Introduction
Colon cancer, a type of malignant tumor that
develops from the large bowel, is one of the most frequently
diagnosed cancer types among males and females, and has
unacceptable high mortality rates worldwide (1–3). In 2014
and 2015, colon cancer caused ~700,000 deaths (2,3).
Treatment outcomes of colon cancer at early stages are generally
satisfactory, and >95% patients can live >5 years following
active treatment (4). However, the
majority of patients with colon cancer are diagnosed with existing
distant tumor metastasis (5), which
lacks radical therapeutic regimens, leading to poor survival
outcomes of those patients. Although several risk factors,
including advanced age and male sex have been identified for colon
cancer, the pathogenesis of this disease remains unclear (1). Therefore, it is important to identify
molecular markers for the diagnosis of colon cancer at early
stages.
Abnormally accelerated glucose metabolism
distinguishes cancer cells from normal cells (6). Glucose metabolism is considered as a
promising target for cancer therapy (7). Glucose transporter 1 (GLUT-1), a
glucose transporter with critical functions in the transport of
glucose across the plasma membranes of mammalian cells, serves a
key role in glucose metabolism (8).
Levels of GLUT-1 have been reported to be elevated in the
development of different types of cancer, including breast cancer
and liver cancer (9,10). Overexpression of GLUT-1 not only
promotes cancer cell proliferation but also protects cancer cells
from stress (11,12). It is understood that GLUT-1, under
certain conditions, interacts with different long non-coding RNAs
(lncRNAs) to participate in cancer progression (13,14).
lncRNA associated with poor prognosis of hepatocellular carcinoma
(AWPPH) is a recently reported lncRNA with oncogenic functions in
bladder cancer (15) and
hepatocellular carcinoma (16). The
present study demonstrated that lncRNA AWPPH could interact with
GLUT-1 to participate in the regulation of colon cancer cell
proliferation.
Materials and methods
Patients
A total of 134 patients with colon cancer were
treated at Liaoning Cancer Hospital and Institute (Shengyang,
China) between May 2015 and January 2018. Among these patients, 46
were included in the present study to serve as the pateint group.
The inclusion criteria were as follows: i) patients with colon
cancer at stage I and II (early stage) diagnosed by a pathological
test; ii) patients with serum samples in the specimen library of
the hospital; ii) patients who could fully understand the
experimental protocol; and iv) patients willing to join the study.
The exclusion criteria were as follows: i) patients with another
severe disease at the time of admission; ii) patients who were
treated within 3 months prior admission; and iii) patients with
chronic diseases. The current study also included 42 healthy
individuals to serve as the control group. The health controls
received physiological examinations at Liaoning Cancer Hospital and
Institute between May 2015 and January 2018, and all had serum
samples in the specimen library of the hospital. The Ethics
Committee of Liaoning Cancer Hospital and Institute approved the
current study prior to the enrollment of participants. All
participants provided written informed consent. No significnat
differences in age and sex were identified between the two groups.
The basic information of all participants is presented in Table I.
 | Table I.Information for the patient group and
control group. |
Table I.
Information for the patient group and
control group.
| Characteristic | Patient group
(n=46) | Control group
(n=42) |
|---|
| Sex |
|
|
| Male,
n | 26 | 24 |
| Female,
n | 20 | 18 |
| Age range, years | 23–70 | 24–72 |
| Mean age, years | 48.3±71.1 | 47.9±6.4 |
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Trizol® reagent (Invitrogen; Thermo
Fisher Scientific, Inc.) was used to extract total RNA from serum
and in vitro cultured cells. Total RNA samples were
subjected to denatured agrose gel electrophoresis to assess the
quality. The RNA concentration was measured using a NanoDrop™ 2000
spectrophotometer (Thermo Fisher Scientific, Inc.). RT was
performed to synthesize cDNA using SuperScript III Reverse
Transcriptase (Thermo Fisher Scientific, Inc.) according to the
following conditions: 5 min at 25°C, 20 min at 55°C and 20 min at
75°C. All qPCR reaction systems were prepared using
SYBR® Green Real-Time PCR Master mix (Thermo Fisher
Scientific, Inc.). The following primers were used: lncRNA-AWPPH
forward, 5′-CTGGATGGTCGCTGCTTTTTA-3′ and reverse,
5′-AGGGGGATGAGTCGTGATTT-3′; and β-actin forward,
5′-ACCTCTATGCCAACACAGT-3′ and reverse, 5′-AGTACTTGCGCTCAGGAGG-3′.
The following thermocycling conditions were used for qPCR: 1 min at
95°C, followed by 40 cycles of 20 sec at 95°C and 40 sec at 58°C.
