Introduction
Breast cancer is a common type of tumor among women,
and the second leading cause of cancer-associated mortality
worldwide in 2017 (1,2). Triple-negative breast cancer (TNBC)
refers to breast cancer lacking the expression of estrogen receptor
(ER), progesterone receptor (PR) and hormone epidermal growth
factor receptor-2 (HER2) (3,4). TNBC accounts for 15% of breast cancer
cases, and is associated with a poorer prognosis compared with that
of other subtypes (3,4). Patients with TNBC have a higher risk of
distant metastasis and a poorer overall survival compared with that
of patients with other types of breast cancer, partly due to the
lack of effective targeted therapies (4). Although patients with TNBC are
sensitive to chemotherapy, they are prone to medical treatment
resistance (4,5). In addition, satisfactory targeted
therapies for TNBC are unavailable due to the lack of expression of
ER, PR and HER2. Therefore, the discovery of new molecular targets
to treat patients with TNBC is required.
MicroRNAs (miRNAs) are endogenous non-coding small
RNAs that consist of 21–22 nucleotides that are abundant in
organisms (6). A number of studies
have revealed that miRNAs are involved in various cell processes,
including differentiation, proliferation, metastasis and apoptosis
(7). Previous studies have
illustrated that miRNAs are aberrantly expressed in some types of
cancer, including breast cancer (8–10).
Although the study of miRNAs is challenging due to the variable
downstream target genes of miRNAs, the application of miRNAs for
TNBC therapy is crucial. Previous studies illustrated that
miRNA-589 (miR-589) serves as a tumor suppressor in non-small cell
lung cancer, glioma and hepatocellular carcinoma (11–14). By
contrast, miR-589 acts as an oncogene in gastric cancer (15). These results revealed that the same
miRNA may serve as an oncogene or tumor suppressor depending on the
tissue type. To date, the expression, biological function and
molecular mechanism of miR-589 in TNBC has not been determined.
The objective of the present study was to determine
the expression and biological role of miR-589 in TNBC, and to
identify the direct target gene(s) that mediate the activity of
miR-589 in TNBC.
Materials and methods
TNBC tissues
Paired TNBC tissues and adjacent non-cancerous
tissues (>5 cm distant from the cancer tissue) were obtained
from the Central Hospital of Zibo (Shandong, China). A total of 20
female patients (age range, 34–71 years; mean age, 61 years)
underwent modified radical mastectomy at Central Hospital of Zibo
between January 2016 and January 2017. None of the patients had
received chemotherapy or radiotherapy prior to surgery. ER and PR
tumor status were determined by immunohistochemistry (IHC). In
brief, paraffin-embedded sections of tumor tissue (4 µm thick) were
deparaffinized in xylene, rehydrated via a graded alcohol series
and blocked in methanol containing 3% hydrogen peroxide for 10 min
at room temperature. The sections were incubated with ERα antibody
(cat. no. sc-8002; 1:200 dilution; Santa Cruz Biotechnology, Inc.,
Dallas, TX, USA) or PR antibody (cat. no. sc-166169; 1:200
dilution; Santa Cruz Biotechnology, Inc.) overnight at 4°C.
Biotinylated goat anti-mouse secondary antibodies (cat. no.
ab64255; 1:1,000 dilution; Abcam, Cambridge, MA, USA) were applied
and incubated for 30 min at 37°C, and then the
streptavidin-peroxidase conjugate (1:200 dilution; Fuzhou Maixin
Biotech Co., Ltd.) was added and incubated at 37°C for 30 min.
Finally, the immunoreactions of sections were visualized by
staining with 3′-diaminobenzidine (Sigma-Aldrich; Merck KGaA,
Darmstadt, Germany) for 3 min at room temperature and
counterstaining with hematoxylin for 30 sec at room temperature.
Subsequently, the IHC-stained sections were observed under a light
microscope (magnification, ×400; Nikon Corporation, Tokyo, Japan).
Cases with breast cancer that had at least 1% of cells staining
positive for ER or PR were considered as ER-positive or
PR-positive. HER2 tumor status was detected by fluorescence in
situ hybridization (FISH). The PathVysion HER2/neu DNA probe
kit II (Abbott Pharmaceutical Co. Ltd., Lake Bluff, IL, USA), which
is designed to detect amplification of the HER2/neu gene via FISH
in formalin-fixed paraffin-embedded human breast cancer tissue
specimens, was used according to the manufacturer's protocol
(16). A high mean copy number of
HER2 (ufacturer's plificawas considered positive regardless of the
HER2/chromosome enumeration probe 17 ratio. Written informed
consent was obtained from all the patients and the study was
approved by the Ethics Committee of the Central Hospital of Zibo
(approval no. ZB 20151229005).
Cell culture
The human TNBC cell lines (MDA-MB-231, HCC-1937 and
MDA-MB-468), the immortal mammary epithelial cell line (MCF-10A)
and 293T cells were obtained from the Cell Bank of Chinese Academy
of Sciences (Shanghai, China). Cell lines were cultured at 37°C in
a humidified incubator with 5% CO2. The TNBC cell lines
and 293T cells were cultured in Dulbecco's modified Eagle's medium
(DMEM; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA USA)
containing 10% fetal bovine serum (FBS; HyClone; GE Healthcare Life
Sciences, Logan, UT, USA). MCF-10A cells were cultured in DMEM/F12
medium (HyClone; GE Healthcare Life Sciences Shanghai, China)
supplemented with 5% horse serum (Gibco; Thermo Fisher Scientific,
Inc.).
