Introduction
Among the different forms of endometrial cancer,
endometrioid endometrial adenocarcinoma (EEA) is the most common
type (85%) (1). Although the
prognosis of EEA is good, extensive heterogeneity has been reported
in a number of studies, particularly in patients with an early
stage of disease, exposing women to recurrent disease (2–4).
Clinically, certain patients with EEA with an advanced stage of
disease have a good prognosis, whereas certain patients with an
early stage of disease can still relapse and succumb (5,6). All
these features indicate that traditional clinical features are not
sufficient to accurately predict the prognosis of EEA. Molecular
biological characteristics and traditional clinical features are
particularly important in the prognosis of EEA. Tumor occurrence
and development are driven by genetic alterations, and the
phenotypic diversity may be accompanied by the corresponding
diversity in the pattern of gene expression (7). Therefore, establishing a predictive
prognostic model on the basis of gene expression profiles and
traditional clinical features, which are different from the
traditional criteria, is of great clinical value.
Machine-learning methods have been used to predict
the prognosis of numerous types of cancer (8,9). In the
machine-learning area of research, the prognosis of cancer is a
typical classification problem. When training a machine-learning
model to undergo a prediction task, the factors relevant to the
prognosis of cancer can be regarded as the features of the data,
and the prognosis results are the class labels. Random Forest (RF)
is a type of machine-learning method, which has been experimentally
proven to be the best classifier (10). RF has a number of advantages and has
already been successfully applied to microarray data classification
(11,12) and numerous other disease
classifications (13,14). Among the different variable selection
methods, variable selection using RF (VSURF) has demonstrated the
best predictive performance thus far (15). VSURF can handle thousands of input
variables and identify the most significant variables (10); thus, it is considered a feature
selection method and has been used to select the genes relevant to
the type of cancer in question (11,16).
However, to the best of our knowledge, there is
currently no RF for predicting EEA prognosis by combining gene
expression and traditional clinical characteristics. Therefore, the
aim of the present study was to establish a prediction model
combining genes and clinical features via RF for the prognosis of
EEA. First, the state-of-the-art method VSURF was used to select
informative factors that are relevant to the prognosis of EEA. The
selected factors were then used to design an accurate predictive
model via RF.
Materials and methods
Patient selection
The present study was performed in the Department of
Obstetrics and Gynecology, Peking University People's Hospital
(PKUPH; Beijing, China). In the training cohort, 154 primary EEA
(PE) samples without neoadjuvant therapy, RNA-sequencing (RNAseq)
expression (combining level 3 data) and clinical data of female
patients with uterine cancer were obtained from The Cancer Genome
Atlas (TCGA) data portal (cancergenome.nih.gov) on January 7, 2018. These data
included 64 PE samples without relapse (≥3 years of clinical
follow-up), without radiation therapy and without additional
pharmaceutical treatment, and 90 samples from relapsed or deceased
PE (R/D-PE) patients with or without postoperative adjuvant
therapy. TCGA samples were sub-stratified into four molecular
subgroups: i) Copy number low (CN-L), ii) copy number high (CN-H),
iii) microsatellite instability (MSI) and iv) catalytic subunit of
DNA polymerase ε involved in nuclear DNA replication and repair
(POLE) ultra-mutated, with different prognoses. Of the 154 cases in
TCGA training cohort, 20.13% were CN-L, 5.19% were CN-H, 33.12%
were MSI and 5.84% were POLE ultra-mutated; in 35.72% of the cases,
the molecular typing information was lacking. The detailed
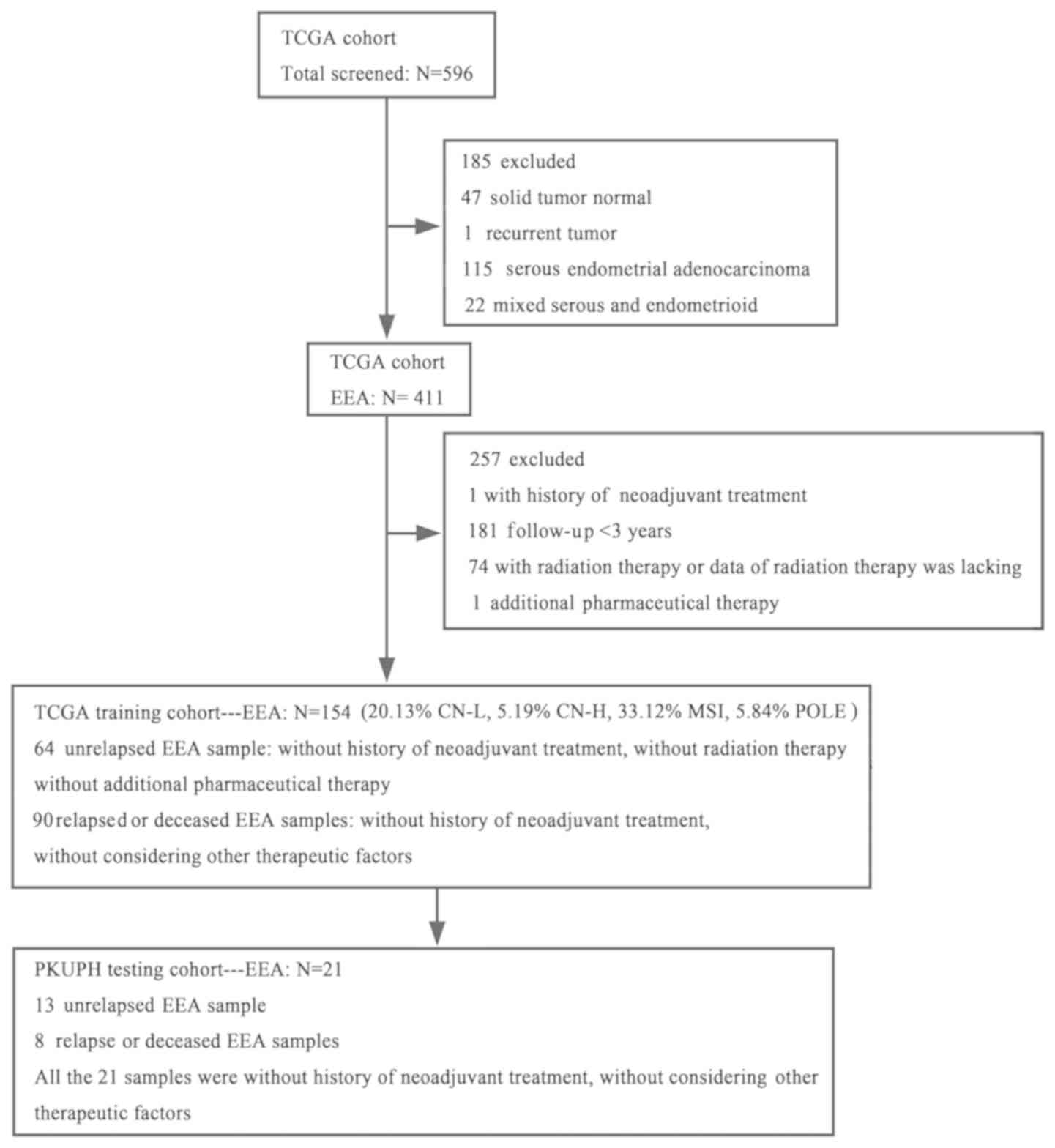
inclusion or exclusion criteria and information on the selection of
these 154 TCGA participants are presented in Fig. 1. In the testing cohort, 21 PE samples
without neoadjuvant therapy, as well as RNAseq expression and
clinical data, were obtained from 21 surgically treated patients at
the Department of Obstetrics and Gynecology PKUPH. All 21 samples
were from patients without neoadjuvant therapy and who underwent
surgical resection between January 2008 and December 2012. The
cohort included 13 PE samples from patients without relapse (≥3
years of clinical follow-up) and 8 R/D-PE samples from patients
with or without adjuvant therapy. The EEA samples were divided into
two groups according to the prognosis. The group with a good
prognosis contained the samples from non-relapsed EEA patients, and
the group with a poor prognosis contained the samples form relapsed
or deceased EEA patients. All deceased patients had succumbed to
EEA. The study was approved by the Institutional Ethics Committee
(Human Research) of the PKUPH.
