Introduction
Hepatocellular carcinoma (HCC) is a high-incidence
malignant tumor worldwide; its incidence rate is ~5.7% of all new
cancer cases, and it has become the third leading cause of
cancer-associated deaths (1). The
chronic persistent infection of hepatitis C virus (HCV) is a major
etiological element of HCC, accounting for 25% of HCC cases
(2). Numerous studies have focused
on the aberrant expression and alteration of genes, a number of
which are involved in the development of HCC. It has been found
that DNA topoisomerase 2α (TOP2A) may act as a biomarker and is a
drug target of nitidine chloride in HCC (3). The increase of vascular endothelial
growth factor plays a key role in the recurrence of HCC (4). Testicular nuclear receptor 4 may affect
the capability of a patient to suppress HCC metastasis (5). In addition, the mRNA overexpression of
the cognate receptors c-Met and epidermal growth factor receptor
were found to be involved in tumor recurrence (6). However, the high mortality rate is due
in part to the lack of early detection of HCC markers (7). Thus, it is vital to identify effective
early diagnostic methods and to understand the molecular mechanism
of HCC development, in order to intervene in the progression of the
disease at the early stages.
Currently, large datasets, which include genomic
sequencing, DNA copy number arrays and protein arrays, are uploaded
to public databases, for example Gene Expression Omnibus (GEO) and
Oncomine. Furthermore, these databases are available for screening
differentially expressed genes (DEGs) and analyzing the molecular
mechanism of carcinogenesis in HCC. Nevertheless, it is difficult
to trust the results from separate microarray analysis. Therefore,
in the present study, three mRNA microarray datasets of HCV-induced
HCC (HCV-HCC) and non-tumor tissues were analyzed to identify DEGs,
using the GEO2R web tool. Subsequently, the DEGs were analyzed
using the Gene Ontology (GO) and the Kyoto Encyclopedia of Genes
and Genomes (KEGG) databases to identify the molecular mechanisms,
biological processes and pathways involved, and a protein-protein
interaction (PPI) network was constructed. Finally, in total, 368
DEGs and 10 hub genes were identified, providing several
possibilities to investigate as potential biomarkers for the
diagnosis of HCV-HCC and for novel drug development.
Materials and methods
Microarray data
GEO (http://www.ncbi.nlm.nih.gov/geo) is a high-throughput
gene expression database. In the present study, three gene
expression datasets (GSE62232 (8)
from France, GSE69715 (9) from Italy
and GSE107170 (10) from Italy) were
downloaded from GEO. GSE62232 comprised nine HCV-HCC samples and 10
adjacent non-tumor tissue samples, GSE69715 comprised 37 HCV-HCC
and 66 normal samples, and GSE107170 included 44 HCV-HCC tissue and
normal tissue (Table I). All of
these datasets were obtained from analyses performed using the
GPL570 Affymetrix Human Genome U133 Plus 2.0 Array (Thermo Fisher
Scientific, Inc.).
 | Table I.Numbers of samples included in three
Gene Expression Omnibus microarray datasets analyzed in the present
study. |
Table I.
Numbers of samples included in three
Gene Expression Omnibus microarray datasets analyzed in the present
study.
| Dataset ID | HCV-HCC, n | Normal, n | Total, n |
|---|
| GSE62232 | 9 | 10 | 19 |
| GSE69715 | 37 | 66 | 103 |
| GSE107170 | 44 | 31 | 75 |
Identification of DEGs
GEO2R (https://www.ncbi.nlm.nih.gov/geo/geo2r/) is an online
analysis tool that compares two or more groups of samples to
identify DEGs. The DEGs of the three gene expression datasets
including HCV-HCC and non-cancer samples were analyzed, and the
|log2 fold-change (FC)| and adjusted P-values were
calculated. Genes that fulfilled the criteria of
|log2FC|≥1.0 and adjusted P<0.05 were considered
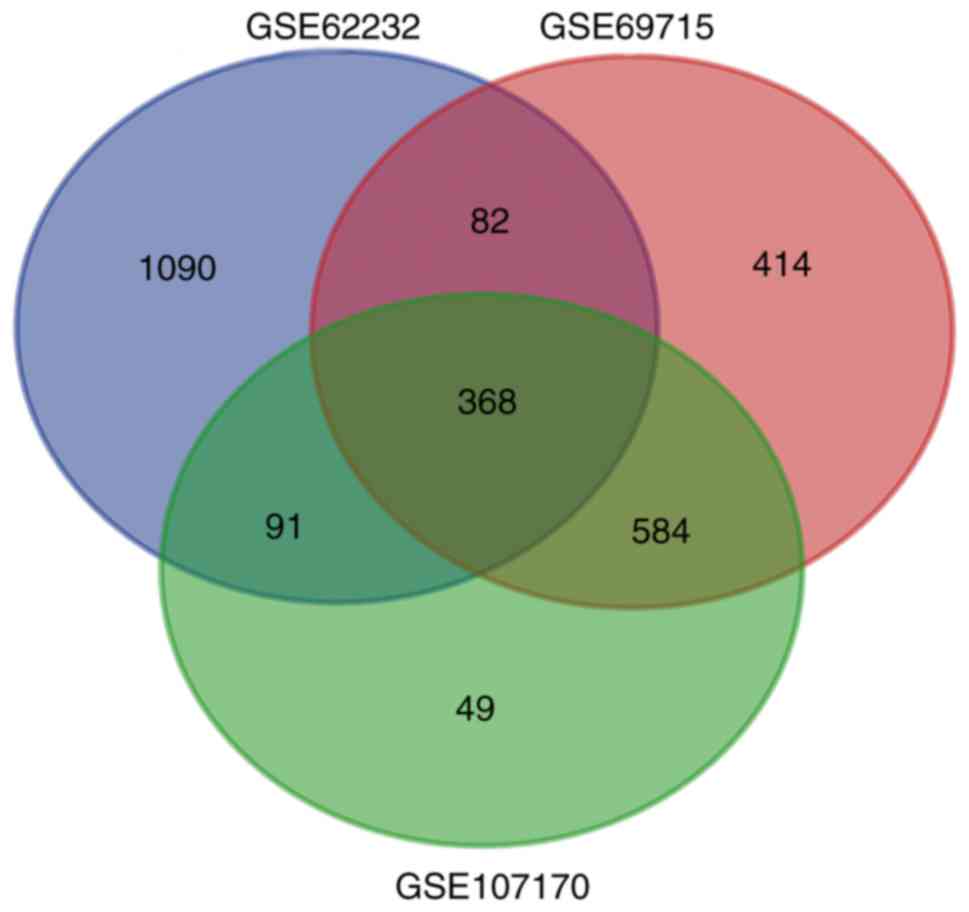
statistically significant and termed DEGs. The Venn diagram web
tool (bioinformatics.psb.ugent.be/webtools/Venn/) was used to
represent the overlapping DEGs from each dataset.