Cell transfection
miR-589 mimic
(5′-UGAGAACCACGUCUGCUCUGAGGGTATTCGCACTGGATACGACGAACTTT-3′), mimic
control (miR-NC) (5′-ACUACUGAGUGACAGUAGA-3′), miR-589 inhibitor
(5′-UGAGAACCACGUCUGCUCUGAG-3′) and inhibitor control (anti-NC)
(5′-CAGUACUUUUGUGUAGUACAA-3′) were purchased from RiboBio Co.,
Ltd., (Guangzhou, China). The small RNAs including
si-metastasis-associated protein 2 (MTA2) and sicontrol were
obtained from Santa Cruz Biotechnology, Inc. The siRNA sequences
were: siMTA2, 5 MTA2ces Cruz Biotechnologyand sicontrol, 5control
Cruz Biotechnology, A total of 5×105 MDA-MB-468 cells
were transfected with miR-NC (2.5 µg) or miR-589 mimic (2.5 µg)
using Lipofectamine® 2000 reagent (Invitrogen; Thermo
Fisher Scientific, Inc.) according to the manufacturer's protocol.
A total of 5×105 MDA-MB-231 cells were transfected with
anti-NC (2.5 µg) or miR-589 inhibitor (2.5 µg) and siMTA2 (100 nM)
or sicontrol (100 nM) using Lipofectamine® 2000 reagent
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol. Following transfection for 48 h, the
efficiency of transfection was analyzed using quantitative
polymerase chain reaction (qPCR) or western blot analysis.
Cell viability assay
Cell viability was assessed using the Cell Counting
Kit-8 (CCK-8) assay (Beyotime Institute of Biotechnology, Haimen,
China), according to the manufacturer's protocol. Briefly, cells
were plated into 96-well plates (1×103/well). At set
time points (0, 24, 48 and 72 h), 10 µl of CCK-8 solution was added
to each well. Following incubation for 3 h, the absorbance of each
well was measured using the Multiskan MK3 microplate photometer
(Thermo Fisher Scientific, Inc.) at 450 nm.
Colony formation assay
The proliferation of cells was measured using the
plate colony formation assay. Each well of the 6-well plates
contained 300 cells/well and were cultured for 10 days in DMEM
containing 10% FBS at 37°C. Subsequently, the cells were washed
with PBS and fixed with 10% methanol for 15 min at room
temperature, and the colonies were stained with crystal violet
(Beyotime Institute of Biotechnology, Haimen, China) for 30 min at
room temperature. The colony images were obtained, and the number
of colonies was counted under a microscope (Olympus Corporation,
Tokyo, Japan).
Transwell and Matrigel assays
The cell migration and invasion abilities were
detected using the Transwell and Matrigel assays, which were
carried out in 24-well Transwell chambers (Corning Inc., Corning
NY, USA). In brief, 5×104 MDA-MB-468 or MDA-MB-231 cells
were plated on the upper Transwell chamber either in the presence
or absence of Matrigel inserts in DMEM. DMEM supplemented with 5%
FBS was added to the lower chamber. The cells were maintained in a
humidified incubator with 5% CO2 for 24 h at 37°C.
Non-migrating cells in the upper chamber were scraped using a
cotton swab. The migrating cells were fixed with 10% methanol for
10 min, and stained with crystal violet (Beyotime Institute of
Biotechnology) for 5 min at room temperature. Finally, the stained
cells from six random fields were counted, and the images were
captured under a light microscope (magnification, ×100).
Western blot analysis
Total proteins were extracted from tissues using the
T-PER Tissue Protein Extraction Reagent (Pierce; Thermo Fisher
Scientific, Inc.). Protein concentrations were determined using a
bicinchoninic acid (BCA) Protein Assay kit (Pierce; Thermo Fisher
Scientific, Inc.). Proteins (20 µg) were separated by SDS-PAGE (10%
gels) and transferred to polyvinylidene fluoride membranes (EMD
Millipore, Billerica, MD, USA). Membranes were blocked at room
temperature with 5% skimmed milk for 1 h and incubated at 4°C
overnight with the following antibodies: MTA2 antibody (cat. no.
sc-55566; 1:500 dilution), N-cadherin (cat. no. sc-8424; 1:500
dilution), vimentin (cat. no. sc-66002; 1:500 dilution), E-cadherin
(cat. no. sc-8426; 1:500 dilution; all Santa Cruz Biotechnology,
Inc.), β-actin antibody (cat. no. AF0003; 1:1,000 dilution;
Beyotime Institute of Biotechnology) and subsequently with
biotinylated goat anti-mouse secondary antibodies (cat. no.
ab64255; 1:1,000 dilution; Abcam) at room temperature for 2 h.
Specific bands of interest were visualized with enhanced
chemiluminescence reagent (Nanjing KeyGen Biotech Co., Ltd.,
Nanjing, China) on an autoradiographic film.
Total mRNA extraction and qPCR
Total RNA was extracted from tissues and MDA-MB-231,
HCC-1937, MDA-MB-468 and MCF-10A cells using the miRcute miRNA
isolation kit (Tiangen Biotech Co., Ltd., Beijing, China),
according to the manufacturer's protocol. Total RNA was reverse
transcribed with oligodT primers using the miRcute Plus miRNA
First-strand cDNA Synthesis kit (Tiangen Biotech Co., Ltd.) at 37°C
for 60 min and 85°C for 1 min. The cDNA was amplified by qPCR using
SYBR® Premix Ex Taq (Takara Biotechnology Co., Ltd.,
Dalian, China). The thermocycling conditions were as follows: 5 min
at 95°C, followed by 40 cycles of 30 sec at 95°C, 60 sec at 60°C
and 30 sec at 72°C; and 1 sec at 99°C; 15 sec at 59°C; 1 sec at
95°C; followed by cooling to 40°C. The relative expression level of
miR-589 and MTA2 was calculated using the 2−ΔΔCq method
(17), and normalized to the
reference gene. The primers used for qPCR were as follows: miR-589
forward, 5′-CGAGGTCAGCGTGATTTCATGG-3′ and reverse
5′-TGTGTCCAAGTCCCAGCCAGAG-3′; U6 forward, 5′-CTCGCTTCGGCAGCACA-3′
and reverse 5′-AACGCTTCACGAATTTGCGT-3′; MTA2 forward,
5′-ATCATTACCAGCCACCCA-3′ and reverse 5′-CGATTATCAGATTCTCCCTC-3′;
N-cadherin forward 5′-ATCAAAGACCCATCCACC-3′ and reverse
5′-CCTCCTCACCACCACTA-3′; vimentin forward, 5′-CTTCCGCGCCTACGCCA-3′
and reverse 5′-GCCCAGGCGACCTACTCC-3′; E-cadherin forward
5′-GTACTTGTAATGACACATCTC-3′ and reverse 5′-TGCCAGTTTCTGCATCTTGC-3′;
and β-actin forward 5′-GATCATTGCTCCTCCTGAGC-3′ and reverse
5′-ACTCCTGCTTGCTGATCCAC-3′.