RNA isolation, RNAseq library
preparation and sequencing of the 21 EEA samples
The total RNA was extracted with TRIzol®
(Tiangen Biotech Co., Ltd., Beijing, China) and assessed with an
Agilent 2100 BioAnalyzer instrument (Agilent Technologies, Inc.,
Santa Clara, CA, USA) and a Qubit™ 4 Fluorometer
(Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA). The
total RNA samples that met the following requirements were used in
subsequent experiments: RNA integrity number >7.0 and a 28S/18S
ratio >1.8. RNAseq libraries were generated and sequenced by
CapitalBio Technology Co., Ltd. (Beijing, China). Triplicate
samples of all assays were constructed in an independent library.
The NEB Next Ultra RNA Library Prep kit for Illumina (New England
BioLabs, Inc., Ipswich, MA, USA) was used to construct the DNA
libraries for sequencing. The NEB Next Poly(A) mRNA Magnetic
Isolation Module kit (New England BioLabs, Inc.) was used to enrich
the poly(A)-tailed mRNA molecules from 1 µg total RNA. The mRNA was
fragmented into ~200-bp pieces. The first-strand cDNA was
synthesized from the mRNA fragments using reverse transcriptase and
random hexamer primers (New England BioLabs, Inc.), and the
second-strand cDNA was synthesized using DNA polymerase I and
RNaseH (New England BioLabs, Inc.). The end of the cDNA fragment
was subjected to an end repair process that included the addition
of a single ‘A’ base, followed by ligation of the adapters,
according to the instructions of the NEB Next Ultra RNA Library
Prep kit (New England BioLabs, Inc.). The end Repair/dA-tail
program was: i) 20°C for 30 min; ii) 65°C for 30 min; iii) Hold at
4°C. The products were purified using Agencourt AMPure XP Beads
(Beckman Coulter, Inc., Brea, CA, USA) according to the
manufacturer's protocol and enriched by polymerase chain reaction
(PCR) to amplify the library DNA. Universal Primer Mix (New England
BioLabs, Inc.) was used for amplification. The thermocycling
conditions were as follows: 98°C for 30 sec; 12 cycles of 98°C for
10 sec, 65°C for 30 sec and 72°C for 30 sec; 72°C for 5 min. The
final libraries were quantified using the KAPA Library
Quantification kit (KAPA Biosystems; Roche Diagnostics, Basel,
Switzerland) and an Agilent 2100 Bioanalyzer (Agilent Technologies,
Inc.). The libraries were validated using reverse
transcription-quantitative PCR, and the thermocycling conditions
were as follows: 95°C for 5 min; 40 cycles of 95°C for 30 sec and
60°C for 45 sec. The libraries were subjected to paired-end
sequencing with a pair-end 150-bp reading length on an Illumina
HiSeq sequencer (Illumina, Inc., San Diego, CA, USA) (17).
Data processing
In total, 18,669 coding genes were included in TCGA
RNAseq data. Fragments per kilobase of exon model per million
mapped fragments (FPKM) gene expression values were used for the
statistical analysis. The format of RNAseq data downloaded from the
TCGA was log2(FPKM+1); thus, the RNAseq data of the TCGA
into FPKM was transformed for the follow-up study. There were a
number of clinical features in TCGA clinical data, including age at
initial pathological diagnosis (age), International Federation of
Gynecology and Obstetrics (FIGO) stage (18), grade (19), peritoneal wash status and lymph node
status. However, only the data for age, FIGO stage and grade were
complete in TCGA cohorts. Thus, of all the clinical features, only
age, FIGO stage and grade were included in the present study. To
improve the generalization of the study results, a numerical value
was given to age, FIGO stage and grade, according to a prior
published study (20) and clinical
experience. The numerical values of age, FIGO stage and grade were
as follows: Age (<60 years, 1; and ≥60 years, 2.55), grade
(I–II, 1; and III, 2.43), and FIGO stage (Ia, 1; Ib, 1.5; II, 2.75;
IIIa-b, 4; IIIc1, 4.21; IIIc2, 4.5; and IV, 6). These numerical
values were used for the establishment of the RF prognostic
prediction model of EEA.
RF
RF is an ensemble of decision trees, which forms
multiple decision trees and then aggregates them to provide a final
prediction. When a new object from an input vector is to be
predicted, the input vector is placed on each of the trees in the
forest simultaneously. Each tree gives a prediction, then, the
forest chooses the classification that has the most votes (out of
all the trees in the forest). RF uses the bagging technique and the
random feature selection technique. RF has two parameters, which
are the number of trees (ntree) and the number of variables
randomly sampled as candidates at each split (mtry).