GO and KEGG pathway analysis of
DEGs
The Database for Annotation, Visualization and
Integrated Discovery (DAVID) tool (version 6.7; http://david.ncifcrf.gov/) is an online biological
information database that integrates biological data and analysis
tools. GO was used to perform bioinformatic analysis and annotate
function enrichment of genes. KEGG is a database containing
large-scale molecular datasets generated from genome sequencing,
used to explore the high-level functions and utilities of the
biological system. DAVID was applied to perform the function
enrichment and biological analyses of these DEGs. P<0.01 and
gene counts ≥10 were considered statistically significant.
PPI network of DEGs
The Search Tool for the Retrieval of Interacting
Genes (STRING) database (version 10.0; http://string-db.org/) was used to construct the PPI
network. Assessing the potential protein interactions enables the
identification of possible mechanisms in the development of
diseases. The PPI pairs with a combined score >0.4 were
considered statistically significant. Cytoscape software (version
3.40; www.cytoscape.org/) was used to
visualize the PPI network. CytoHubba is an application of
Cytoscape, and was used for calculating the degree of each protein
node.
Hub gene identification and
analysis
In the present study, the top 10 genes with a high
degree of connectivity from the PPI network were identified as hub
genes. Hierarchical clustering of hub genes was constructed using
University of California Santa Cruz Cancer genomics browser
(version hg19; http://genome-cancer.ucsc.edu). The overall survival
and disease-free survival time analyses with the hub genes were
performed by constructing Kaplan-Meier curves in cBioPortal
(version 2.2.1; http://www.cbioportal.org). The datasets of Wurmbach
Liver (11) and Mas Liver (12) were obtained from the online database
Oncomine (http://www.oncomine.com) and used to
analyze the relationship between expression patterns and HCV
status. Gene Expression Profiling Interactive Analysis (GEPIA;
http://gepia.cancer-pku.cn/detail.php) is an online
tool for analyzing gene expression data from cancer and normal
tissues from The Cancer Genome Atlas and genotype-tissue expression
projects. GEPIA and The Human Protein Atlas (https://www.proteinatlas.org), which originated in
Sweden and is designed to map all the human proteins in tissues,
cells and organs, were used to verify the expression of hyaluronan
mediated motility receptor (HMMR), cyclin B1 (CCNB1) and kinesin
family member 20A (KIF20A) in HCV-HCC (13,14).
Results
Identification of DEGs in HCV-HCC
In the present study, three mRNA microarray datasets
(GSE62232, GSE69715 and GSE107170) were selected, and DEGs (1,631
in GSE62232, 1,448 in GSE69715 and 1,092 in GSE107170) were
identified. The overlap among the three gene expression profiles
included 368 DEGs, displayed using a Venn diagram (Fig. 1), including 264 downregulated genes
and 104 upregulated genes.
GO and KEGG analyses of DEGs
Functional enrichment analyses of DEGs, including GO
and KEGG pathways, were performed using DAVID (Table II). The resulting GO terms were
enriched in biological processes, including ‘organic acid catabolic
process’, ‘carboxylic acid catabolic process’, ‘secondary metabolic