Prediction of miRNA targets
The hypothetical targets of miR-589 were predicted
using TargetScan human version 7.1 (http://www.targetscan.org/vert_71/) (18). This revealed that the 3′-untranslated
region (3′-UTR) of MTA2 may be complementarily paired with the seed
sequences of miR-589.
Dual-luciferase assay
To validate the direct target gene of miR-589, an
online bioinformatics search using TargetScan was performed.
MTA2-3′-UTR wild-type and MTA2-3′-UTR mutant were synthesized by
Shanghai GenePharma Co., Ltd. (Shanghai, China) and cloned into the
psiCHECK2 vector (Promega Corporation, Madison, WI, USA). The
psiCHECK2-3′-UTR-wild-type or psiCHECK2-3′-UTR-mutant luciferase
plasmids were transfected into 293T cells using Lipofectamine 2000
(Invitrogen; Thermo Fisher Scientific, Inc.). At 24 h following
transfection, luciferase activity was measured using the
dual-luciferase reporter assay kit (Promega Corporation), according
to the manufacturer's protocol. The activity of firefly luciferase
was normalized to the corresponding Renilla luciferase
activity.
Statistical analysis
All experiments were performed three times, and all
values are presented as the mean ± standard deviation (SD).
Statistical analysis was performed using SPSS (11.0; SPSS, Inc.,
Chicago, IL, USA). The differences between 2 groups were assessed
using a two-tailed unpaired Student's t-test. Data of >2 groups
were analyzed using one-way analysis of variance with the Tukey's
post hoc test. Pearson's correlation analysis was used to analyze
the correlation between the expression of miR-589 and MTA2.
P<0.05 was considered to indicate a statistically significant
difference.
Results
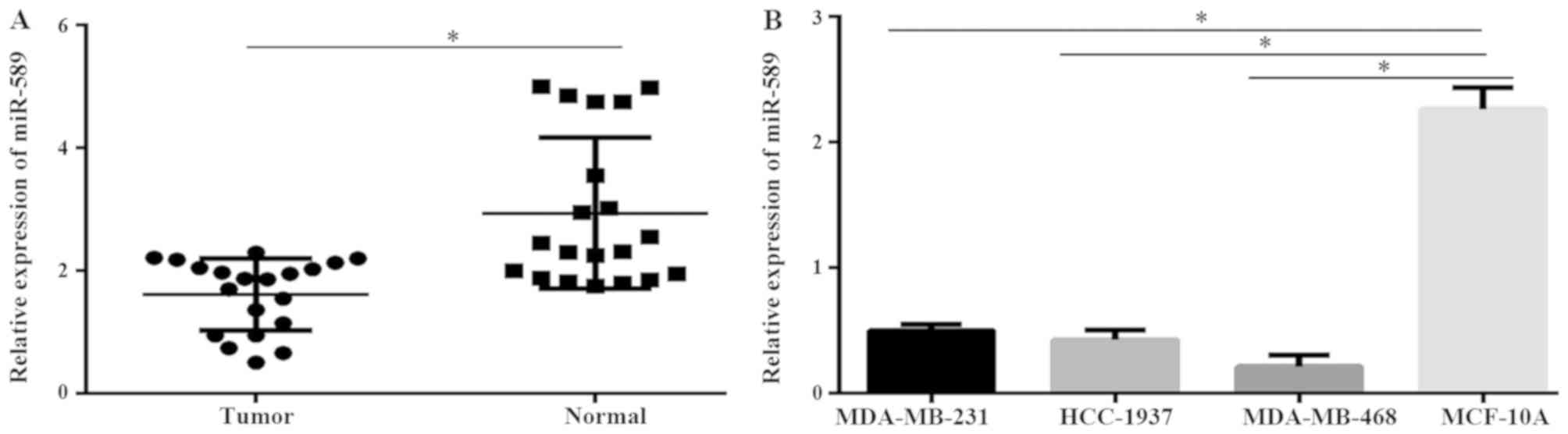
Expression of miR-589 decreases in
TNBC tissues and cell lines
To investigate the role of miR-589 in TNBC, the
expression of miR-589 in TNBC patient samples and adjacent
non-tumor tissues were evaluated. Results from the qPCR
demonstrated that miR-589 was downregulated in TNBC tissues
compared with that in the adjacent normal tissues (P<0.05;
Fig. 1A). The results also revealed
that the expression level of miR-589 was significantly
downregulated in the MDA-MB-231, MDA-MB-468 and HCC-1937 TNBC cell
lines compared with that in the normal breast epithelial cell line,
MCF-10A (P<0.05; Fig. 1B). These
data revealed that miR-589 may serve a critical role in the
progression of TNBC. The expression level of miR-589 was highest in
the MDA-MB-231 cells, while it was lowest in the MDA-MB-468 cells.
Subsequently, these 2 cancer cell lines were used in further
investigations.