VSURF
VSURF is a three-step feature selection method based
on RF. The first step is dedicated to removing irrelevant features
from the dataset. The second step aims to select important features
relevant to the class labels for interpretation purposes. The third
step refines the selection by removing redundancy in the set of
features selected by the second step, for prediction purposes. The
ntree parameter was set to its default value of 2,000, and the mtry
parameter was set to its default value (if nvm, the number of
variables in the model is not greater than the number of
observations; otherwise it is set to nvm/3). The VSURF results of
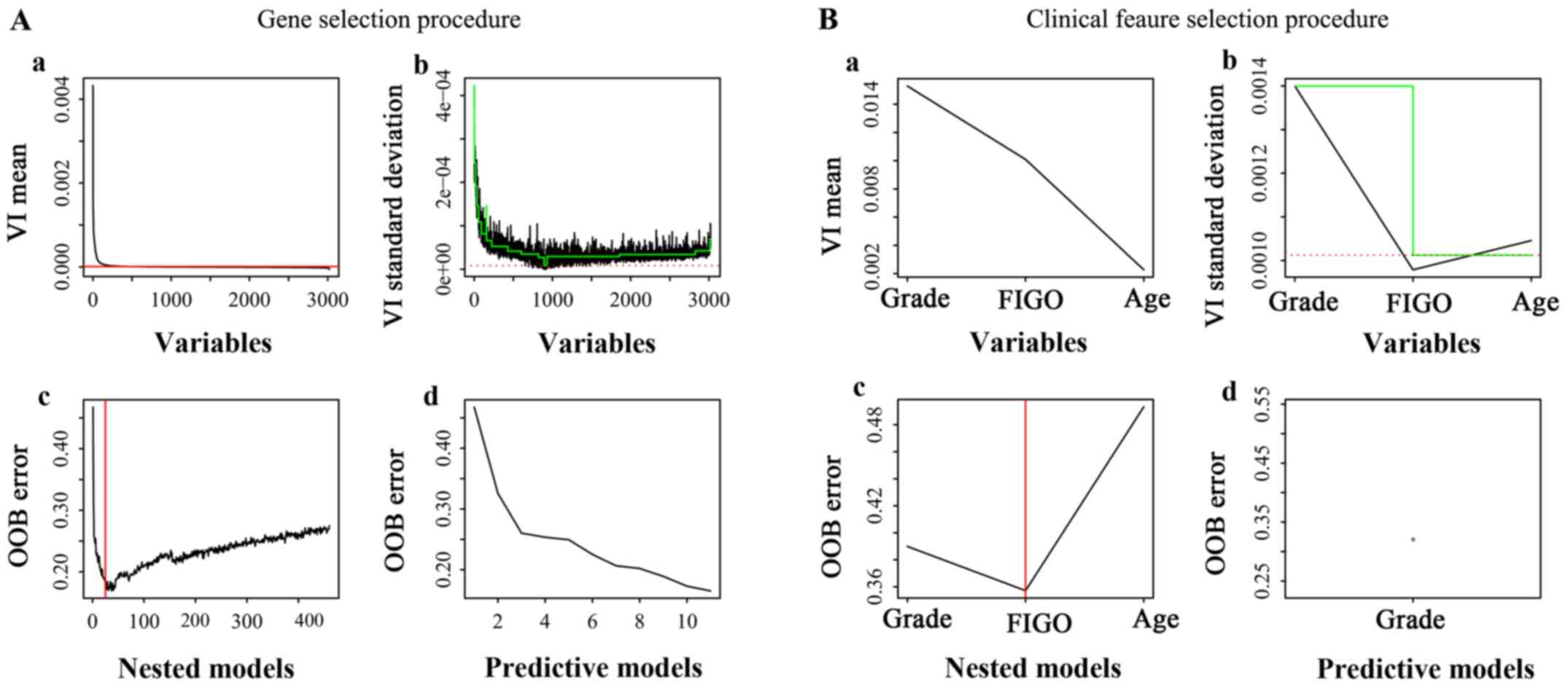
the genes and clinical features are summarized in Fig. 2, with Fig.
2Aa-b, and 2Ba-b corresponding
to the ‘thresholding step’, Fig. 2Ac
and 2Bc corresponding to the
‘interpretation step’, and Fig. 2Ad
and 2Bd corresponding to the
‘prediction step’. The features from the ‘interpretation step’ had
a strong association with EEA prognosis and were determined as the
most important factors that affect the prognosis of EEA.
Prediction experiment
RF parameter setting. The ntree parameter was set to
2,000, i.e., the RF included 2,000 decision trees, and the mtry
parameter was set to its default value.
Statistical analysis
All the model-associated data analyses were
performed using R software (version 3.2.4; http://www.r-project.org). The VSURF method was used
to select the most relevant prognostic genes and clinical
characteristics. RF was used to build the predictive model for
separating (relapsed or deceased) and unrelapsed patients. SPSS
software (version 13.0; SPSS Inc., Chicago, IL, USA) was used to
perform the statistical analysis. The associations between
clinicopathological characteristics and outcomes were calculated
using the χ2 test and Fisher's exact test. Survival
curves for the 154 PE samples from TCGA cohort (Fig. 3) were plotted using the Kaplan-Meier
method and the differences between survival curves were calculated
using a log-rank test. P<0.05 was considered to indicate a
statistically significant difference.
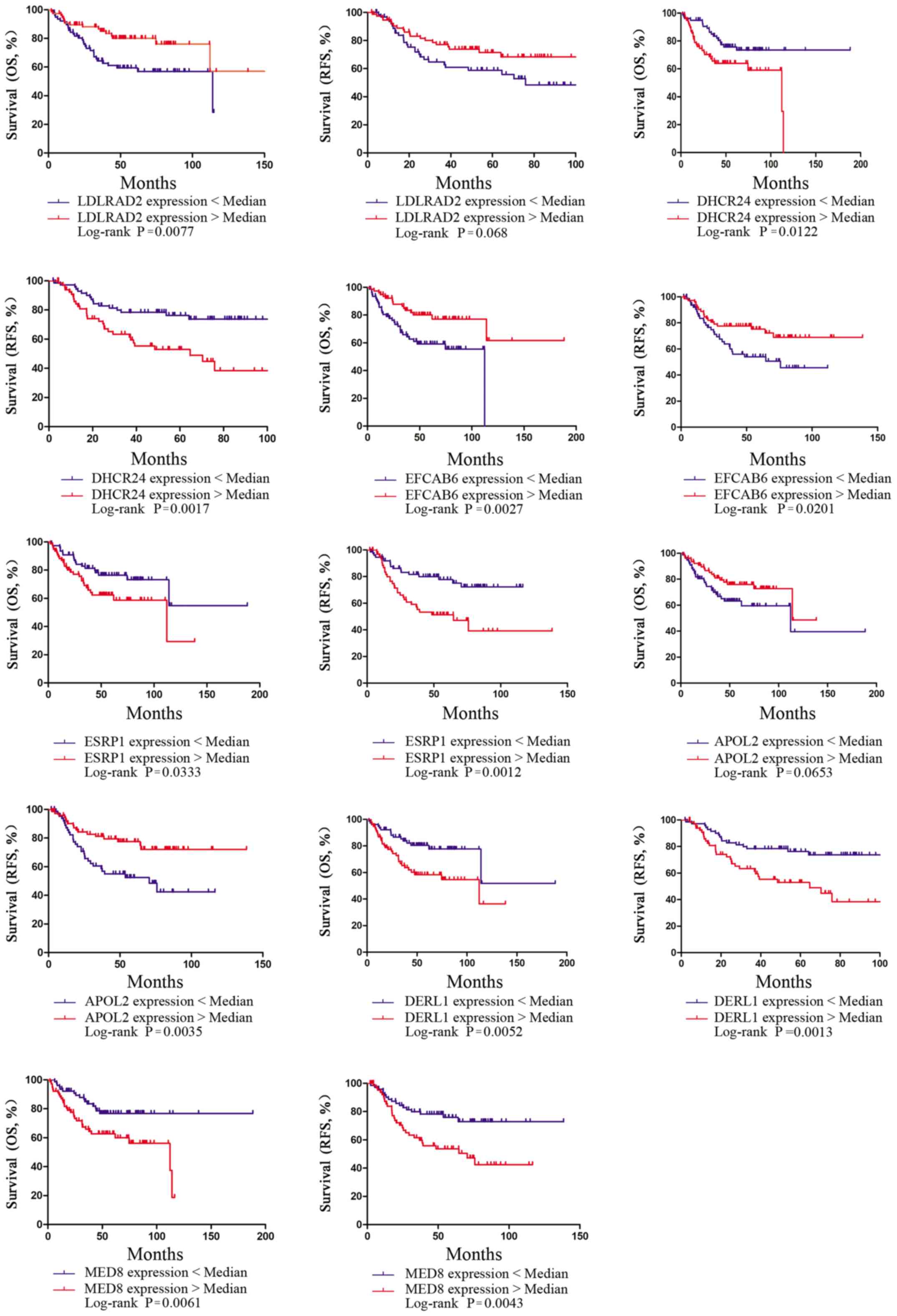 | Figure 3.Kaplan-Meier survival curves
presenting the effects of expression of the 11 genes on the overall
survival and relapse-free survival in patients with EEA in TCGA
cohort. OS, overall survival; RFS, relapse-free survival; EEA,
endometrioid endometrial adenocarcinoma; TCGA, The Cancer Genome
Atlas; LDLRAD2, low density lipoprotein receptor class A
domain-containing 2; DHCRAD2, 24-dehydrocholesterol reductase;
EFCAB6, EF-hand calcium-binding domain-containing protein 6; ESRP1,
epithelial-splicing-regulatory-protein 1; APOL2, apolipoprotein L2;
DERL1, derlin-1; MED8, mediator complex subunit 8. |
Results
Patient characteristics
In total, 154 PE samples meeting the inclusion
criteria from TCGA cohort were selected for the training set, and
21 PE samples from the PKUPH cohort were included in the testing
set. The median age of diagnosis for the samples in TCGA cohort was
64 years (range, 35–90 years). No significant difference was
observed in the diagnostic age and menopause status between the PE
samples and the R/D-PE samples, whereas significant differences
existed in the grade, FIGO stage, lymph node status, adjuvant
radiotherapy and body mass index. The median age of diagnosis for
the samples in the PKUPH cohort was 55 years (range, 31–75 years).