process’, ‘cellular amino acid catabolic process’, ‘amine catabolic
process’, ‘cellular amino acid derivative metabolic process’, and
‘response to steroid hormone stimulus’. Molecular function analysis
results indicated that the DEGs were enriched in ‘carbohydrate
binding’. For the results of cell component analysis, the DEGs were
mainly enriched in ‘extracellular region’, ‘extracellular region
part’, and ‘extracellular space’. Moreover, KEGG pathway analysis
revealed that the DEGs were primarily enriched in ‘metabolic
pathways’.
 | Table II.GO and KEGG pathway functional
enrichment analysis of the differentially expressed genes. |
Table II.
GO and KEGG pathway functional
enrichment analysis of the differentially expressed genes.
| Category | Term | Description | Count | FDR |
|---|
| GOTERM_BP_FAT | GO:0016054 | Organic acid
catabolic process | 16 |
4.38×10−06 |
| GOTERM_BP_FAT | GO:0046395 | Carboxylic acid
catabolic process | 16 |
4.38×10−06 |
| GOTERM_BP_FAT | GO:0019748 | Secondary metabolic
process | 12 |
4.47×10−04 |
| GOTERM_BP_FAT | GO:0009063 | Cellular amino acid
catabolic process | 11 |
9.39×10−04 |
| GOTERM_BP_FAT | GO:0009310 | Amine catabolic
process | 11 |
3.44×10−03 |
| GOTERM_BP_FAT | GO:0006575 | Cellular amino acid
derivative metabolic process | 15 |
5.54×10−03 |
| GOTERM_BP_FAT | GO:0048545 | Response to steroid
hormone stimulus | 16 |
6.39×10−03 |
| GOTERM_CC_FAT | GO:0005576 | Extracellular
region | 73 |
1.08×10−05 |
| GOTERM_CC_FAT | GO:0044421 | Extracellular
region part | 43 |
1.74×10−04 |
| GOTERM_CC_FAT | GO:0005615 | Extracellular
space | 33 |
1.65×10−03 |
| GOTERM_MF_FAT | GO:0030246 | Carbohydrate
binding | 25 |
6.44×10−05 |
| KEGG_PATHWAY | hsa01100 | Metabolic
pathways | 53 |
3.08×10−03 |
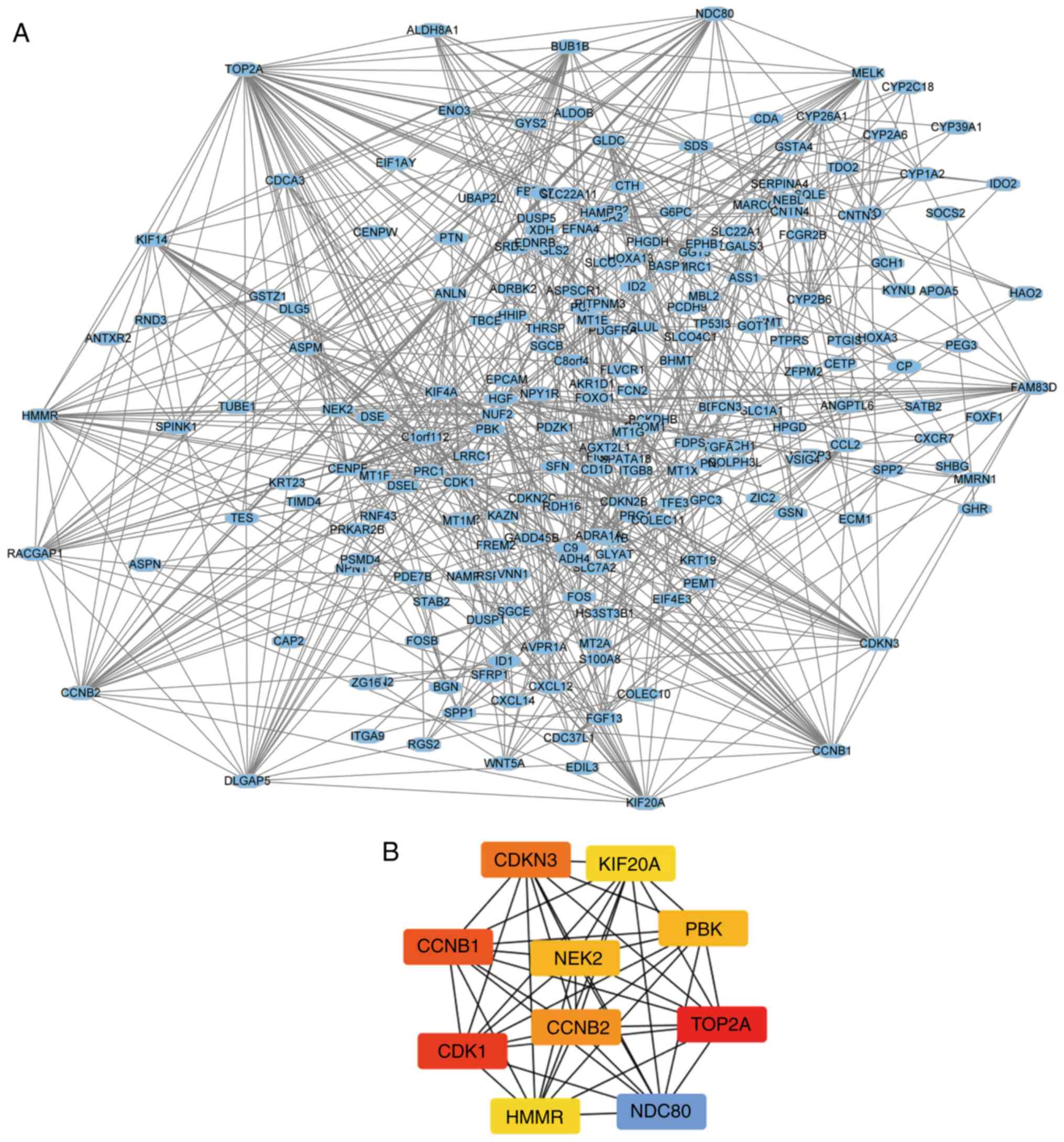
PPI network construction and hub gene
selection
The PPI network of the DEGs was constructed using
STRING (Fig. 2A) and the top ten
genes based on connectivity degree were identified using Cytoscape
(Fig. 2B). The 10 genes with the
highest degree of connectivity were identified as hub genes in the
PPI network (Table III) 4.
 | Table III.Top 10 hub genes based on degree of
connectivity. |
Table III.
Top 10 hub genes based on degree of
connectivity.
| Gene symbol | Gene name | Degree of
connectivity |
|---|
| TOP2A | DNA topoisomerase
2α | 46 |
| CDK1 | Cyclin dependent
kinase 1 | 34 |
| CCNB1 | Cyclin B1 | 31 |
| CDKN3 | Cyclin-dependent
kinase inhibitor 3 | 28 |
| NDC80 | Kinetochore complex
component | 27 |
| CCNB2 | Cyclin B2 | 27 |
| NEK2 | NIMA-related kinase
2 | 25 |
| PBK | PDZ-binding
kinase | 25 |
| KIF20A | Kinesin family
member 20A | 24 |
| HMMR | Hyaluronan-mediated
motility receptor | 24 |
Analysis of hub genes
The hierarchical clustering of hub genes inferred
that there is a difference in gene expression between HCV-HCC
samples and normal samples (Fig. 3).
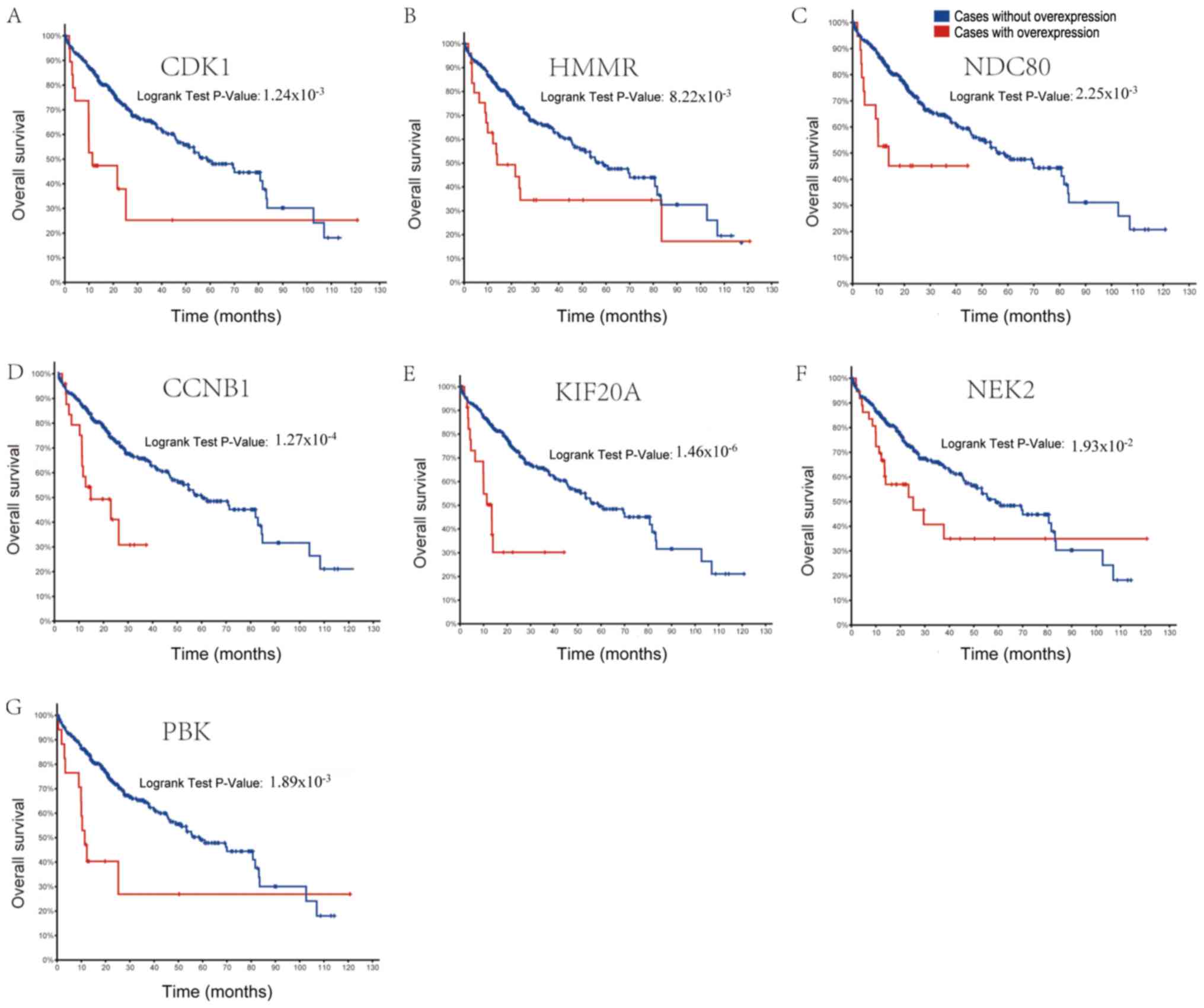
Subsequently, in order to identify and understand the prognostic
values of the hub genes, overall and disease-free survival time
analyses were performed using the Kaplan-Meier method. The results
of the overall survival analysis are displayed in Fig. 4. A significant association was
observed between poor prognosis and overexpression of genes CDK1,
HMMR, NDC80, CCNB1, KIF20A, NEK2 and PBK, in patients with HCV-HCC.