The qPCR products were all subjected to agrose gel electrophoresis
and some were sequenced to make sure the correct products were
obtained. Data normalizations were performed using the
2−ΔΔCq method (17).
ELISA
Serum levels of GLUT-1 were measured by ELISA using
a GLUT-1 ELISA kit (cat. no. MBS721715; MyBioSource).
Cell lines, cell culture and
transfection
The normal human colon cell line FHC (cat. no.
CRL-1831) and colorectal adenocarcinoma cell line HT-29 (cat. no.
HTB-38™) were purchased from the American Type Culture Colletcion
(ATCC). Cells were cultivated with McCoy's 5a Medium Modified
(ATCC) containing 10% fetal bovine serum (ATCC) and placed at 37°C
in a humidified incubator containing 5% CO2. FHC is a
normal colon epithelial cell line collected at 13 weeks gestation.
lncRNA-AWPPH short hairpin RNA (shRNA; 5′-GGAATGCAGCTGAAAGATTCC-3′)
and scrambled shControl (5′-UUUCCGAACGUGUCACGUdTdT-3′) were
synthesized by Shanghai GenePharma Co., Ltd.. Full length GLUT-1
cDNA was amplified by PCR reaction. GLUT-1 cDNA was inserted into a
pIRSE2 vector (Clontech Laboratories, Inc.) to generate GLUT-1
expression vector. Vectors (10 nM) and shRNA (50 mM) were
transfected into 1×106 FHC and HT-29 cells using
Lipofectamine® 2000 reagent (cat. no. 11668-019;
Invitrogen; Thermo Fisher Scientific, Inc,). Untransfected cells
were used as control cells. Cells transfected with scrambled
shControl or empty vector were used as negative control cells.
GLUT-1 overexpression and lncRNA AWPPH-silencing were confirmed 12
h after transfection. Subsequent experiments were performed when
the overexpression rate was >200% and the knockdown rate was
<50% compared with the control cells.
Cell proliferation assay
Following transfection and confirmation, cells were
harvested and cell suspensions were prepared with a final cell
density of 4×104 cells/well. Each well of a 96-well
plate was filled with 0.1 ml cell suspension containing
4×103 cells. The plate was incubated at 37°C in a 5%
CO2 incubator, followed by addition of 10 µl Cell
Counting Kit-8 (CCK-8) reagent at 24, 48, 72 and 96 h. Cells were
cultured for an additional 4 h, followed by measurement of the
optical density values at 450 nm using a microplate reader.
Western blot
RIPA lysis and extraction buffer (Thermo Fisher
Scientific, Inc.) was used to extract total protein from FHC and
HT-29 cells, followed by measurement of protein concentrations
using a BCA assay (Sigma-Aldrich; Merck KGaA). Proteins (20 µg)
were separated by 12% SDS-PAGE. After gel transfer onto
polyvinylidene difluoride membranes, the membranes were blocked in
5% skimmed milk for 1 h at room temperature. Subsequently, the
membranes were incubated with the following primary antibodies at
4°C overnight: Rabbit anti-human GLUT-1 (1:2,000; cat. no. ab15309;
Abcam) and GAPDH antibody (1:1,000; cat. no. ab9485; Abcam).
Membranes were then incubated with goat anti-rabbit IgG-HRP
secondary antibody (1:1,000; cat. no. MBS435036; MyBioSource) for
2.5 h at 25°C. ECL Western Blotting Substrate (Thermo Fisher
Scientific, Inc.) was used to develop signal. Data were normalized
using ImageJ v1.48 software (National Institutes of Health).
Statistical analysis
All experiments were performed in triplicate.
GraphPad Prism 6 software (GraphPad Software Inc.) was used for
data analyses and comparisons. Gene expression and cell
proliferation data were compared using unpaired Student's t-test
for two groups and one-way analysis of variance followed by
Fisher's least significant difference test for multiple groups.
Correlations between serum lncRNA AWPPH and GLUT-1 were analyzed by
Pearson's correlation coefficient. χ2 test was used for
the analysis between serum levels of lncRNA AWPPH and the clinical
data of patients with colon cancer. Data are presented as the mean
± standard deviation. Receiver operating characteristic curve (ROC)
analysis was used to evaluate the diagnostic value of serum lncRNA
AWPPH in colon cancer. For this analysis, patients with colon
cancer represented the true positive cases and healthy controls
represented the true negative cases. P<0.05 was considered to
indicate a statistically significant difference.