There was no significant difference observed in all the stated
clinical characteristics, perhaps due to the limited sample size in
the PKUPH cohort. The detailed data are presented in Table I.
 | Table I.Clinicopathological characteristics
of patients with EEA in TCGA and PKUPH cohorts. |
Table I.
Clinicopathological characteristics
of patients with EEA in TCGA and PKUPH cohorts.
|
| TCGA cohort |
| PKUPH cohort |
|
|---|
|
|
|
|
|
|
|---|
| Variable | Overall
(n=154) | PE (n=64) | R/D-PE (n=90) | P-value | Overall (n=21) | PE (n=13) | R/D-PE (n=8) | P-value |
|---|
| Age, years |
|
|
| 0.09 |
|
|
| 0.631 |
| Median
(range) | 64 (35–90) | 62 (35–89) | 65 (35–90) |
| 55 (31–75) | 51 (41–75) | 56 (31–63) |
|
| <60,
n | 55 | 28 | 27 |
| 13 | 10 | 5 |
|
| ≥60,
n | 99 | 36 | 63 |
| 8 | 3 | 3 |
|
| Grade, n |
|
|
| 0.005 |
|
|
| 0.930 |
|
1–2 | 80 | 42 | 38 |
| 18 | 11 | 7 |
|
| 3 | 74 | 22 | 52 |
| 3 | 2 | 1 |
|
| FIGO, n |
|
|
| 0.005 |
|
|
| 0.203 |
| I | 105 | 52 | 53 |
| 12 | 9 | 3 |
|
|
II–IV | 49 | 12 | 37 |
| 9 | 4 | 5 |
|
| Menopause status,
n |
|
|
| 0.789 |
|
|
| 0.131 |
|
Premenopausal | 12 | 4 | 8 |
| 4 | 4 | 0 |
|
|
Postmenopausal | 129 | 54 | 75 |
| 17 | 9 | 8 |
|
|
Unknown | 13 | 6 | 7 |
| 0 | 0 | 0 |
|
| ER status, n |
|
|
|
|
|
|
| 0.133 |
|
Positive | NA | 19 | 13 | 6 |
|
|
|
|
|
Negative |
|
|
|
| 2 | 0 | 2 |
|
| PR status, n |
|
|
|
|
|
|
| 0.381 |
|
Positive | NA | 20 | 13 | 7 |
|
|
|
|
|
Negative |
|
|
|
| 1 | 0 | 1 |
|
| Lymph node status,
n |
|
|
| 0.008 |
|
|
| 0.716 |
|
Positive | 22 | 4 | 18 |
| 2 | 1 | 1 |
|
|
Negative | 46 | 25 | 21 |
| 19 | 12 | 7 |
|
|
Unknown | 86 | 35 | 51 |
| 0 | 0 | 0 |
|
| Adjuvant
radiotherapy, n |
|
|
| <0.001 |
|
|
| 0.67 |
|
Yes | 41 | 0 | 41 |
| 11 | 6 | 5 |
|
| No | 110 | 64 | 46 |
| 9 | 6 | 3 |
|
|
Unknown | 3 | 0 | 3 |
| 1 | 1 | 0 |
|
| Adjuvant
chemotherapy, n |
|
|
|
|
|
|
| 0.599 |
|
Yes | NA | 13 | 7 | 6 |
|
|
|
|
| No |
|
|
|
| 6 | 4 | 2 |
|
|
Unknown |
|
|
|
| 2 | 2 | 0 |
|
| BMI, n |
|
|
| <0.001 |
|
|
| 0.659 |
|
<28 | 40 | 40 | 0 |
| 10 | 7 | 3 |
|
|
≥28 | 108 | 24 | 84 |
| 11 | 6 | 5 |
|
|
Unknown | 6 | 0 | 6 |
| 0 | 0 | 0 |
|
Establishing an RF prediction model on
the basis of the selected genes
The VSURF method was used to select genes from
18,669 coding genes of TCGA RNAseq data for the establishment of RF
prediction models, and ultimately, 11 genes were selected (Fig. 2Ab). First, 19 genes that had the most
relevance to the prognosis of EEA were selected (Fig. 2Ac). To further reduce the number of
genes for the RF models, 11 genes (Table II) were selected from these 19 genes
as the input factors. For seven of the 11 genes [low density
lipoprotein receptor class A domain-containing 2 (LDLRAD2) (OS,
P<0.05; RFS, P>0.05), 24-dehydrocholesterol reductase
(DHCR24) (OS, P<0.05; RFS, P<0.05), EF-hand calcium-binding
domain-containing protein 6 (EFCAB6) (OS, P<0.05; RFS,
P<0.05), epithelial-splicing-regulatory-protein 1 (ESRP1) (OS,
P<0.05; RFS, P<0.05), apolipoprotein L2 (APOL2) (OS,
P>0.05; RFS, P<0.05), derlin-1 (DERL1) (OS, P<0.05; RFS,
P<0.05) and mediator complex subunit 8 (MED8) (OS, P<0.05;
RFS, P<0.05), the gene expression was significantly associated
with the survival of EEA (P<0.05; Fig. 3). The classification ability of the
11 genes (Fig. 2Ad) was
approximately equal to the 19 genes (Fig. 2Ac). In the training set, the
out-of-bag (OOB) error of RF model-1 established by the 11 genes
was 15% (Fig. 2Ad). In the testing
set, when RF model-1 was used to validate the 21 EEA samples from
the PKUPH cohort, its classification accuracy was 71.43%.
 | Table II.Genes selected by Random Forest
feature selection that may contribute to the prognosis of
endometrial adenocarcinoma. |
Table II.