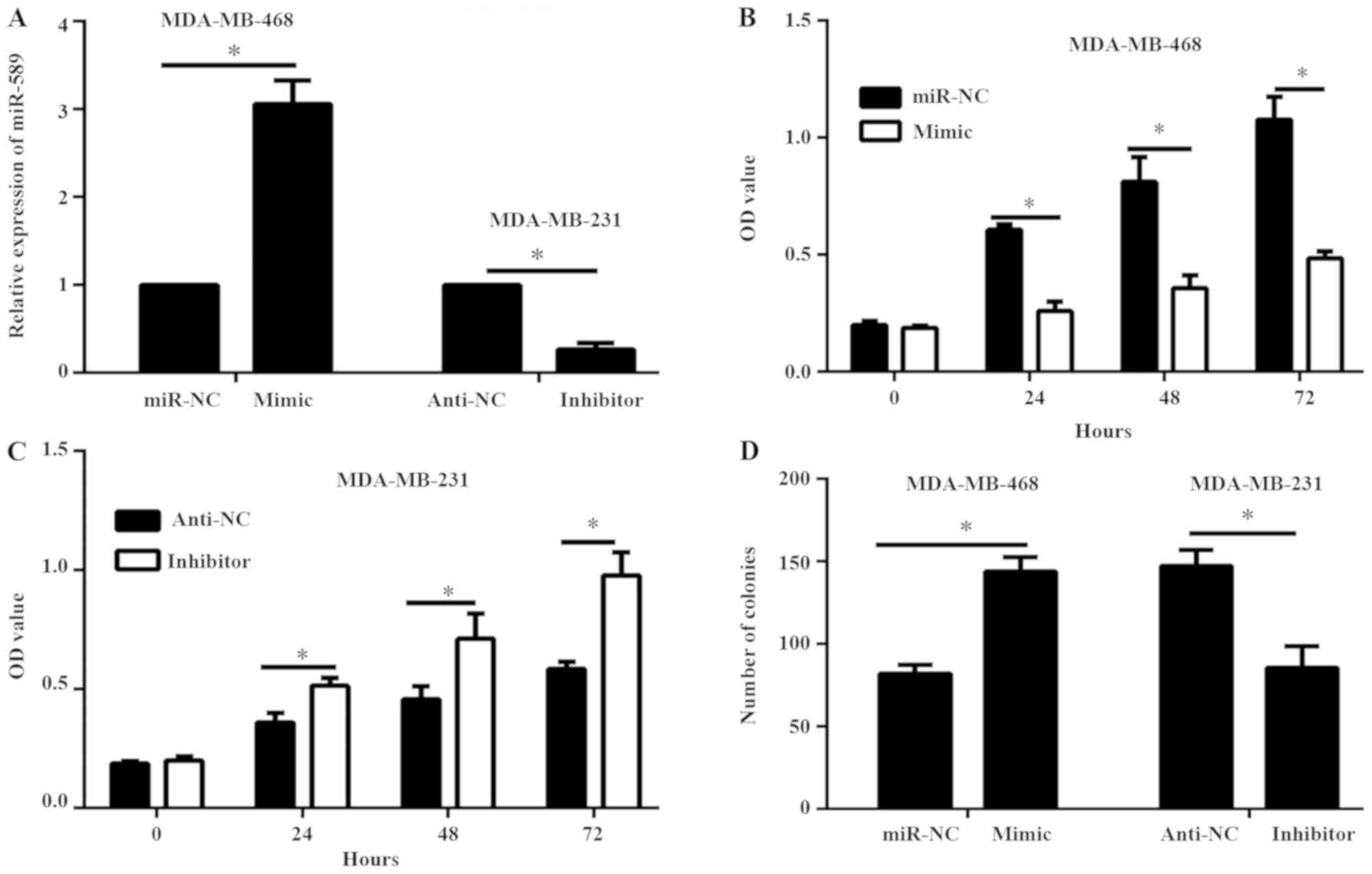
miR-589 decreases TNBC cell viability
and proliferation
Subsequently, the role of miR-589 in TNBC cell
viability and proliferation was determined. miR-589 mimic and
miR-NC were transfected into the MDA-MB-468 cells, while the
miR-589 inhibitor and anti-NC were transfected into the MDA-MB-231
cells. Results of the qPCR demonstrated that the expression level
of miR-589 was enhanced in MDA-MB-468 cells transfected with
miR-589 mimic compared with MDA-MB-468 cells containing miR-NC;
while the expression level of miR-589 was reduced in the MDA-MB-231
cells transfected with miR-589 inhibitor compared with that in the
anti-NC group (P<0.05; Fig. 2A).
Results of the CCK-8 and colony formation assays demonstrated that
miR-589 overexpression decreased the cell viability and
proliferation of MDA-MB-468 cells, while miR-589 silencing
remarkably enhanced the cell viability and proliferation of
MDA-MB-231 cells (P<0.05; Fig.
2B-D).