Results
Serum levels of lncRNA AWPPH and
GLUT-1 are significantly higher in patients with colon cancer
compared with healthy controls
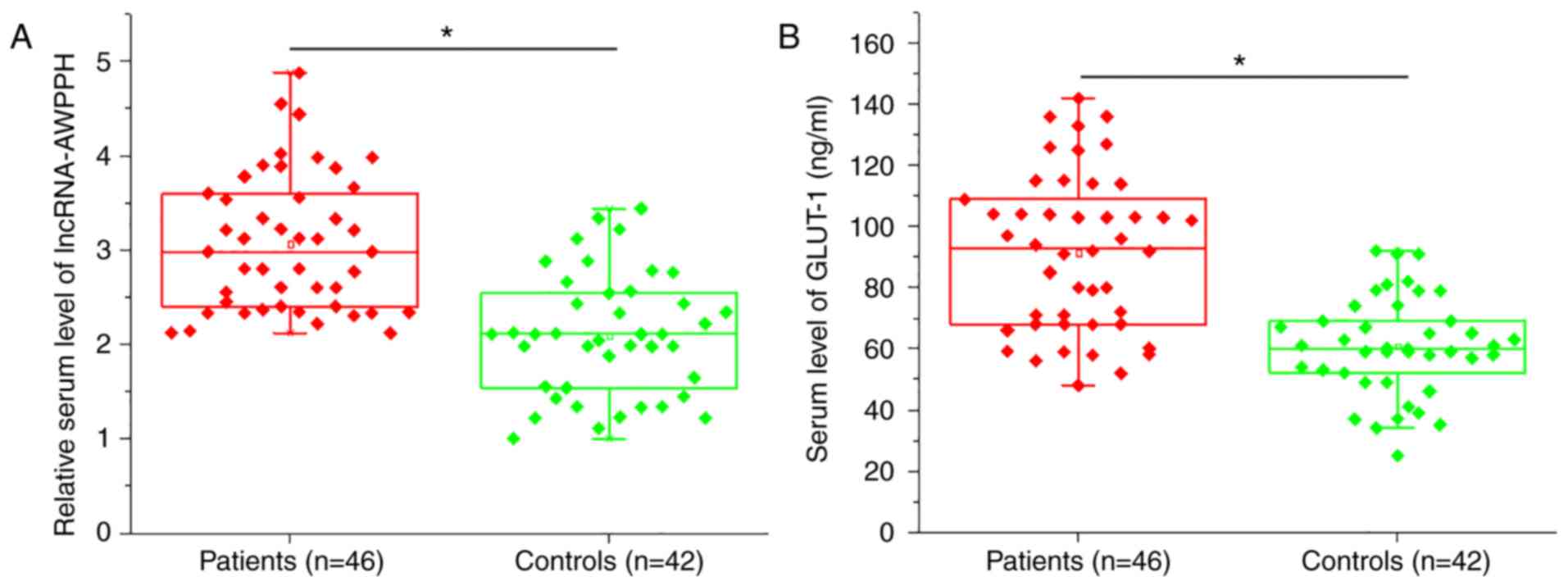
RT-qPCR and ELISA demonstrated significantly
upregulated serum levels of lncRNA AWPPH (P<0.05; Fig. 1A) and GLUT-1 (P<0.05; Fig. 1B) in patients with colon cancer
compared with healthy controls.
Serum levels of AWPPH and GLUT-1 are
significantly positively correlated in patients with colon cancer
but not in healthy controls
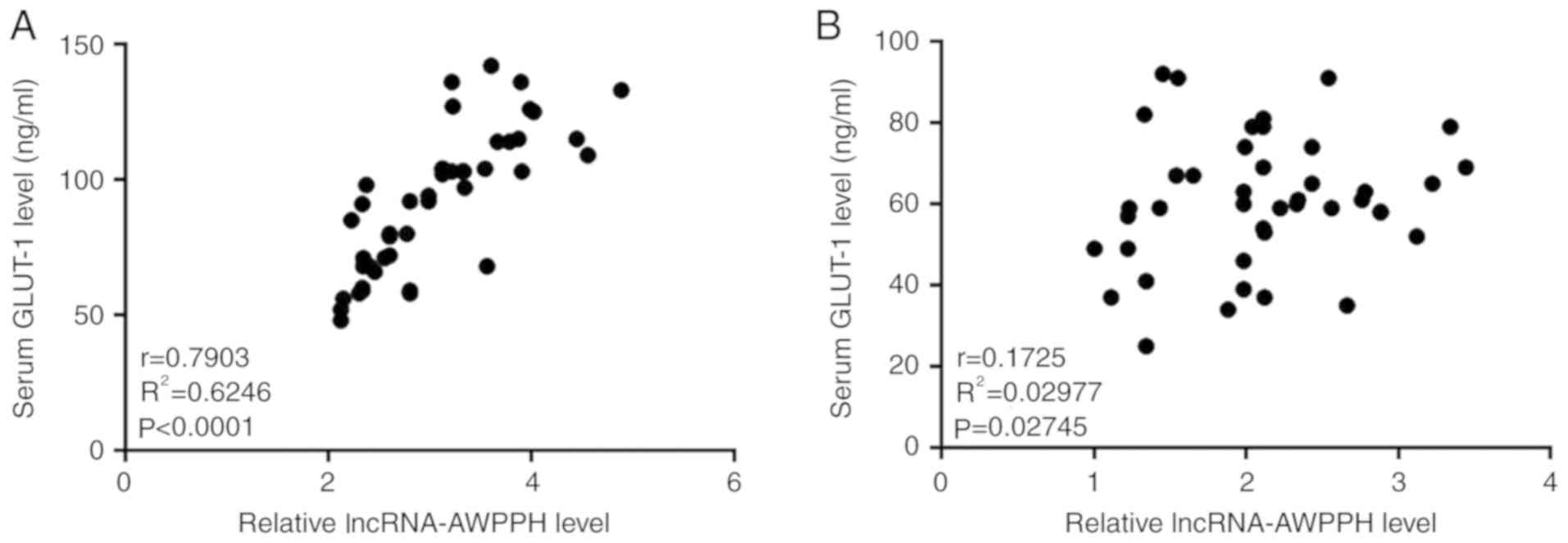
Pearsons correlation coefficient analysis was