Genes selected by Random Forest
feature selection that may contribute to the prognosis of
endometrial adenocarcinoma.
| Gene | Chromosome no. | Definition |
|---|
| LDLRAD2 | 1 | Low density
lipoprotein receptor class A domain containing 2 |
| DHCR24 | 1 |
24-dehydrocholesterol reductase |
| EFCAB6 | 22 | EF-hand
calcium-binding domain 6 |
| ESRP1 | 8 | Epithelial splicing
regulatory protein 1 |
| APOL2 | 22 | Apolipoprotein
L2 |
| DERL1 | 8 | Derlin 1 |
| MED8 | 1 | Mediator complex
subunit 8 |
| PGAP3 | c7 | Post-GPI attachment
to proteins 3 |
| ELAVL4 | 1 | Embryonic lethal
abnormal visual system-like neuron-specific RNA binding protein
4 |
| TMEM27 | X | Transmembrane
protein 27 |
| ATF7IP2 | 16 | Activating
transcription factor 7 interacting protein 2 |
Establishing an RF prediction model on
the basis of the clinical features
The VSURF method was used to select clinical
features for establishing RF prediction models, and the grade was
selected (Fig. 2B). The results
indicated that grade and FIGO stage were the most relevant to the
EEA prognosis (Fig. 2Bc). To further
reduce the number of clinical factors in the RF models, grade was
finally chosen as the input factor (Fig.
2Bd). Grade had an almost equal ability to assign a
classification compared with the ‘grade combined with FIGO stage’
(Fig. 2Bc, 2Bd). In the training
set, the OOB error of the RF model-2 established by grade was 0.39
(Fig. 2Bd). When RF model-2 was used
to validate the 21 EEA samples from the PKUPH cohort, the
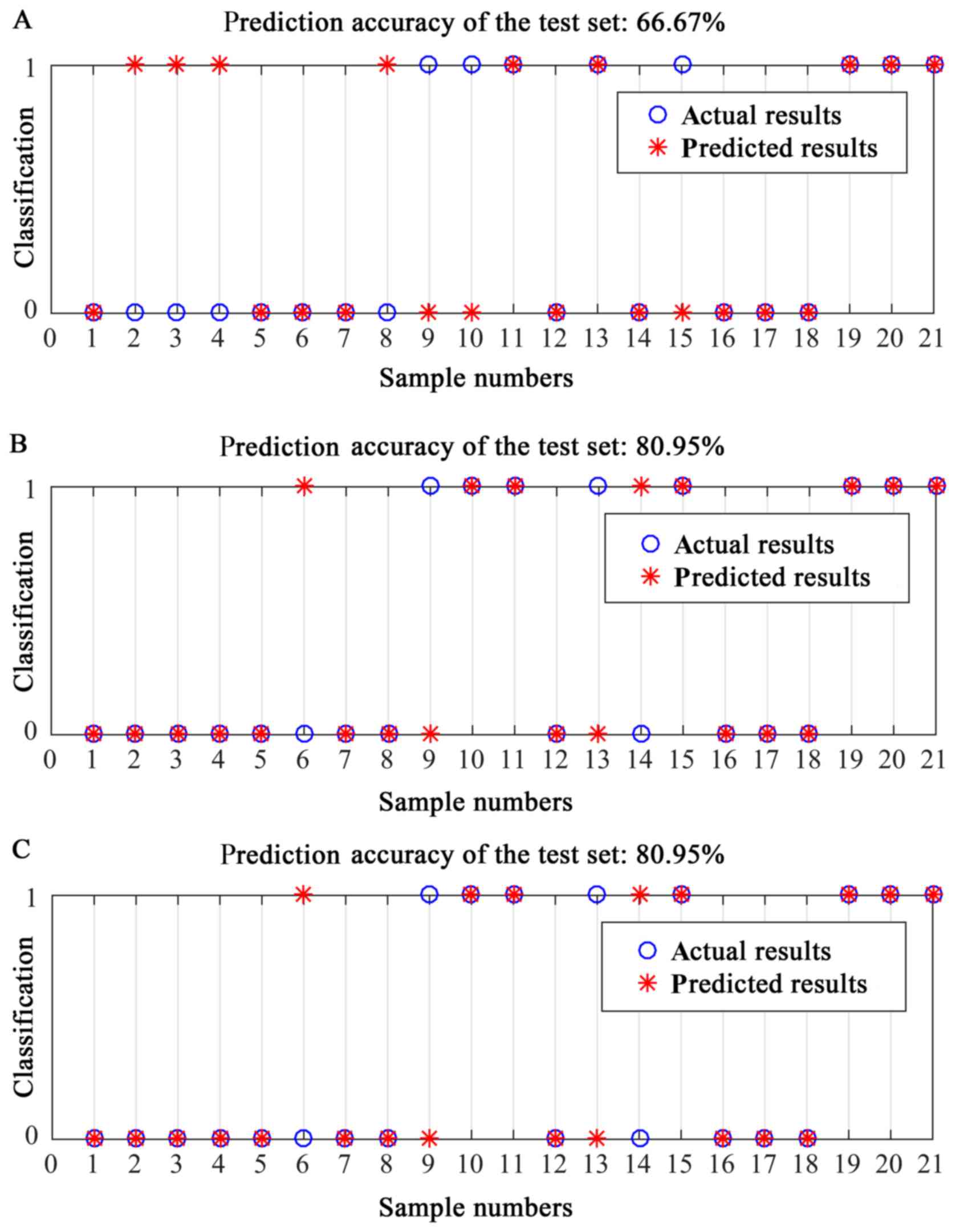
classification accuracy was 66.67% (Fig.
4A).
Establishing a RF combined model on
the basis of the ‘genes and clinical features’
Molecular biological characteristics and traditional
features serve important roles in EEA prognosis. Thus, a
RF-combined model-3 for EEA prognosis was established by combining
‘11 genes and grade’. In the training set, the OOB error of RF
model-3 established by ‘11 genes and grade’ was 0.15. When RF
model-3 was used to validate the 21 EEA samples from the PKUPH
cohort, its classification accuracy was 80.95% (Fig. 4B). The classification accuracy of the
RF model established by ‘11 genes, grade and stage’ was 80.95%
(Fig. 4C), further proving that
grade alone had equal classification ability compared with ‘grade
combined with FIGO stage’.
