikov SI, Lovat
F, et al: MicroRNA-135b promotes cancer progression by acting as a
downstream effector of oncogenic pathways in colon cancer. Cancer
Cell. 25:469–483. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhang J, Raju GS, Chang DW, Lin SH, Chen Z
and Wu X: Global and targeted circulating microRNA profiling of
colorectal adenoma and colorectal cancer. Cancer. 124:785–796.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Jia L, Luo S, Ren X, Li Y, Hu J, Liu B,
Zhao L, Shan Y and Zhou H: miR-182 and miR-135b mediate the
tumorigenesis and invasiveness of colorectal cancer cells via
targeting ST6GALNAC2 and PI3K/AKT pathway. Dig Dis Sci.
62:3447–3459. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Liu B, Liu Y, Zhao L, Pan Y, Shan Y, Li Y
and Jia L: Upregulation of microRNA-135b and microRNA-182 promotes
chemoresistance of colorectal cancer by targeting ST6GALNAC2 via
PI3K/AKT pathway. Mol Carcinog. 56:2669–2680. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yau TO, Wu CW, Dong Y, Tang CM, Ng SS,
Chan FK, Sung JJ and Yu J: microRNA-221 and microRNA-18a
identification in stool as potential biomarkers for the
non-invasive diagnosis of colorectal carcinoma. Br J Cancer.
111:1765–1771. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Inoue A, Yamamoto H, Uemura M, Nishimura
J, Hata T, Takemasa I, Ikenaga M, Ikeda M, Murata K, Mizushima T,
et al: MicroRNA-29b is a novel prognostic marker in colorectal
cancer. Ann Surg Oncol. 22 (Suppl 3):S1410–S1418. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang B, Li W, Liu H, Yang L, Liao Q, Cui
S, Wang H and Zhao L: miR-29b suppresses tumor growth and
metastasis in colorectal cancer via downregulating Tiam1 expression
and inhibiting epithelial-mesenchymal transition. Cell Death Dis.
5:e13352014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang JY, Huang JC, Chen G and Wei DM:
Expression level and potential target pathways of miR-1-3p in
colorectal carcinoma based on 645 cases from 9 microarray datasets.
Mol Med Rep. 17:5013–5020. 2018.PubMed/NCBI
|
|
21
|
Xu W, Zhang Z, Zou K, Cheng Y, Yang M,
Chen H, Wang H, Zhao J, Chen P, He L, et al: MiR-1 suppresses tumor
cell proliferation in colorectal cancer by inhibition of
Smad3-mediated tumor glycolysis. Cell Death Dis. 8:e27612017.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Xu L, Zhang Y, Wang H, Zhang G, Ding Y and
Zhao L: Tumor suppressor miR-1 restrains epithelial-mesenchymal
transition and metastasis of colorectal carcinoma via the MAPK and
PI3K/AKT pathway. J Transl Med. 12:2442014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Sun K, Deng HJ, Lei ST, Dong JQ and Li GX:
miRNA-338-3p suppresses cell growth of human colorectal carcinoma
by targeting smoothened. World J Gastroenterol. 19:2197–2207. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sun K, Su G, Deng H, Dong J, Lei S and Li
G: Relationship between miRNA-338-3p expression and progression and
prognosis of human colorectal carcinoma. Chin Med J (Engl).
127:1884–1890. 2014.PubMed/NCBI
|
|
25
|
Liu M, Yin K, Guo X, Feng H, Yuan M, Liu
Y, Zhang J, Guo B, Wang C, Zhou G, et al: Diphthamide biosynthesis
1 is a novel oncogene in colorectal cancer cells and is regulated
by MiR-218-5p. Cell Physiol Biochem. 44:505–514. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ilm K, Fuchs S, Mudduluru G and Stein U:
MACC1 is post-transcriptionally regulated by miR-218 in colorectal
cancer. Oncotarget. 7:53443–53458. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li PL, Zhang X, Wang LL, Du LT, Yang YM,
Li J and Wang CX: MicroRNA-218 is a prognostic indicator in
colorectal cancer and enhances 5-fluorouracil-induced apoptosis by
targeting BIRC5. Carcinogenesis. 36:1484–1493. 2015.PubMed/NCBI
|
|
28
|
Zhang X, Shi H, Tang H, Fang Z, Wang J and
Cui S: miR-218 inhibits the invasion and migration of colon cancer
cells by targeting the PI3K/Akt/mTOR signaling pathway. Int J Mol
Med. 35:1301–1308. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Sun Y, Liu WZ, Liu T, Feng X, Yang N and
Zhou HF: Signaling pathway of MAPK/ERK in cell proliferation,
differentiation, migration, senescence and apoptosis. J Recept
Signal Transduct Res. 35:600–604. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Bogaert J and Prenen H: Molecular genetics
of colorectal cancer. Ann Gastroenterol. 27:9–14. 2014.PubMed/NCBI
|