performed to investigate the potential correlation between serum
lncRNA AWPPH and GLUT-1. The results revealed that serum lncRNA
AWPPH and GLUT-1 were significantly positively correlated in
patients with colon cancer (P<0.0001; Fig. 2A), but not in healthy controls
(P=0.2745; Fig. 2B).
Serum levels of AWPPH distinguish
patients with colon cancer from healthy controls
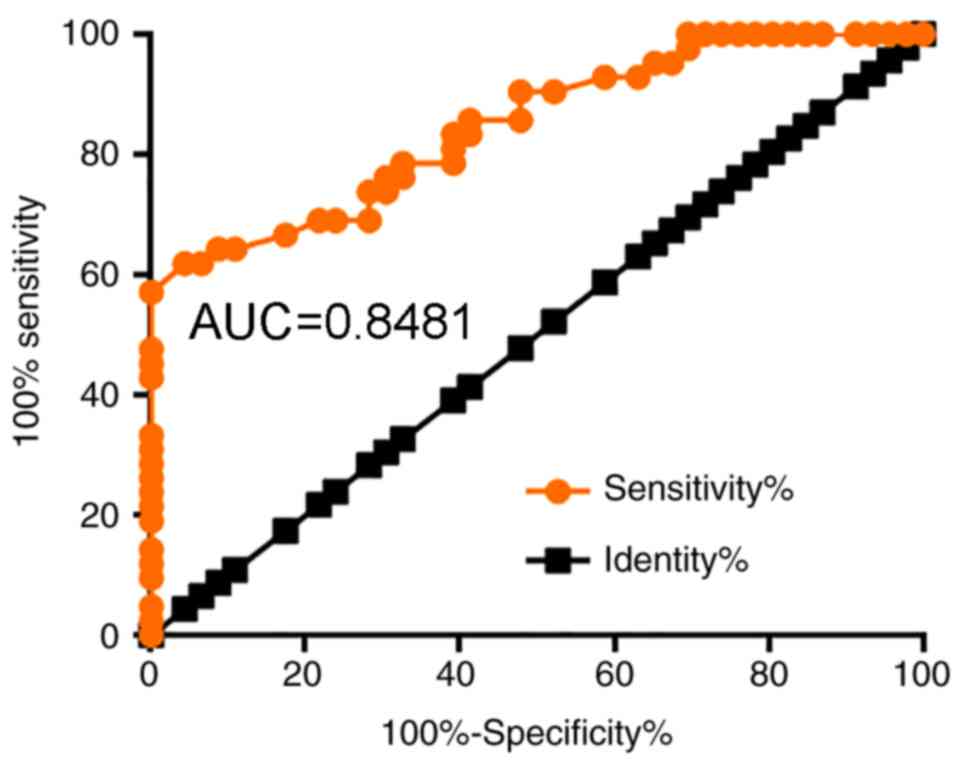
ROC analysis was used to evaluate the diagnostic
value of serum lncRNA AWPPH for colon cancer. As presented in
Fig. 3, the area under the curve
(AUC) was 0.8481, with a 95% confidence interval of 0.7687–09275
and a standard error of 0.04052.