Discussion
Although the prognosis of EEA is good, extensive
heterogeneity can expose patients to recurrent disease and poor
prognosis (3,4). Treatments for EEA have become more
complicated, as the histological classification, adjuvant
therapies, indications and modalities for lymphadenectomy, and the
classifications used to predict relapse risk factors have all
changed (21). Traditional clinical
criteria are not enough to predict EEA prognosis accurately,
although studies have demonstrated that a number of clinical
factors, including tumor grade, age, comorbidities, tumor diameter,
American Society of Anesthesiologists score (22), lymphovascular space involvement and
postoperative complications at 30 days, serve important roles in
the prognosis of endometrial cancer (23–25). For
the limits of conventional traditional methods used for
histological classification of endometrial cancer subtypes, Barlin
et al suggested a combination of molecular and conventional
characteristics as classifications for better appraisal of
prognostic and predictive factors (26). Combining traditional clinical factors
and molecular biological characteristics for the prognosis of EEA
is important.
Machine-learning methods (27,28) can
provide increased prediction accuracy and can account for complex
interactions among predictors. In addition, machine-learning
approaches tend to be more suitable than traditional statistical
methods for certain situations, such as cancer prognostic
prediction, which involves a certain number of potential predictors
(28). In machine learning,
traditional classifiers are usually desired for prediction accuracy
and easily fit in with clinical norms, whereas RF stands out for
its own inherent characteristics, which include a better
generalization performance and excellent classification results
(10,29). RF has also been demonstrated to be
highly suitable for reducing the dimensionality of the data
(29); it has been successfully used
in numerous scientific realms, such as evaluating cancer-associated
cognitive impairment, disease prediction, genetics, proteomics and
informatics (29–31), but currently has no application in
the prediction of EEA prognosis. Not only are RF good classifiers,
but they are also increasingly used as feature-selection methods.
In the present study, the VSURF method was used to identify
informative factors that were relevant to the prognosis of EEA. The
selected factors were then used to design a good RF predictive
model.
In the present study, grade and 11 genes were
selected for the establishment of an RF model. The selected 11
genes were involved in a number of important biological processes
and potentially affect the prognosis of EC. LDLRAD2 is an integral
component of the cell membrane. The present study indicated that
LDLRAD2 was associated with the prognosis of EEA, but the definite
biological significance of LDLRAD2 remains to be investigated. In
addition, DHCR24 serves important roles in anti-apoptosis, cell
cycle arrest, the negative regulation of cell death and the
regulation of caspase activity; these biological processes are
associated with poor prognosis (32). Dai et al (33) identified that insulin-induced
cholesterol synthetase DHCR24 aggravates the invasion of cancer and
the resistance to progesterone in endometrial carcinoma. A previous
study also demonstrated that DHCR24 is able to predict poor
clinicopathological features of patients with bladder cancer, and
that its expression may promote bladder cancer cell proliferation
via several oncogenesis-associated biological processes (for
example, via estrogen response, heme metabolism, the p53 pathway,
cholesterol homeostasis, mammalian target of rapamycin complex 1
signaling, peroxisomes, xenobiotic metabolism, glycolysis and
protein secretion) (34). EFCAB6 and
MED8 genes serve important roles in the transcription of genes,
including certain prognosis-associated genes. ESRP1 and embryonic
lethal abnormal visual system-like neuron-specific RNA-binding
protein 4 (ELAVL4) participates in RNA processing, mRNA processing
and mRNA metabolic processing. Li et al (35) demonstrated that ESRP1 inhibited the
invasion and metastasis of lung adenocarcinoma, and served a role
in regulating proteins involved in the epithelial-to-mesenchymal
transition. ESRP1 was associated with prognosis in epithelial
ovarian cancer (36) and human
colorectal cancer (37). Expression
of the ELAVL4 gene was demonstrated to be a diagnostic and
prognostic marker of bone marrow lesions in patients with
neuroblastoma and male patients with meningioma (38,39).
APOL2 serves important roles in the acute inflammatory response,
lipid transport, the steroid metabolic process and the cholesterol
metabolic process. APOL2 was found to be overexpressed in
ovarian/peritoneal carcinoma and may provide a molecular basis for
therapeutic target discovery (40).
DERL1 participated in the endoplasmic reticulum (ER)-nuclear
signaling pathway, the ER-associated protein catabolic process and
the ER to cytosol process. The results of a previous study have
indicated that the expression of DERL1 distinguishes malignant from
benign canine mammary tumors (41).
Post-glycosylphosphatidylinositol attachment to proteins 3 (PGAP3)
is involved in protein amino acid lipidation, the
glycerophospholipid metabolic process, the lipid biosynthetic
process, the lipoprotein metabolic process and the lipoprotein
biosynthetic process. Previous studies demonstrated that lipid
metabolism disorders serve an important role in endometrial cancer
(42,43). PGAP3 may affect the prognosis of EEA
by regulating the lipid metabolism process. Transmembrane protein
27 (TMEM27) serve important roles in proteolysis. Javorhazy et
al (44) demonstrated that a
lack of TMEM27 expression in conventional renal cell carcinoma
defines a group of patients as at a high risk of cancer-associated
mortality.
The use of the RF model for the prediction of EEA
prognosis when deciding whether to recommend adjuvant therapies is
of great importance, particularly for patients with FIGO stage I
disease. Those who have a low risk of relapse according to
traditional clinicopathological risk factors may not have to
receive postoperative adjuvant chemoradiotherapy. The results from
previous studies have indicated that a large proportion of patients
with EEA, who were at a low risk of relapse according to the
traditional criteria and had not received postoperative adjuvant
chemoradiotherapy, eventually relapsed or deceased (5,6). The RF
prediction model derived on the basis of clinical features and gene
expression is promising for providing an individualized and more
accurate prediction for patients with EEA. Combining the predictive
results of the RF model and traditional criteria could also be used
for better stratification of patients in clinical trials, as well
as for providing more accurate counseling ideas for patients.
Two nomograms (45,46)
established by traditional characteristics for the predictive
survival of EC have been produced, and their training accuracies
were between 0.71 and 0.78. The first nomogram consists of five
simple clinical features, including FIGO stage, age at diagnosis,
final histological grade, negative lymph nodes and histological
subtype (45). The second nomogram
was validated in randomly selected patients (46) and indicated that tumor grade, age and
lymphovascular space involvement were highly predictive for all
outcomes. The establishment of the two nomograms was based on Cox
regression analyses. Previous studies have demonstrated that
machine-learning approaches appear to be more suitable than
traditional statistical methods for some situations, such as the
prediction of cancer prognosis, which involves a certain number of
potential predictors (10,11); thus, it may be more suitable to build
such nomograms with machine-learning methods such as RF. In
addition, biological characteristics and clinical features were
particularly important in the prognosis of EEA. Using a combination
of molecular and conventional characteristics as classifications
would provide a better appraisal of prognostic and predictive
factors, and the combination of traditional clinical factors and
molecular biological characteristics is very important for the
prognosis of EEA.