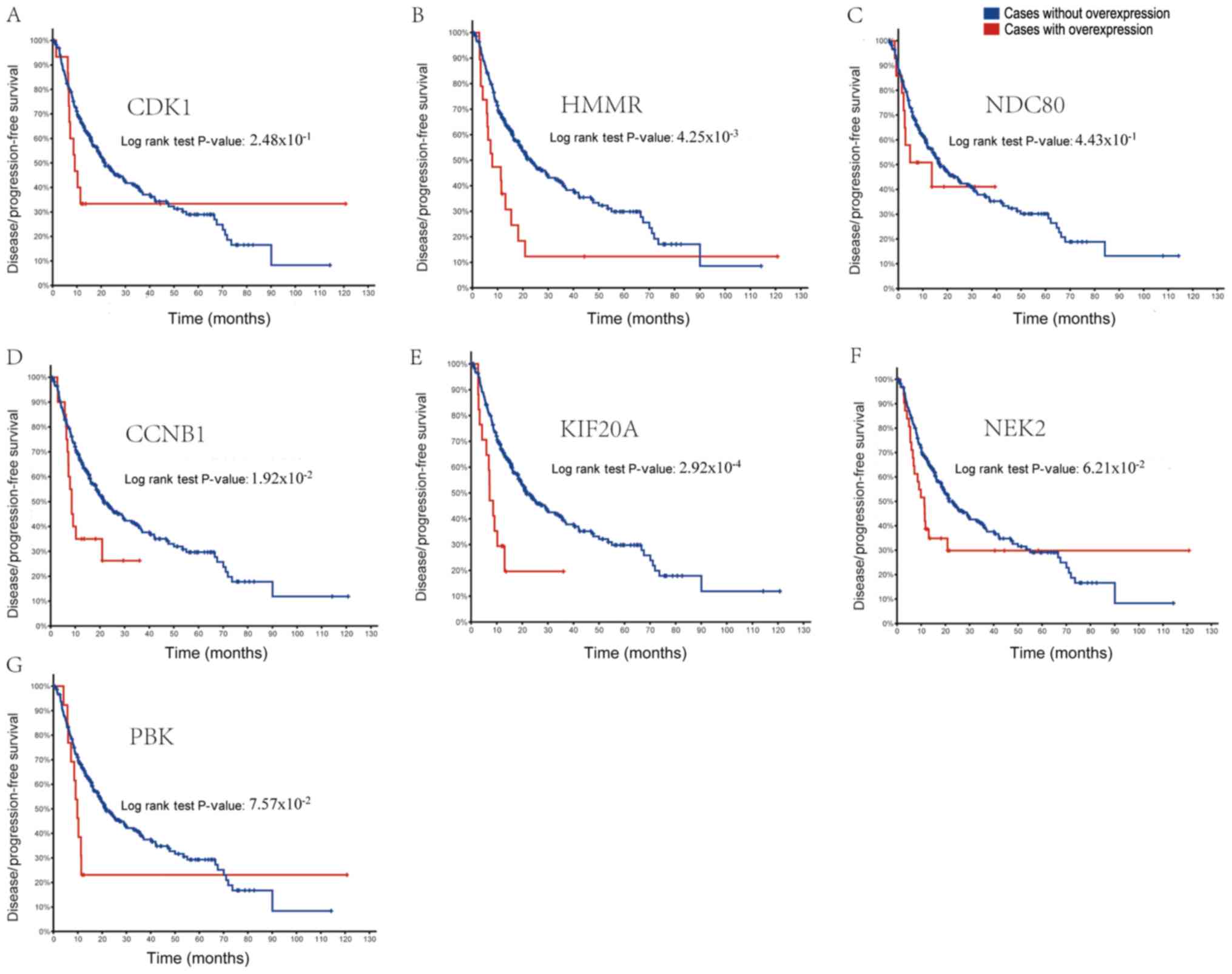
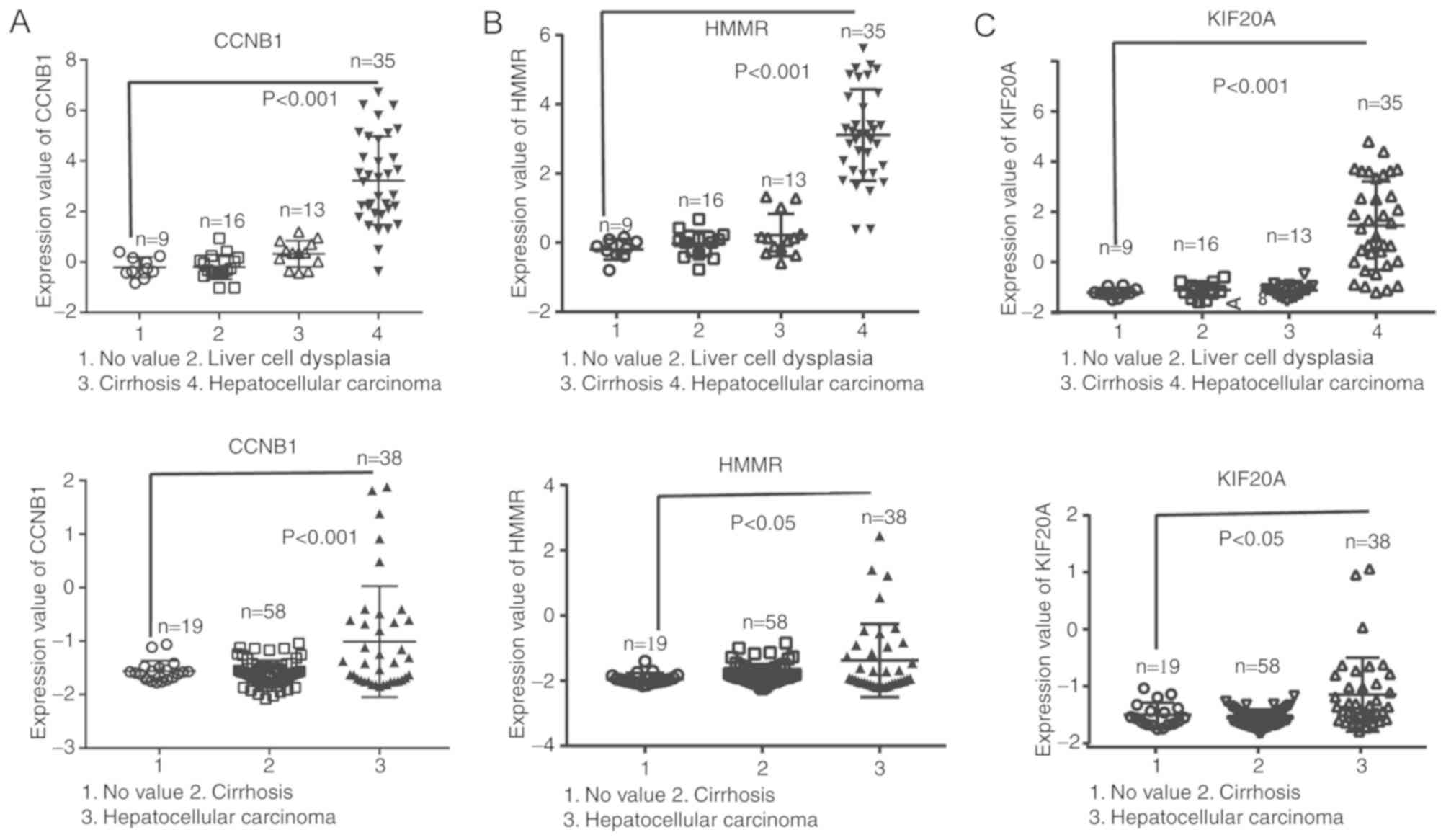
Furthermore, as shown in Fig. 5,
patients with HCC and overexpression of HMMR, CCNB1 and KIF20A
presented with lower disease-free survival rates. Analysis of the
Wurmbach Liver and Mas Liver datasets from the Oncomine database
showed that higher mRNA levels of CCNB1, KIF20A and HMMR were
significantly associated with HCV-HCC (Fig. 6). GEPIA analysis of tumor vs. normal
tissue demonstrated that CCNB1, KIF20A and HMMR were significantly
overexpressed in HCC (Fig. 7A-C). In
addition, the analysis from The Human Protein Atlas indicated that
the expression of CCNB1, KIF20A and HMMR is enhanced in HCC tissue
(Fig. 7D-I) (14).
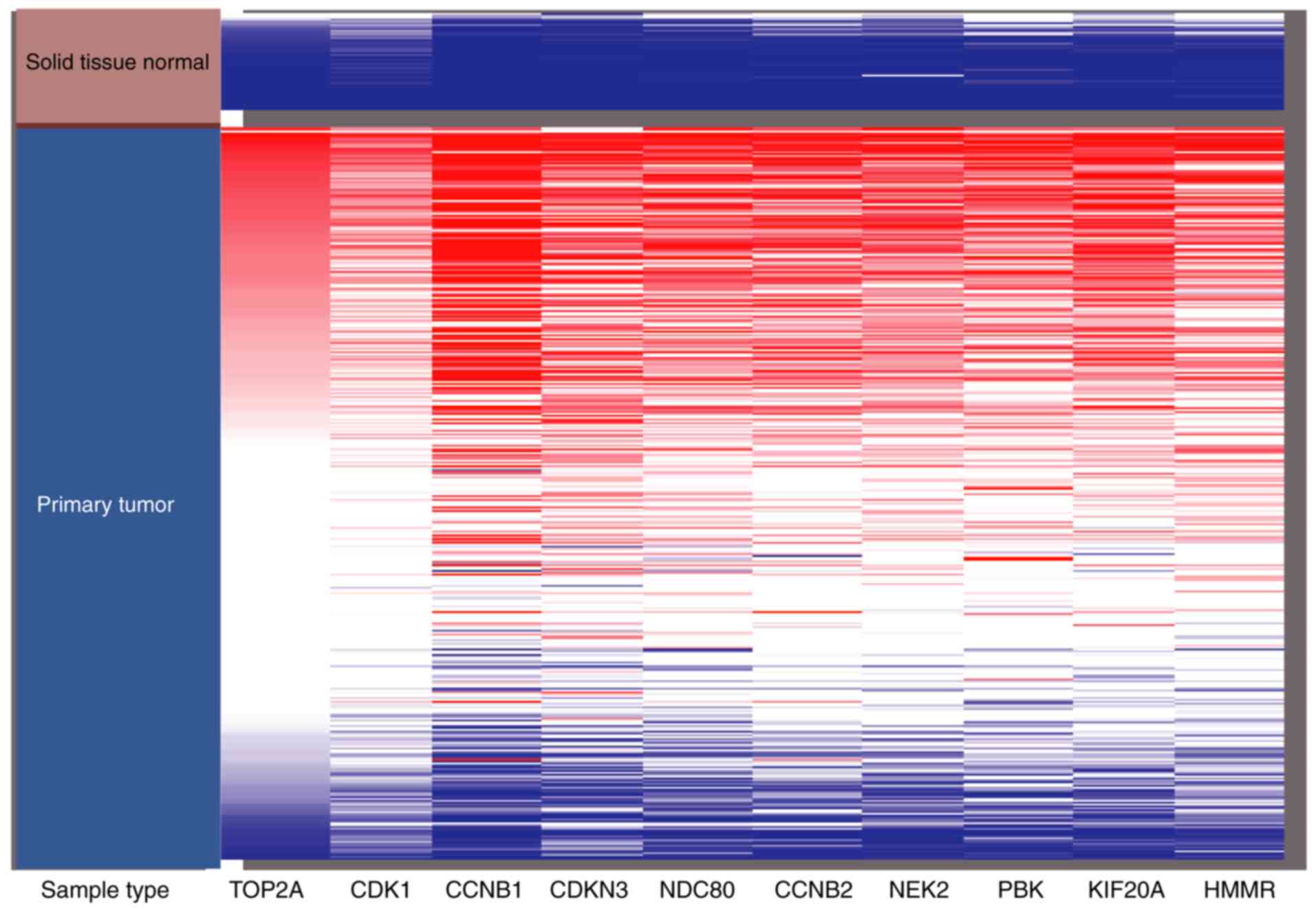 | Figure 3.Hierarchical clustering of the hub
genes was constructed using University of California Santa Cruz
genomics browser. Upregulation of genes is marked in red, whereas
downregulation of genes is marked in blue. TOP2A, DNA topoisomerase
2α; CDK1, cyclin dependent kinase 1; CCNB1, cyclin B1; CDKN3,
cyclin dependent kinase inhibitor 3; NDC80, kinetochore complex
component; CCNB2, cyclin B2; NEK2, NIMA related kinase 2; PBK, PDZ
binding kinase; KIF20A, kinesin family member 20A; HMMR, hyaluronan
mediated motility receptor. |
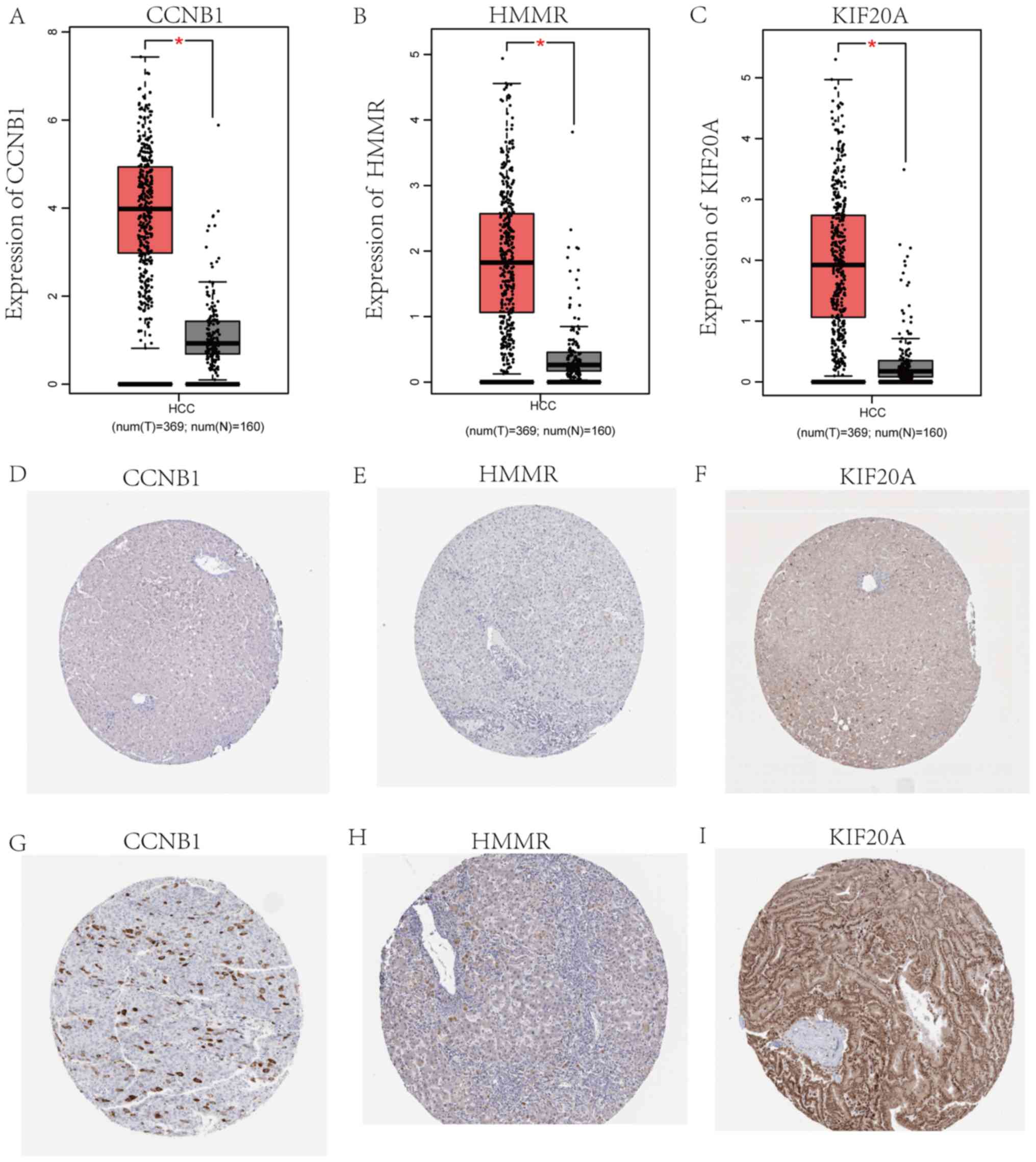 | Figure 7.mRNA and protein expression of CCNB1,
HMMR and KIF20A in HCV-HCC. mRNA expression of (A) CCNB1, (B) HMMR
and (C) KIF20A in HCV-HCC (red; n=369) compared with normal (gray;
n=160) tissues. Immunohistochemistry of the CCNB1, HMMR and KIF20A
based on the Human Protein Atlas (https://www.proteinatlas.org). (D) Protein expression