Serum levels of lncRNA AWPPH are
associated with tumor size
χ2 test was used for the analysis between
serum levels of lncRNA AWPPH and clinical data of patients with
colon cancer. As presented in Table
II, serum levels of lncRNA AWPPH were significantly associated
with tumor size (P=0.04), but not age, sex, smoking and drinking
habits (P>0.05).
 | Table II.Associations between serum levels of
lncRNA AWPPH and clinical data of patients with colon cancer. |
Table II.
Associations between serum levels of
lncRNA AWPPH and clinical data of patients with colon cancer.
| Variable | Total, n | High-expression,
n | Low-expression,
n | χ2 | P-value |
|---|
| Sex |
|
|
| 0.35 | 0.55 |
| Male | 26 | 12 | 14 |
|
|
|
Female | 20 | 11 | 9 |
|
|
| Age, years |
|
|
| 0.79 | 0.37 |
| ≥50 | 21 | 9 | 12 |
|
|
|
<50 | 25 | 14 | 11 |
|
|
| Tumor size, cm |
|
|
| 4.39 | 0.04 |
|
>5 | 19 | 13 | 6 |
|
|
| ≤5 | 27 | 10 | 17 |
|
|
| Smoking |
|
|
| 0.79 | 0.37 |
| Yes | 21 | 9 | 12 |
|
|
| No | 25 | 14 | 11 |
|
|
| Drinking |
|
|
| 0.35 | 0.55 |
| Yes | 22 | 10 | 12 |
|
|
| No | 24 | 13 | 11 |
|
|
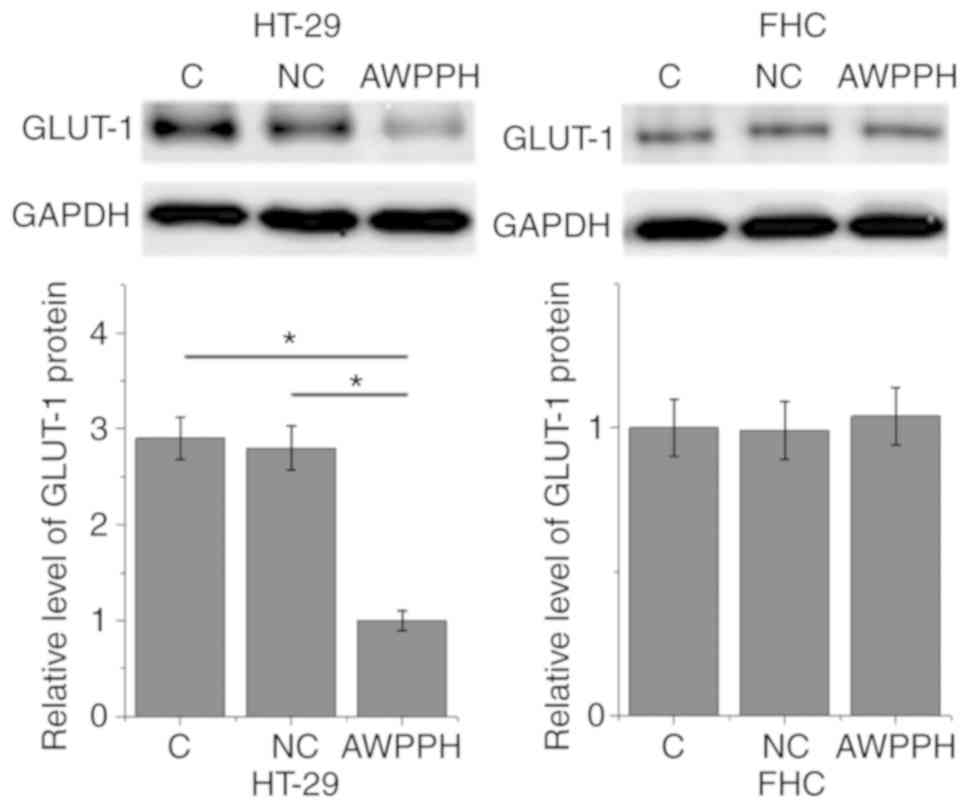
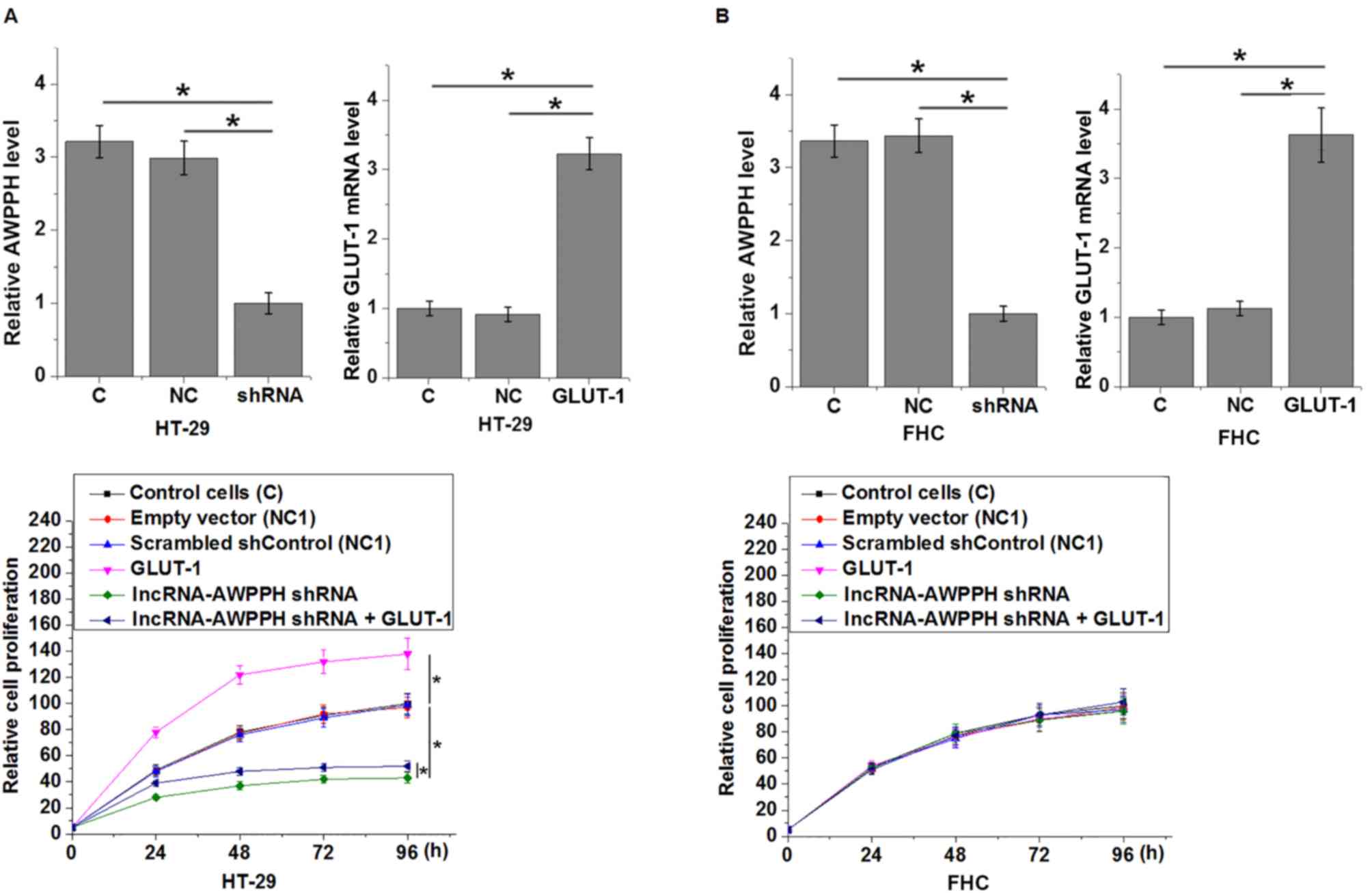
lncRNA AWPPH-silencing inhibits GLUT-1
expression in colon cancer cells but not in normal colon cells