The classification accuracy of the RF prediction
model combined with traditional clinicopathological features and
gene expression was markedly higher than that of the RF models that
were based on the traditional clinicopathological features or gene
expression alone, indicating that traditional clinicopathological
features and gene expression were important factors for the
prognosis of EEA. The inclusion of numerous patients and
prognosis-associated clinical features in the establishment of the
RF prediction models is vital, and unfortunately, the number of
samples in the present study was limited. In future research, more
clinical samples and more clinical features will be collected for
RF model establishment. The RF prediction model presented within
the present study could provide a more individualized and accurate
estimation of relapse and/or mortality for patients diagnosed with
EEA following primary therapy. The RF model could also be used for
better stratification of patients in clinical trials and for
providing more accurate counseling ideas for patients.
To the best of our knowledge, the present study is
the first to establish an EEA predictive model that combines genes
and traditional features using RF. The RF model derived on the
basis of the ‘11 genes and grade’ achieved better predictive
performances than RF models established by either the 11 genes or
grade alone, indicating that the RF model derived on the basis of
the ‘genes and clinical features’ had a stronger predictive ability
for the prognosis of EEA.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National
Natural Science Foundation of China (grant nos. 81502237, 81272869
and 81672571), the National Key Technology Research and Development
Program of the Ministry of Science and Technology of China (grant
no. 2015BAI13B06), and the Basic research project of Peking
University (grant no. BMU2018JC005).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding authors upon reasonable
request.
Authors' contributions
FY and XS wrote the manuscript, collected the
clinical information and performed the statistical analyses. LZ,
XL, YC, JZ, XH and JL designed the study and revised the
manuscript. SL analyzed the data. JW conceived and supervised the
study and approved the final manuscript. All authors read and
approved the manuscript and agree to be accountable for all aspects
of the research to ensure that the accuracy or integrity of any
part of the work is appropriately investigated and resolved.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Peking University People's Hospital (Beijing, China).
All participating patients received and provided written informed
consent prior to joining the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
EEA
|
endometrioid endometrial
adenocarcinoma
|
|
TCGA
|
The Cancer Genome Atlas
|
|
PKUPH
|
Peking University People's
Hospital
|
|
VSURF
|
variable selection method using Random
Forests
|
|
PE
|
primary EEA
|
|
R/D-PE
|
relapsed or deceased primary EEA
|
|
RF
|
Random Forest
|
References
|
1
|
Albertini AF, Devouassoux-Shisheboran M
and Genestie C: Pathology of endometrioid carcinoma. Bull Cancer.
99:7–12. 2012.PubMed/NCBI
|
|
2
|
Piulats JM, Guerra E, Gil-Martin M,
Roman-Canal B, Gatius S, Sanz-Pamplona R, Velasco A, Vidal A and
Matias-Guiu X: Molecular approaches for classifying endometrial
carcinoma. Gynecol Oncol. 145:200–207. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bendifallah S, Ouldamer L, Lavoue V,
Canlorbe G, Raimond E, Coutant C, Graesslin O, Touboul C, Collinet
P, Darai E, et al: Patterns of recurrence and outcomes in
surgically treated women with endometrial cancer according to
ESMO-ESGO-ESTRO consensus conference risk groups: Results from the
FRANCOGYN study group. Gynecol Oncol. 144:107–112. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Cancer Genome Atlas Research Network, ;
Kandoth C, Schultz N, Cherniack AD, Akbani R, Liu Y, Shen H,
Robertson AG, Pashtan I, Shen R, et al: Integrated genomic
characterization of endometrial carcinoma. Nature. 497:67–73. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Marnitz S and Kohler C: Current therapy of
patients with endometrial carcinoma. A critical review.
Strahlenther Onkol. 188:12–20. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wright JD, Barrena Medel NI, Sehouli J,
Fujiwara K and Herzog TJ: Contemporary management of endometrial
cancer. Lancet. 379:1352–1360. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Perou CM, Sorlie T, Eisen MB, van de Rijn
M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA,
et al: Molecular portraits of human breast tumours. Nature.
406:747–752. 2000. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yu KH, Zhang C, Berry GJ, Altman RB, Re C,
Rubin DL and Snyder M: Predicting non-small cell lung cancer
prognosis by fully automated microscopic pathology image features.
Nat Commun. 7:124742016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kleppe A, Albregtsen F, Vlatkovic L,
Pradhan M, Nielsen B, Hveem TS, Askautrud HA, Kristensen GB,
Nesbakken A, Trovik J, et al: Chromatin organisation and cancer
prognosis: A pan-cancer study. Lancet Oncol. 19:356–369. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Fernandez-Delgado M, Cernadas E, Barro S
and Amorim D: Do we need hundreds of classifiers to solve real
world classification problems? J Mach Learn Res. 15:3133–3181.
2014.
|
|
11
|
Diaz-Uriarte R and Alvarez de Andres S:
Gene selection and classification of microarray data using Random
Forest. BMC Bioinformatics. 7:32006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Moorthy K and Mohamad MS: Random Forest
for gene selection and microarray data classification.
Bioinformation. 7:142–146. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Eshaghi A, Wottschel V, Cortese R,
Calabrese M, Sahraian MA, Thompson AJ, Alexander DC and Ciccarelli
O: Gray matter MRI differentiates neuromyelitis optica from
multiple sclerosis using Random Forest. Neurology. 87:2463–2470.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Llorens-Rico V, Lluch-Senar M and Serrano
L: Distinguishing between productive and abortive promoters using a
Random Forest classifier in Mycoplasma pneumoniae. Nucleic Acids
Res. 43:3442–3453. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Cadenas JM, Garrido MC and Martinez R:
Feature subset selection Filter-Wrapper based on low quality data.
Expert Syst Appl. 40:6241–6252. 2013. View Article : Google Scholar
|
|
16
|
Diaz-Uriarte R: GeneSrF and varSelRF: A
web-based tool and R package for gene selection and classification
using Random Forest. BMC Bioinformatics. 8:3282007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kwon SG, Hwang JH, Park DH, Kim TW, Kang
DG, Kang KH, Kim IS, Park HC, Na CS, Ha J and Kim CW: Associated
with litter size in ber identification of differentially expressed
genes kshire pig placenta. PLoS One. 11:e01533112016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Pecorelli S: Revised FIGO staging for
carcinoma of the vulva, cervix, and endometrium. Int J Gynecol
Obstet. 105:103–104. 2009. View Article : Google Scholar
|
|
19
|
Shepherd JH: Revised FIGO staging for
gynaecological cancer. Br J Obstet Gynaecol. 96:889–892. 1989.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ouldamer L, Bendifallah S, Body G, Touboul
C, Graesslin O, Raimond E, Collinet P, Coutant C, Lavoue V, Leveque
J, et al: Predicting poor prognosis recurrence in women with
endometrial cancer: A nomogram developed by the FRANCOGYN study
group. Br J Cancer. 115:1296–1303. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Morice P, Leary A, Creutzberg C,
Abu-Rustum N and Darai E: Endometrial cancer. Lancet.