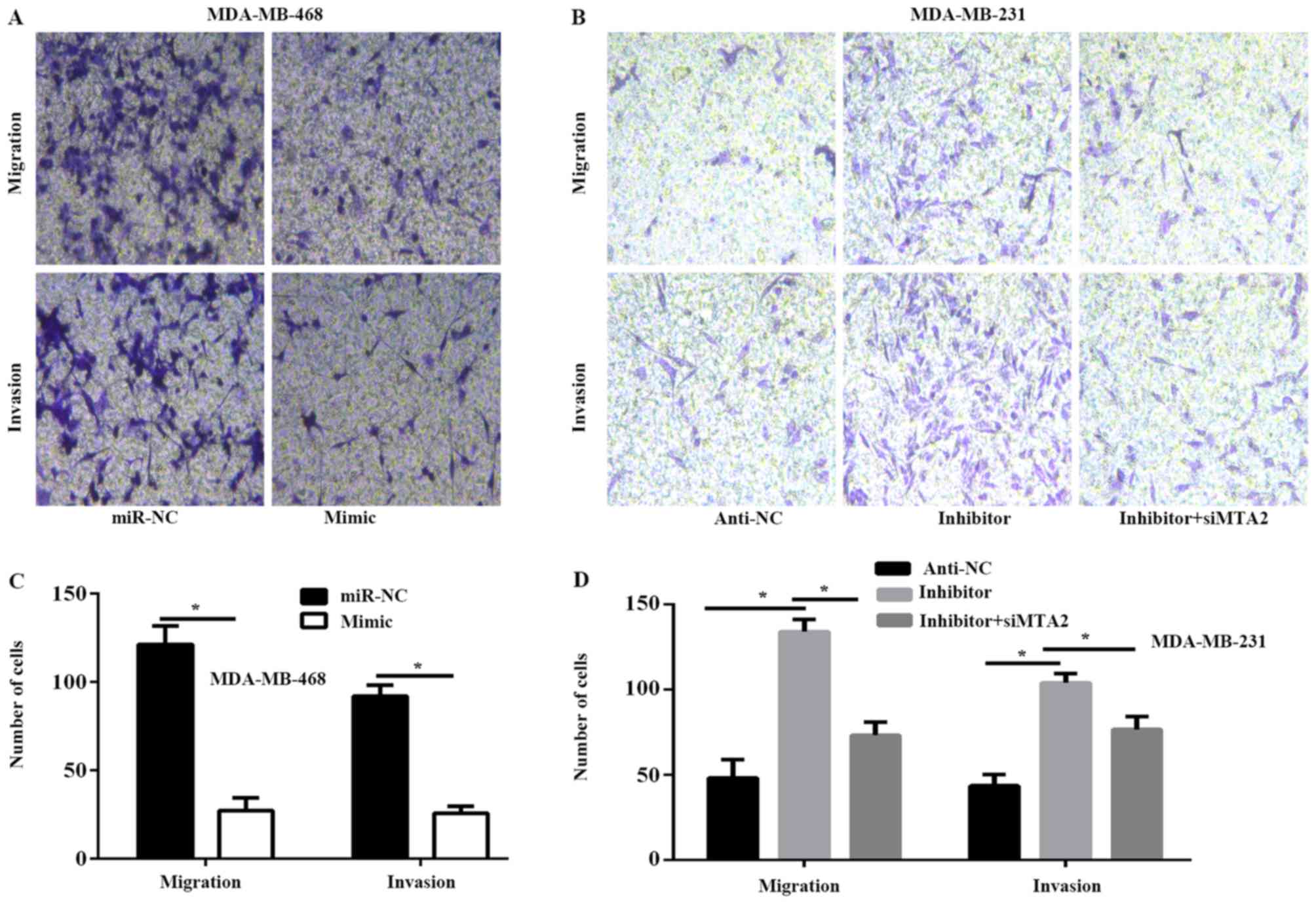
miR-589 inhibits TNBC cell migration
and invasion in vitro
Furthermore, the ability of miR-589 to regulate TNBC
cell migration and invasion was determined. The results of the
migration and invasion assays revealed that exogenous miR-589
expression significantly decreased the capability of migration and
invasion of MDA-MB-468 cells, whereas miR-589 silencing produced
the opposite results in MDA-MB-231 cells (P>0.05; Fig. 3A-D). In addition, the results of the
Transwell assay revealed that MTA2 silencing decreased the
migration and invasion capability of MDA-MB-231 cells, which was
initially enhanced by miR-589 inhibitor (P<0.05; Fig. 3B and D).
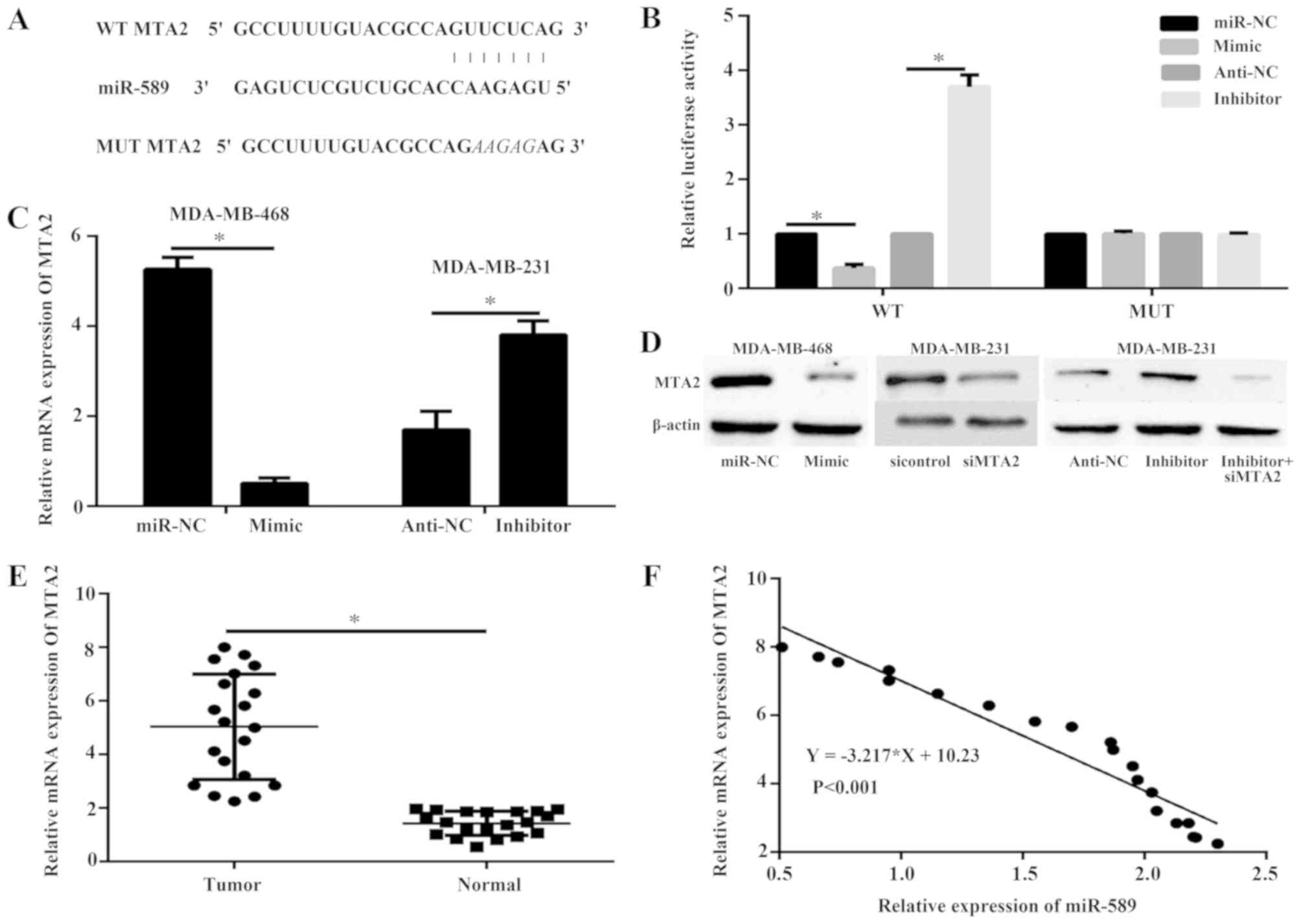
Identification of MTA2 as a target of
miR-589
The potential targets of miR-589 were predicted via
bioinformatics analysis to investigate the underlying mechanism by
which miR-589 decreases TNBC cell progression. MTA2 serves as an
oncogene in TNBC and was thus selected for further confirmation
(Fig. 4A). The 293T cells were
co-transfected with miR-589 mimic or miR-589 inhibitor and either
the wild-type or mutated MTA2 3′-UTR reporter and subsequently
subjected to dual-luciferase assay. The results illustrated that
miR-589 mimic decreased the luciferase intensity of MTA2 3′-UTR,
whereas a mutation within the binding site abolished the inhibitory
role of miR-589 in MTA2 3′-UTR (P<0.05; Fig. 4B). The miR-589 inhibitor increased
the luciferase intensity of MTA2 3′-UTR (P<0.05; Fig. 4B). Furthermore, qPCR (P<0.05;
Fig. 4C) and western blot analysis
(Fig. 4D) revealed that the
expression levels of MTA2 mRNA and protein were inhibited in the
MDA-MB-468 cells transfected with miR-589 mimic, while miR-589
silencing produced higher expression levels in the MDA-MB-231
cells. In addition, the mRNA expression level of MTA2 was
upregulated in the TNBC tissues compared with that in the adjacent
normal tissues (P<0.05; Fig. 4E),
and was negatively correlated with miR-589 expression in TNBC
tissues (P<0.05; Fig. 4F).
MTA2 is a functional target of
miR-589
The MDA-MB-231 cells were transfected with miR-589
inhibitor or miR-589 inhibitor and siMTA2 to determine if MTA2 is a
functional target of miR-589 in TNBC. The protein expression of
MTA2 was detected using western blot analysis. The results showed
that the expression of MTA2 was decreased in response to siMTA2
compared to control. In addition, the expression of MTA2 was
markedly reduced when cells were treated with siMTA2 and miR-589
inhibitor compared with miR-589 inhibitor only (Fig. 4D).