of CCNB1 in normal tissue (not detected). (E) Protein expression of
HMMR in normal tissue (not detected). (F) Protein expression of
KIF20A in normal tissue (staining, low; intensity, weak; quantity,
>75%). (G) Protein expression of CCNB1 in tumor tissue
(staining, medium; intensity, strong; quantity, <25%). (H)
Protein expression of HMMR in tumor tissue (staining, medium;
intensity, strong; quantity, <25%). (I) Protein expression of
KIF20A in tumor tissue (staining, high; intensity, strong,
quantity, >75%). *P<0.001. CCNB1, cyclin B1; KIF20A, kinesin
family member 20A; HMMR, hyaluronan mediated motility receptor;
HCV-HCC, hepatitis C virus-induced hepatocellular carcinoma. |
Discussion
Persistent HCV infection has been considered to
significantly increase the risk of developing HCC (15). However, following a review of the
literature, few studies have attempted to describe the molecular
mechanism of HCV-HCC (16–18). Due to an absence of appropriate
diagnostic markers and targeted treatments, early diagnosis is
challenging, and high mortality rates are observed amongst,
patients with this condition (7).
Consequently, it is vital to explore definitive targets for
HCV-HCC.
In the present study, 10 hub genes were identified
among 368 DEGs in HCV-HCC. Most notably, overexpression of CCNB1,
KIF20A and HMMR were associated with poorer prognosis. CCNB1 is an
important modulator of cell cycle progression (19), and it has been reported that its
overexpression contributes to the regulation of invasive growth,
metastasis and apoptosis of tumor cells (20,21).
Overexpression of CCNB1 has also been demonstrated in a number of