Cells with lncRNA AWPPH-knockdown were generated and
the effects on GLUT-1 expression were investigated. As presented in
Fig. 4, compared with the control
cells and negative control cells, lncRNA AWPPH-silencing
significantly downregulated GLUT-1 in the colorectal adenocarcinoma
cell line HT-29 (P<0.05), but not in the normal FHC colon cell
line.
GLUT-1 overexpression attenuated the
inhibitory effects of lncRNA AWPPH-silencing on the proliferation
of colon cancer cells
Data in Table II
revealed the potential involvement of lncRNA AWPPH in the
regulation of tumor growth of colon cancer. To further investigate
the role of lncRNA AWPPH in the proliferation of colon cancer
cells, a CCK-8 assay was performed to detect cell proliferation
following lncRNA AWPPH-silencing. AWPPH-silencing and GLUT-1
overexpression were confirmed (Fig.
5). Compared with the control cells, lncRNA AWPPH-silencing
significantly inhibited the proliferation of the colorectal
adenocarcinoma HT-29 cell line (P<0.05), but not the normal
colon cell line FHC (Fig. 5). In
addition, GLUT-1 overexpression promoted cancer cell proliferation
and significantly attenuated the inhibitory effects of lncRNA
AWPPH-silencing on the proliferation of the colorectal
adenocarcinoma cell line HT-29 (P<0.05).