miR-589 inhibits the
epithelial-mesenchymal transition (EMT) progression via MTA2
A previous study has revealed that MTA2 promoted EMT
progression (19). To further
discover the mechanism of miR-589, whether miR-589 regulates EMT in
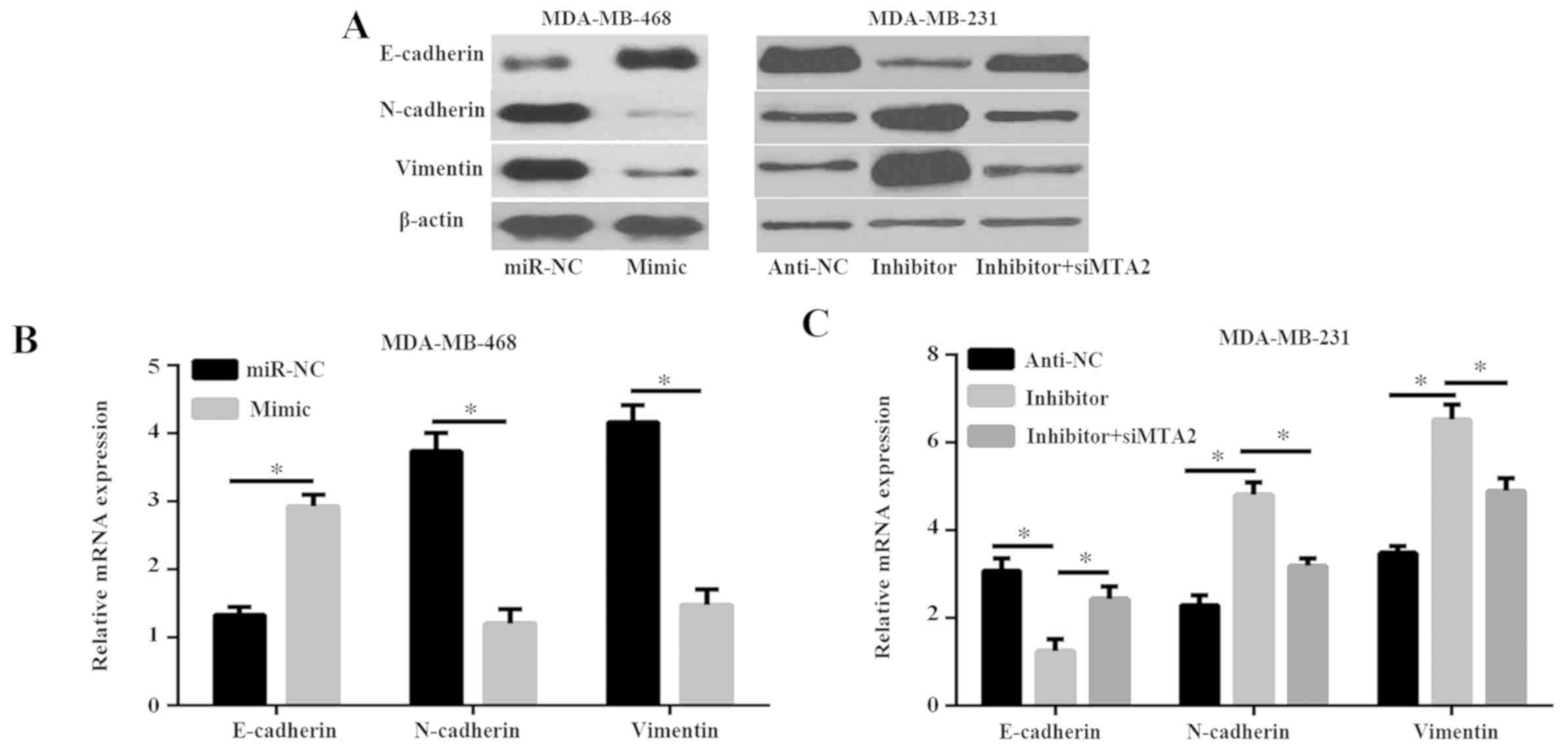
breast cancer cells was investigated. As revealed in Fig. 5A and B, miR-589 mimic reduced the
expression of the mesenchymal cell markers N-cadherin and vimentin,
but enhanced the expression levels of the epithelial cell marker
E-cadherin. However, miR-589 silencing resulted in increased
expression of N-cadherin and vimentin, and decreased the expression
of E-cadherin in the MDA-MB-231 cells (Fig. 5A and C). These data revealed that
miR-589 silencing triggered more aggressive EMT, and promoted cell
migration and invasion. Furthermore, the present study demonstrated
that the expression of N-cadherin and vimentin in the siMTA2 and
miR-589 inhibitor group was significantly decreased compared with
the miR-589 inhibitor group, while the expression of E-cadherin was
markedly enhanced in the siMTA2 and miR-589 inhibitor group
compared with the miR-589 inhibitor group (P<0.05; Fig. 5A and C). These results suggested that
miR-589 inhibited the process of EMT via MTA2.
Discussion
miRNA is a non-coding RNA consisting of 21–22
nucleotides in length in eukaryotes, and its primary function is to
regulate the expression of target genes (6,7). miRNAs
serve in a number of crucial roles in the development and
progression of many types of cancer (6,7). As a
novel cancer-associated miRNA, miR-589 serves as an oncogene in
gastric cancer (15). However,
miR-589 may act as a tumor suppressor in hepatocellular carcinoma,
glioma and lung cancer (11–14). The role of miR-589 in TNBC has yet to
be established. In the present study, the expression level of
miR-589 in tumor and adjacent normal tissues from TNBC patients was
measured using qPCR and identified that miR-589 is significantly
downregulated in TNBC tissues compared with that in normal tissue.
In addition, miR-589 was downregulated in 3 TNBC cell lines.
Furthermore, miR-589 inhibited the proliferation and metastasis of
breast cancer cells. MTA2 was identified as a direct target gene of
miR-589, and MTA2 silencing inhibited the cell proliferation and
metastasis induced by miR-589 silencing.
miR-589 serves as an oncogene or tumor suppressor
via a number of molecular mechanisms in different types of cancer.
A recent study revealed that miR-589 is overexpressed and promotes
the progression of gastric cancer via a leukemia inhibitory factor