types of tumors, for example breast (22), colorectal (23) and laryngeal cancer (24). Regarding HCC, numerous studies have
indicated that overexpressed CCNB1, an independent prognostic
factor, is associated with shorter recurrence-free survival time
and is a potential therapeutic target (25,26). In
the present study, CCNB1 was found to be significantly
overexpressed in HCV-HCC, and this was associated with a lower
disease-free survival rate. Analogous results were revealed for
KIF20A, a member of the kinesin family (27). Previous studies have characterized
KIF20A as a prognostic indicator for a number of cancer types,
including ovarian carcinoma, pancreatic cancer and nasopharyngeal
cancer (28–30). Some of these studies suggested that
overexpression of KIF20A could promote tumor cell proliferation and
invasion. Shi et al (31)
showed that KIF20A is important for proliferation of HCC cells. In
the present study, overexpression of KIF20A was significantly
associated with poor prognosis, and it was identified as a hub gene
in HCV-HCC. HMMR is a multifunctional oncogenic protein (32) and many studies have reported that it
is overexpressed in human leukemia types and breast cancer
(33–35). Tilghman et al (36) considered that HMMR is a candidate
therapeutic target in glioblastoma, and Wang et al (37) suggested that overexpression of HMMR
led to a poor clinical outcome. However, few studies have reported
on the role of HMMR in HCC (38),
whereas the present study suggests that the gene is overexpressed
in patients with HCV-HCC, and that this overexpression is
significantly associated with the overall and disease-free
survival. Therefore, HMMR is a potential novel candidate biomarker
for HCV-HCC.