387:1094–1108. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Moreno RP, Pearse R and Rhodes A; European
Surgical Outcomes Study (EuSOS) Group of the European Society of
Intensive Care Medicine and European Society of Anaesthesiology
Trials Groups, : American society of anesthesiologists score: Still
useful after 60 years? Results of the EuSOS study. Rev Bras Ter
Intensiva. 27:105–112. 2015.(In English, Portuguese). View Article : Google Scholar : PubMed/NCBI
|
|
23
|
AlHilli MM, Mariani A, Bakkum-Gamez JN,
Dowdy SC, Weaver AL, Peethambaram PP, Keeney GL, Cliby WA and
Podratz KC: Risk-scoring models for individualized prediction of
overall survival in low-grade and high-grade endometrial cancer.
Gynecol Oncol. 133:485–493. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
AlHilli MM, Podratz KC, Dowdy SC,
Bakkum-Gamez JN, Weaver AL, McGree ME, Keeney GL, Cliby WA and
Mariani A: Risk-scoring system for the individualized prediction of
lymphatic dissemination in patients with endometrioid endometrial
cancer. Gynecol Oncol. 131:103–108. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Creutzberg CL, van Putten WL, Koper PC,
Lybeert ML, Jobsen JJ, Warlam-Rodenhuis CC, De Winter KA, Lutgens
LC, van den Bergh AC, van de Steen-Banasik E, et al: Surgery and
postoperative radiotherapy versus surgery alone for patients with
stage-1 endometrial carcinoma: Multicentre randomised trial. PORTEC
study group. Post operative radiation therapy in endometrial
carcinoma. Lancet. 355:1404–1411. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Barlin JN, Zhou Q, St Clair CM, Iasonos A,
Soslow RA, Alektiar KM, Hensley ML, Leitao MM Jr, Barakat RR and
Abu-Rustum NR: Classification and regression tree (CART) analysis
of endometrial carcinoma: Seeing the forest for the trees. Gynecol
Oncol. 130:452–456. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Jordan MI and Mitchell TM: Machine
learning: Trends, perspectives, and prospects. Science.
349:255–260. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Burki TK: Predicting lung cancer prognosis
using machine learning. Lancet Oncol. 17:e4212016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Sarica A, Cerasa A and Quattrone A: Random
Forest algorithm for the classification of neuroimaging data in
Alzheimer's disease: A systematic review. Front Aging Neurosci.
9:3292017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wang X, Lin P and Ho JW: Discovery of
cell-type specific DNA motif grammar in cis-regulatory elements
using Random Forest. BMC Genomics. 19:9292018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Taherzadeh G, Zhou Y, Liew AW and Yang Y:
Structure-based prediction of protein-peptide binding regions using
Random Forest. Bioinformatics. 34:477–484. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Dong W, Guan FF, Zhang X, Gao S, Liu N,
Chen W, Zhang LF and Lu D: Dhcr24 activates the PI3K/Akt/HKII
pathway and protects against dilated cardiomyopathy in mice. Animal
Model Exp Med. 1:40–52. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Dai M, Zhu XL, Liu F, Xu QY, Ge QL, Jiang
SH, Yang XM, Li J, Wang YH, Wu QK, et al: Cholesterol synthetase
DHCR24 induced by insulin aggravates cancer invasion and
progesterone resistance in endometrial carcinoma. Sci Rep.
7:414042017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Liu XP, Yin XH, Meng XY, Yan XH, Cao Y,
Zeng XT and Wang XH: DHCR24 predicts poor clinicopathological
features of patients with bladder cancer: A STROBE-compliant study.
Medicine. 97:e118302018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Li L, Qi L, Qu T, Liu C, Cao L, Huang Q,
Song W, Yang L, Qi H, Wang Y, et al: Epithelial splicing regulatory
protein 1 inhibits the invasion and metastasis of lung
adenocarcinoma. Am J Pathol. 188:1882–1894. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Chen L, Yao Y, Sun L, Zhou J, Miao M, Luo
S, Deng G, Li J, Wang J and Tang J: Snail driving alternative
splicing of CD44 by ESRP1 enhances invasion and migration in
epithelial ovarian cancer. Cell Physiol Biochem. 43:2489–2504.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Fagoonee S, Picco G, Orso F, Arrigoni A,
Longo DL, Forni M, Scarfo I, Cassenti A, Piva R, Cassoni P, et al:
The RNA-binding protein ESRP1 promotes human colorectal cancer
progression. Oncotarget. 8:10007–10024. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Druĭ AE, Tsaur GA, Popov AM, Tuponogov SN,
Shorikov EV, Tsvirenko SV, Savel'ev LI and Fechina LG: The TH,
ELAVL4 and GD2 gene expression as diagnostic markers of bone marrow
lesions in patients with neuroblastoma. Vopr Onkol. 58:514–520.
2012.(In Russian). PubMed/NCBI
|
|
39
|
Stawski R, Piaskowski S,
Stoczynska-Fidelus E, Wozniak K, Bienkowski M, Zakrzewska M,
Witusik-Perkowska M, Jaskolski DJ, Och W, Papierz W, et al: Reduced
expression of ELAVL4 in male meningioma patients. Brain Tumor
Pathol. 30:160–166. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Davidson B, Stavnes HT, Holth A, Chen X,
Yang Y, Shih IM and Wang TL: Gene expression signatures
differentiate ovarian/peritoneal serous carcinoma from breast
carcinoma in effusions. J Cell Mol Med. 15:535–544. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Klopfleisch R, Klose P and Gruber AD: The
combined expression pattern of BMP2, LTBP4, and DERL1 discriminates
malignant from benign canine mammary tumors. Vet Pathol.
47:446–454. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Hirasawa A, Makita K, Akahane T, Yokota M,
Yamagami W, Banno K, Susumu N and Aoki D: Hypertriglyceridemia is
frequent in endometrial cancer survivors. Jpn J Clin Oncol.
43:1087–1092. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Trousil S, Lee P, Pinato DJ, Ellis JK,
Dina R, Aboagye EO, Keun HC and Sharma R: Alterations of choline
phospholipid metabolism in endometrial cancer are caused by choline
kinase alpha overexpression and a hyperactivated deacylation
pathway. Cancer Res. 74:6867–6877. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Javorhazy A, Farkas N, Beothe T, Pusztai
C, Szanto A and Kovacs G: Lack of TMEM27 expression is associated
with postoperative progression of clinically localized conventional
renal cell carcinoma. J Cancer Res Clin Oncol. 142:1947–1953. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Abu-Rustum NR, Zhou Q, Gomez JD, Alektiar
KM, Hensley ML, Soslow RA, Levine DA, Chi DS, Barakat RR and
Iasonos A: A nomogram for predicting overall survival of women with
endometrial cancer following primary therapy: Toward improving
individualized cancer care. Gynecol Oncol. 116:399–403. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Creutzberg CL, van Stiphout RG, Nout RA,
Lutgens LC, Jurgenliemk-Schulz IM, Jobsen JJ, Smit VT and Lambin P:
Nomograms for prediction of outcome with or without adjuvant
radiation therapy for patients with endometrial cancer: A pooled
analysis of PORTEC-1 and PORTEC-2 trials. Int J Radiat Oncol Biol
Phys. 91:530–539. 2015. View Article : Google Scholar : PubMed/NCBI
|