receptor (LIFR)-phosphoinositide 3-kinase/protein kinase B-c-Jun
regulatory feedback loop (15).
However, in non-small lung cancer, miR-589 serves as a tumor
suppressor and negatively regulates histone deacetylases (HDAC)
(13). These studies revealed that
the same miRNA may serve as different roles in the progression of
cancer via different mechanisms depending on the tissue type. In
addition, the same miRNA may serve as different roles in different
biological functions in the same cancer tissue. In hepatocellular
carcinoma, miR-589 serves a contrary role in the progression of
cancer cells, including inhibiting the proliferation of cancer cell
proliferation and maintaining the stemness and enhancing the
chemoresistance to doxorubicin (11,14,20). The
same miRNA may regulate different genes in cancer, and the gene may
be regulated by different miRNAs in different tissues. Previous
studies revealed that miR-589 could regulate many genes, including
LIFR, zinc finger MYND-type containing 19, signal transducer and
activator of transcription 3, HDAC5 and mitogen-activated protein
kinase 8 (11–15,20). To
explore the molecular mechanism of miR-589 in inhibiting TNBC
progression, publicly available bioinformatic algorithms were used
to analyze the target gene of miR-589. MTA2 was identified as a
direct and functional target of miR-589 using bioinformatics,
dual-luciferase reporter assays, qPCR and western blot analysis.
MTA2 is a member of a small protein family (including MTA1, MTA2
and MTA3) (21). A previous study
revealed that MTA2 promotes tumor progression (22). A study in 2013 revealed that MTA2
enhances the metastasis of TNBC cells by inhibiting Rho pathway
signaling (23). In the present
study, a significant negative correlation was observed between MTA2
and miR-589 expression in patients with TNBC. More importantly, the
silencing of MTA2 could abolish the inducing effect of miR-589
silencing on the progression of TNBC cells. These data suggest that
miR-589 inhibits TNBC, at least in part, by the downregulation of
MTA2.
However, the present study has limitations. The
number of patients is small and does not include Luminal A, Luminal
B and HER-2 positive subtypes of breast cancer. Thus, the
association of miR-589 and the other types of breast cancer was not
determined. Future studies, would aim to recruit more patients and
investigate additional subtypes of breast cancer. In addition,
other target genes or signaling pathways via which miR-589 inhibits
breast cancer require further investigation.