According to epidemiological studies, nearly 170
million people worldwide are carriers of HCV, and have a 17-fold
increased risk of developing HCC (39). HCV infection is the main cause of HCC
in Latin America, Europe and Japan (40). Therefore, performing survival
analyses and exploring the prognostic value of the 10 hub genes
identified from the present study in cohorts with various ethnic
backgrounds is important in future studies. The results of the
Oncomine analysis showed that overexpression of CCNB1, KIF20A and
HMMR mRNA were significantly associated with HCC progression,
suggesting these genes are important factors in the occurrence and
development of HCV-HCC. mRNA expression of CCNB1, KIF20A and HMMR
from the GEPIA database was analyzed, indicating that they were all
overexpressed in HCV-HCC. However, the difference between the
numbers of HCC and normal tissue samples in the GEPIA is a limiting
factor in the present study. Finally, the Human Protein Atlas
database, providing information on the expression of 24,000 human
proteins in cells and tissues, was utilized to explore the protein
expression of CCNB1, KIF20A and HMMR in HCC tissues, demonstrating
significant upregulation of all of them.
In conclusion, in the present study, 368 DEGs and 10
hub gens were identified and are potential diagnostic biomarkers
and therapeutic targets for HCV-HCC. Specifically, the findings
suggest CCNB1, KIF20A and HMMR as candidate targets for HCV-HCC
diagnosis and therapy. However, in vitro and in vivo
experiments are necessary for validation of these results.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The dataset of GSE62232, GSE69715 and GSE107170 are
available from the Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo). The dataset of
Wurmbach Liver and Mas Liver are available from Oncomine
(http://www.oncomine.com).
Authors' contributions
WLL, JL and ZZM designed the study. JL analyzed the
data and wrote the manuscript. ZWH, YML and LYW contributed to data
collection, analysis and figure preparation. ZWH wrote parts of the
manuscript and revised the manuscript. All authors read and
approved the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Zhu RX, Seto WK, Lai CL and Yuen MF:
Epidemiology of hepatocellular carcinoma in the Asia-Pacific
region. Gut Liver. 10:332–339. 2016. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hiotis SP, Rahbari NN, Villanueva GA,
Klegar E, Luan W, Wang Q and Yee HT: Hepatitis B vs. hepatitis C
infection on viral hepatitis-associated hepatocellular carcinoma.
BMC Gastroenterol. 12:642012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Liu LM, Xiong DD, Lin P, Yang H, Dang YW
and Chen G: DNA topoisomerase 1 and 2A function as oncogenes in
liver cancer and may be direct targets of nitidine chloride. Int J
Oncol. 53:1897–1912. 2018.PubMed/NCBI
|
|
4
|
Faillaci F, Marzi L, Critelli R, Milosa F,
Schepis F, Turola E, Andreani S, Vandelli G, Bernabucci V, Lei B,
et al: Liver Angiopoietin-2 is a key predictor of de novo or
recurrent hepatocellular cancer after hepatitis C virus direct
acting antivirals. Hepatology. 68:1010–1024. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Jin R, Lin H, Li G, Xu J, Shi L, Chang C
and Cai X: TR4 nuclear receptor suppresses HCC cell
invasion via downregulating the EphA2 expression. Cell Death Dis.
9:2832018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Daveau M, Scotte M, François A, Coulouarn
C, Ros G, Tallet Y, Hiron M, Hellot MF and Salier JP: Hepatocyte
growth factor, transforming growth factor alpha, and their
receptors as combined markers of prognosis in hepatocellular
carcinoma. Mol Carcinog. 36:130–141. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Song PP, Xia JF, Inagaki Y, Hasegawa K,
Sakamoto Y, Kokudo N and Tang W: Controversies regarding and
perspectives on clinical utility of biomarkers in hepatocellular
carcinoma. World J Gastroenterol. 22:262–274. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Schulze K, Imbeaud S, Letouzé E,
Alexandrov LB, Calderaro J, Rebouissou S, Couchy G, Meiller C,
Shinde J, Soysouvanh F, et al: Exome sequencing of hepatocellular
carcinomas identifies new mutational signatures and potential
therapeutic targets. Nat Genet. 47:505–511. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sekhar V, Pollicino T, Diaz G, Engle RE,
Alayli F, Melis M, Kabat J, Tice A, Pomerenke A, Altan-Bonnet N, et
al: Infection with hepatitis C virus depends on TACSTD2, a
regulator of claudin-1 and occludin highly downregulated in
hepatocellular carcinoma. PLoS Pathog. 14:e10069162018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Diaz G, Engle RE, Tice A, Melis M,
Montenegro S, Rodriguez-Canales J, Hanson J, Emmert-Buck MR, Bock
KW, Moore IN, et al: Molecular signature and mechanisms of
hepatitis D virus-associated hepatocellular carcinoma. Mol Cancer
Res. 16:1406–1419. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wurmbach E, Chen YB, Khitrov G, Zhang W,
Roayaie S, Schwartz M, Fiel I, Thung S, Mazzaferro V, Bruix J, et
al: Genome-wide molecular profiles of HCV-induced dysplasia and
hepatocellular carcinoma. Hepatology. 45:938–947. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Mas VR, Maluf DG, Archer KJ, Yanek K, Kong
X, Kulik L, Freise CE, Olthoff KM, Ghobrial RM, McIver P and Fisher
R: Genes involved in viral carcinogenesis and tumor initiation in
hepatitis C virus-induced hepatocellular carcinoma. Mol Med.
15:85–94. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res. 45:W98–W102.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Uhlén M, Fagerberg L, Hallström BM,
Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C,
Sjöstedt E, Asplund A, et al: Proteomics. Tissue-based map of the
human proteome. Science. 347:12604192015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Saito I, Miyamura T, Ohbayashi A, Harada
H, Katayama T, Kikuchi S, Watanabe Y, Koi S, Onji M, Ohta Y, et al:
Hepatitis C virus infection is associated with the development of
hepatocellular carcinoma. Proc Natl Acad Sci USA. 87:6547–6599.