Discussion
The present study used human samples to investigate
the involvement of lncRNA AWPPH in colon cancer. The key finding of
the current study is that lncRNA AWPPH is involved in the growth of
colon cancer. Furthermore, the involvement of lncRNA AWPPH in colon
cancer is at least partially mediated by its interactions with
GLUT-1.
Treatment of colon cancer is challenged by the fact
that the majority of patients are diagnosed with existing distant
tumor metastasis (18). Therefore,
early diagnosis and treatment is critical. The present study only
included patients with stage I or II colon cancer, which are
considered early stages of cancer development. It was observed that
serum levels of lncRNA AWPPH were significantly higher in patients
with colon cancer compared with healthy controls. In addition, a
high expression of lncRNA AWPPH could distinguish patients with
colon cancer from healthy controls. These data suggest that plasma
lncRNA AWPPH may serve as a biomarker for the early diagnosis of
colon cancer. However, lncRNA AWPPH is a recently identified lncRNA
with an unknown expression pattern in the majority of human
diseases. Therefore, multiple diagnostic markers should be combined
with lncRNA AWPPH to improve the diagnostic specificity.
Overexpression of GLUT-1 has been observed in
different types of human malignancy and inhibition of GLUT-1 is
considered to be a promising target for the treatment of human
cancer (19,20). Consistent with previous studies, the
current study also observed significantly elevated plasma levels of
GLUT-1 in patients with colon cancer. Notably, a significant
positive correlation was also observed between plasma levels of
GLUT-1 and lncRNA AWPPH in patients with colon cancer. In view of
the critical functions of GLUT-1 in cancer cell proliferation
(21) and the significant
association between plasma levels of lncRNA AWPPH and tumor size of
colon cancer observed in the present study, it can be hypothesised
that lncRNA AWPPH may interact with GLUT-1 to participate in colon
cancer cell proliferation. The current in vitro cell
experimental data also demonstrated that lncRNA AWPPH and GLUT-1
are involved in the proliferation of colon cancer cells. It can be
suggested that lncRNA AWPPH may serve as an upstream activator of
GLUT-1 in this process for the following reasons: i) lncRNA
AWPPH-silencing downregulated GLUT-1 expression; and ii) GLUT-1
overexpression attenuated the inhibitory effects of lncRNA
AWPPH-silencing on colon cancer cell proliferation.
In the present study, lncRNA AWPPH-silencing
demonstrated no significant effects on the behaviors of normal
colon cells. Notably, GLUT-1 overexpression only promoted the
proliferation of colon cancer cells but not the normal colon cells,
which may be due to the specific normal colon cancer cell line used
in the current study. The present study only revealed a lncRNA
AWPPH-GLUT-1 sequential signalling transduction in colon cancer.
The interaction between lncRNA-AWPPH and GLUT-1 is likely to be
indirect due to the facts that: i) no significant correlation was
identified between plasma lncRNA AWPPH and GLUT-1 in healthy
controls; and ii) lncRNA AWPPH-silencing demonstrated no
significant effects on GLUT-1 in normal colon cells.
Two previous studies have reported that lncRNA AWPPH
regulates bladder cancer progression by inhibiting SMAD family
member 4 via enhancer of zeste homolog 2 (16) and promotes hepatocellular carcinoma
progression through Y-box binding protein 1 (18). The present study first reported the
involvement of lncRNA AWPPH in cancer biology through the
regulation of GLUT-1. This suggests that lncRNA AWPPH may interact
with different pathways to participate in different types of
cancer.
In conclusion, lncRNA AWPPH and GLUT-1 were
demonstrated to be upregulated in patients with colon cancer. In
addition, a downregulation of lncRNA AWPPH may inhibit colon cancer
cell proliferation by downregulating GLUT-1. However, glucose
uptake was not directly measured in the present study. In addition,
the sample size was small. Future investigation is needed to solve
those problems.
Acknowledgements
Not applicable.
Funding
The present study was supported by a National
Natural Science Fund from the National Natural Science Foundation
of China (grant no. 81672427).
Availability of data and materials
The analyzed datasets generated during the present
study are available from the corresponding author on reasonable
request.