In summary, the present study is the first to report
that miR-589 inhibits TNBC progression by directly targeting MTA2.
These findings indicate that miR-589 is a potential tumor
suppressor in TNBC through enhancing cellular proliferation and
metastasis. The present study suggests that miR-589 is a novel
biomarker in improving the therapeutic care of patients with
TNBC.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
JC conceived, designed and conducted all
experiments, and wrote and revised the manuscript.
Ethics approval and consent to
participate
Written informed consent was obtained from all the
patients and the study was approved by the Ethics Committee of the
Central Hospital of Zibo (approval no: ZB 20151229005).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
DeSantis CE, Ma J, Goding Sauer A, Newman
LA and Jemal A: Breast cancer statistics, 2017, racial disparity in
mortality by state. CA Cancer J Clin. 67:439–448. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bauer KR, Brown M, Cress RD, Parise CA and
Caggiano V: Descriptive analysis of estrogen receptor
(ER)-negative, progesterone receptor (PR)-negative, and
HER2-negative invasive breast cancer, the so-called triple-negative
phenotype: A population-based study from the California cancer
Registry. Cancer. 109:1721–1728. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Dent R, Trudeau M, Pritchard KI, Hanna WM,
Kahn HK, Sawka CA, Lickley LA, Rawlinson E, Sun P and Narod SA:
Triple-negative breast cancer: Clinical features and patterns of
recurrence. Clin Cancer Res. 13:4429–4434. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wahba HA and El-Hadaad HA: Current
approaches in treatment of triple-negative breast cancer. Cancer
Biol Med. 12:106–116. 2015.PubMed/NCBI
|
|
6
|
Iorio MV and Croce CM: MicroRNA
dysregulation in cancer: Diagnostics, monitoring and therapeutics.
A comprehensive review. EMBO Mol Med. 9:8522017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Pasquinelli AE: MicroRNAs and their
targets: Recognition, regulation and an emerging reciprocal
relationship. Nat Rev Genet. 13:271–282. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Christodoulatos GS and Dalamaga M:
Micro-RNAs as clinical biomarkers and therapeutic targets in breast
cancer: Quo vadis? World J Clin Oncol. 5:71–81. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lü L, Mao X, Shi P, He B, Xu K, Zhang S
and Wang J: MicroRNAs in the prognosis of triple-negative breast
cancer: A systematic review and meta-analysis. Medicine
(Baltimore). 96:e70852017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Piasecka D, Braun M, Kordek R, Sadej R and
Romanska H: MicroRNAs in regulation of triple-negative breast
cancer progression. J Cancer Res Clin Oncol. 144:1401–1411. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Xu M, Wang Y, He HT and Yang Q: MiR-589-5p
is a potential prognostic marker of hepatocellular carcinoma and
regulates tumor cell growth by targeting MIG-6. Neoplasma.
65:753–761. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cesarini V, Silvestris DA, Tassinari V,
Tomaselli S, Alon S, Eisenberg E, Locatelli F and Gallo A:
ADAR2/miR-589-3p axis controls glioblastoma cell
migration/invasion. Nucleic Acids Res. 46:2045–2059. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Liu C, Lv D, Li M, Zhang X, Sun G, Bai Y
and Chang D: Hypermethylation of miRNA-589 promoter leads to
upregulation of HDAC5 which promotes malignancy in non-small cell
lung cancer. Int J Oncol. 50:2079–2090. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhang X, Jiang P, Shuai L, Chen K, Li Z,
Zhang Y, Jiang Y and Li X: miR-589-5p inhibits MAP3K8 and
suppresses CD90+ cancer stem cells in hepatocellular
carcinoma. J Exp Clin Cancer Res. 35:1762016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang F, Li K, Pan M, Li W, Wu J, Li M,
Zhao L and Wang H: miR-589 promotes gastric cancer aggressiveness
by a LIFR-PI3K/AKT-c-Jun regulatory feedback loop. J Exp Clin
Cancer Res. 37:1522018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Patil Okaly GV, Panwar D, Lingappa KB,
Kumari P, Anand A, Kumar P, Chikkalingaiah MH and Kumar RV: FISH
and HER2/neu equivocal immunohistochemistry in breast carcinoma.
Indian J Cancer. 56:119–123. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Agarwal V, Bell GW, Nam JW and Bartel DP:
Predicting effective microRNA target sites in mammalian mRNAs.
Elife. 4:2015. View Article : Google Scholar
|
|
19
|
Fu J, Qin L, He T, Qin J, Hong J, Wong J,
Liao L and Xu J: The TWIST/Mi2/NuRD protein complex and its
essential role in cancer metastasis. Cell Res. 21:275–289. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Long J, Jiang C, Liu B, Dai Q, Hua R, Chen
C, Zhang B and Li H: Maintenance of stemness by miR-589-5p in
hepatocellular carcinoma cells promotes chemoresistance via STAT3
signaling. Cancer Lett. 423:113–126. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kumar R, Wang RA and Bagheri-Yarmand R:
Emerging roles of MTA family members in human cancers. Semin Oncol
30 (5 Suppl). S30–S37. 2003. View Article : Google Scholar
|
|
22
|
Covington KR and Fuqua SA: Role of MTA2 in
human cancer. Cancer Metastasis Rev. 33:921–928. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Covington KR, Brusco L, Barone I,
Tsimelzon A, Selever J, Corona-Rodriguez A, Brown P, Kumar R,
Hilsenbeck SG and Fuqua SA: Metastasis tumor-associated protein 2
enhances metastatic behavior and is associated with poor outcomes
in estrogen receptor-negative breast cancer. Breast Cancer Res
Treat. 141:375–384. 2013. View Article : Google Scholar : PubMed/NCBI
|