1999. View Article : Google Scholar
|
|
16
|
McGivern DR and Lemon SM: Virus-specific
mechanisms of carcinogenesis in hepatitis C virus associated liver
cancer. Oncogene. 30:1969–1983. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lupberge J, Croonenborghs T, Roca Suarez
AA, Van Renne N, Jühling F, Oudot MA, Virzì A, Bandiera S, Jamey C,
Meszaros G, et al: Combined analysis of metabolomes, proteomes and
transcriptomes of HCV-infected cells and liver to identify pathways
associated with disease development. Gastroenterology.
pii:S0016-S5085(19)35670-7. 2019.
|
|
18
|
Wu SY, Lan SH and Liu HS: Degradative
autophagy selectively regulates CCND1 (cyclin D1) and MIR224, two
oncogenic factors involved in hepatocellular carcinoma
tumorigenesis. Autophagy. 15:729–730. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sartor H, Ehlert F, Grzeschik KH, Müller R
and Adolph S: Assignment of two human cell cycle genes, CDC25C and
CCNB1, to 5q31 and 5q12, respectively. Genomics. 13:911–912. 1992.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Matthess Y, Raab M, Sanhaji M, Lavrik IN
and Strebhardt K: Cdk1/cyclin B1 controls Fas-mediated apoptosis by
regulating caspase-8 activity. Mol Cell Biol. 30:5726–5740. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Song Y, Zhao C, Dong L, Fu M, Xue L, Huang
Z, Tong T, Zhou Z, Chen A, Yang Z, et al: Overexpression of cyclin
B1 in human esophageal squamous cell carcinoma cells induces tumor
cell invasive growth and metastasis. Carcinogenesis. 29:307–315.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Agarwal R, Gonzalez-Angulo AM, Myhre S,
Carey M, Lee JS, Overgaard J, Alsner J, Stemke-Hale K, Lluch A,
Neve RM, et al: Integrative analysis of cyclin protein levels
identifies cyclin b1 as a classifier and predictor of outcomes in
breast cancer. Clin Cancer Res. 15:3654–3662. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Li JQ, Kubo A, Wu F, Usuki H, Fujita J,
Bandoh S, Masaki T, Saoo K, Takeuchi H, Kobayashi S, et al: Cyclin
B1, unlike cyclin G1, increases significantly during colorectal
carcinogenesis and during later metastasis to lymph nodes. Int J
Oncol. 22:1101–1110. 2003.PubMed/NCBI
|
|
24
|
Dong Y, Sui L, Watanabe Y, Sugimoto K and
Tokuda M: Clinical relevance of cyclin B1 overexpression in
laryngeal squamous cell carcinoma. Cancer Lett. 177:13–19. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Weng L, Du J, Zhou Q, Cheng B, Li J, Zhang
D and Ling C: Identification of cyclin B1 and Sec62 as biomarkers
for recurrence in patients with HBV-related hepatocellular
carcinoma after surgical resection. Mol Cancer. 11:392012.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li L, Lei Q, Zhang S, Kong L and Qin B:
Screening and identification of key biomarkers in hepatocellular
carcinoma: Evidence from bioinformatic analysis. Oncol Rep.
38:2607–2618. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Miki H, Setou M, Kaneshiro K and Hirokawa
N: All kinesin superfamily protein, KIF, genes in mouse and human.
Proc Natl Acad Sci USA. 98:7004–7011. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kawai Y, Shibata K, Sakata J, Suzuki S,
Utsumi F, Niimi K, Sekiya R, Senga T, Kikkawa F and Kajiyama H:
KIF20A expression as a prognostic indicator and its possible
involvement in the proliferation of ovarian clear-cell carcinoma
cells. Oncol Rep. 40:195–205. 2018.PubMed/NCBI
|
|
29
|
Taniuchi K, Furihata M and Saibara T:
KIF20A-mediated RNA granule transport system promotes the
invasiveness of pancreatic cancer cells. Neoplasia. 16:1082–1093.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Liu SL, Lin HX, Qiu F, Zhang WJ, Niu CH,
Wen W, Sun XQ, Ye LP, Wu XQ, Lin CY, et al: Overexpression of
kinesin family member 20A correlates with disease progression and
poor prognosis in human nasopharyngeal cancer: A retrospective
analysis of 105 patients. PLoS One. 12:e01692802017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Shi C, Huang D, Lu N, Chen D, Zhang M, Yan
Y, Deng L, Lu Q, Lu H and Luo S: Aberrantly activated Gli2-KIF20A
axis is crucial for growth of hepatocellular carcinoma and predicts
poor prognosis. Oncotarget. 7:26206–26219. 2016.PubMed/NCBI
|
|
32
|
Fagerberg L, Hallström BM, Oksvold P,
Kampf C, Djureinovic D, Odeberg J, Habuka M, Tahmasebpoor S,
Danielsson A, Edlund K, et al: Analysis of the human
tissue-specific expression by genome-wide integration of
transcriptomics and antibody-based proteomics. Mol Cell Proteomics.
13:397–406. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Maxwell CA, McCarthy J and Turley E:
Cell-surface and mitotic-spindle RHAMM: Moonlighting or dual
oncogenic functions? J Cell Sci. 121:925–932. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Telmer PG, Tolg C, McCarthy JB and Turley
EA: How does a protein with dual mitotic spindle and extracellular
matrix receptor functions affect tumor susceptibility and
progression? Commun Integr Biol. 4:182–185. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Turley EA, Noble PW and Bourguignon LY:
Signaling properties of hyaluronan receptors. J Biol Chem.
277:4589–4592. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Tilghman J, Wu H, Sang Y, Shi X,
Guerrero-Cazares H, Quinones-Hinojosa A, Eberhart CG, Laterra J and
Ying M: HMMR maintains the stemness and tumorigenicity of
glioblastoma stem-like cells. Cancer Res. 74:3168–3179. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wang C, Thor AD, Moore DH II, Zhao Y,
Kerschmann R, Stern R, Watson PH and Turley EA: The overexpression
of RHAMM, a hyaluronan-binding protein that regulates ras
signaling, correlates with overexpression of mitogen-activated
protein kinase and is a significant parameter in breast cancer
progression. Clin Cancer Res. 4:567–576. 1998.PubMed/NCBI
|
|
38
|
He X, Liao W, Li Y, Wang Y, Chen Q, Jin J
and He S: Upregulation of hyaluronan-mediated motility receptor in
hepatocellular carcinoma predicts poor survival. Oncol Lett.
10:3639–3646. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Ferenci P, Fried M, Labrecque D, Bruix J,
Sherman M, Omata M, Heathcote J, Piratsivuth T, Kew M, Otegbayo JA,
et al: World gastroenterology organisation guideline.
Hepatocellular carcinoma (HCC): A global perspective. J
Gastrointestin Liver Dis. 19:311–317. 2010.PubMed/NCBI
|
|
40
|
Gurtsevitch VE: Human oncogenic viruses:
Hepatitis B and hepatitis C viruses and their role in
hepatocarcinogenesis. Biochemistry (Mosc). 73:504–513. 2008.
View Article : Google Scholar : PubMed/NCBI
|