Authors contributions
JB and RZ designed experiments; JB, JX and JZ
performed experiments an collected data. RZ analyzed data and
drafted manuscript. All authors approved the manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Cancer Hospital of China Medical University
(Shengyang, China). All patients and healthy volunteers provided
written informed consent prior to their inclusion in the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Marisa L, de Reyniès A, Duval A, Selves J,
Gaub MP, Vescovo L, Etienne-Grimaldi MC, Schiappa R, Guenot D,
Ayadi M, et al: Gene expression classification of colon cancer into
molecular subtypes: Characterization, validation, and prognostic
value. PLoS Med. 10:e10014532013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel R, Desantis C and Jemal A:
Colorectal cancer statistics, 2014. CA Cancer J Clin. 64:104–117.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lacy AM, García-Valdecasas JC, Delgado S,
Castells A, Taurá P, Piqué JM and Visa J: Laparoscopy-assisted
colectomy versus open colectomy for treatment of non-metastatic
colon cancer: A randomised trial. Lancet. 359:2224–2229. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Benson AB, Bekaii-Saab T, Chan E, Chen YJ,
Choti MA, Cooper HS, Engstrom PF, Enzinger PC, Fakih MG, Fenton MJ,
et al: Metastatic colon cancer, version 3.2013: Featured updates to
the NCCN guidelines. J Natl Compr Canc Netw. 11:141–152. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Annibaldi A and Widmann C: Glucose
metabolism in cancer cells. Curr Opin Clin Nutr Metab Care.
13:466–470. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hamanaka RB and Chandel NS: Targeting
glucose metabolism for cancer therapy. J Exp Med. 209:211–215.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Olson AL and Pessin JE: Structure,
function, and regulation of the mammalian facilitative glucose
transporter gene family. Annu Rev Nutr. 16:235–256. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Pinheiro C, Sousa B, Albergaria A, Paredes
J, Dufloth R, Vieira D, Schmitt F and Baltazar F: GLUT1 and CAIX
expression profiles in breast cancer correlate with adverse
prognostic factors and MCT1 overexpression. Histol Histopathol.
26:1279–1286. 2011.PubMed/NCBI
|
|
10
|
Carvalho KC, Cunha IW, Rocha RM, Ayala FR,
Cajaíba MM, Begnami MD, Vilela RS, Paiva GR, Andrade RG and Soares
FA: GLUT1 expression in malignant tumors and its use as an
immunodiagnostic marker. Clinics (Sao Paulo). 66:965–972. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Oh S, Kim H, Nam K and Shin I: Glut1
promotes cell proliferation, migration and invasion by regulating
epidermal growth factor receptor and integrin signaling in
triple-negative breast cancer cells. BMB Rep. 50:132–137. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gonzalez-Menendez P, Hevia D, Alonso-Arias
R, Alvarez-Artime A, Rodriguez-Garcia A, Kinet S, Gonzalez-Pola I,
Taylor N, Mayo JC and Sainz RM: GLUT1 protects prostate cancer
cells from glucose deprivation-induced oxidative stress. Redox
Biol. 17:112–127. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang Y, Zhang X, Wang Z, Hu Q, Wu J, Li Y,
Ren X, Wu T, Tao X, Chen X, et al: LncRNA-p23154 promotes the
invasion-metastasis potential of oral squamous cell carcinoma by
regulating Glut1-mediated glycolysis. Cancer Lett. 434:172–183.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wei S, Fan Q, Yang L, Zhang X, Ma Y, Zong
Z, Hua X, Su D, Sun H, Li H and Liu Z: Promotion of glycolysis by
HOTAIR through GLUT1 upregulation via mTOR signaling. Oncol Rep.
38:1902–1908. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhu F, Zhang X, Yu Q, Han G, Diao F, Wu C
and Zhang Y: LncRNA AWPPH inhibits SMAD4 via EZH2 to regulate
bladder cancer progression. J Cell Biochem. 119:4496–4505. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhao X, Liu Y and Yu S: Long noncoding RNA
AWPPH promotes hepatocellular carcinoma progression through YBX1
and serves as a prognostic biomarker. Biochim Biophys Acta.
1863:1805–1816. 2017. View Article : Google Scholar
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Riihimäki M, Hemminki A, Sundquist J and
Hemminki K: Patterns of metastasis in colon and rectal cancer. Sci
Rep. 6:297652016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kanjanapan Y, Deb S, Young RJ, Bressel M,
Mileshkin L, Rischin D, Hofman MS, Narayan K and Siva S: Glut-1
expression in small cervical biopsies is prognostic in cervical
cancers treated with chemoradiation. Clin Transl Radiat Oncol.
2:53–58. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Sharen G, Peng Y, Cheng H, Liu Y, Shi Y
and Zhao J: Prognostic value of GLUT-1 expression in pancreatic
cancer: Results from 538 patients. Oncotarget. 8:19760–19767. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Noguchi C, Kamitori K, Hossain A,
Hoshikawa H, Katagi A, Dong Y, Sui L, Tokuda M and Yamaguchi F:
D-Allose inhibits cancer cell growth by reducing GLUT1 expression.
Tohoku J Exp Med. 238:131–141. 2016. View Article : Google Scholar : PubMed/NCBI
|