Introduction
Cervical cancer is one of the leading causes of
cancer-associated mortality among females worldwide (1,2). Despite
the existence of a safe vaccine against human papillomavirus (HPV)
and an early screening test for cervical cancer, the frequency of
females undergoing the test is not sufficient (3,4).
Furthermore, an insufficient number of HPV vaccines are available
in countries that exhibit a high prevalence rate of cervical cancer
and the worldwide incidence rate of cervical cancer has not
significantly decreased (5,6). Therefore, there is a requirement to
explore the underlying molecular mechanisms of cervical cancer.
Estrogen receptor 1 (ESR1) is a transcription factor
that can be activated by estrogen and other growth factors in a
ligand-dependent manner; activated ESR1 dimerizes and regulates the
transcription of numerous target genes (7,8).
Previous comprehensive genomic studies have revealed frequent ESR1
mutations in metastatic types of breast cancer (9,10).
Subsequent studies also confirmed these observations (11–13) and
demonstrated that ESR1-mutated samples often exhibit increased
resistance to aromatase inhibitors (14). In addition, ESR1 mutations have been
identified in endometrial (15) and
colorectal cancer (16) at low
frequencies. However, to the best of our knowledge, it remains
unknown whether ESR1 mutations exist in other types of cancer,
including cervical cancer.
Cervical cancer shares certain genetic aberrations
with breast cancer, including frequent mutations in the PIK3CA,
TP53, PTEN and ARID1A genes (17–21), and
the development of cervical and breast cancer can be affected by
estrogen action (22–25). These similarities suggest that
cervical cancer may also harbor ESR1 mutations. To test this
hypothesis, a total of 260 samples from Chinese patients with
distinct subtypes of cervical cancer were investigated for the
presence of ESR1 mutations. A total of three heterozygous missense
ESR1 mutations were identified in 207 samples of cervical squamous
cell carcinoma (3/207, 1.4%), whereas no mutations were detected in
the 27 adenosquamous carcinoma or 26 adenocarcinomas samples. The
identified ESR1 mutations could have predictive values and may
provide insights into the diagnosis and molecular therapy of
cervical cancer.
Patients and methods
Formalin-fixed, paraffin-embedded
(FFPE) samples
The sample cohort has been previously described
(26). Briefly, a total of 260 FFPE
cancerous and paired adjacent non-cancerous tissue sections (10
µm), including squamous cell carcinoma (n=207), adenosquamous
carcinoma (n=27) and adenocarcinoma (n=26) tissues, were fixed in
10% neutral buffered formalin for 36 h at room temperature, and
collected from the archives of the Department of Pathology at the
Jiangxi Provincial Maternal and Child Health Hospital (Nanchang,
China) between July 2008 and August 2013. The median age of
patients was 43 years old (range, 22–74 years). The present study
was approved by the Institutional Review Board of Jiangxi
Provincial Maternal and Child Health Hospital (Nanchang, China),
and performed according to the Declaration of Helsinki. All
patients provided written informed consent prior to the study.
Mutation analysis
Genomic DNA was isolated using QIAamp DNA FFPE
Tissue kit (Qiagen GmbH, Hilden, Germany). The entire coding exons
and the corresponding intron/exon boundaries of the ESR1 gene were
amplified with a set of primer pairs (Table I). The polymerase chain reaction
(PCR) amplification was performed in a 50 µl reaction volume
containing 0.2 µM deoxyribonucleotide triphosphate, 5 µl 10X PCR
buffer, 0.5 U rTaq DNA Polymerase (Takara Biotechnology Co., Ltd.,
Dalian, China) and 200 ng genomic DNA. PCR amplifications were
performed in a Bio-Rad iCycler thermal cycler (Bio-Rad
Laboratories, Inc., Hercules, CA, USA) with the following
conditions: 94°C for 3 min, 35 cycles of 94°C for 30 sec, 50–58°C
for 30 sec and 72°C for 30 sec, and a final extension step at 72°C
for 10 min. The amplicons were sequenced bidirectionally on an ABI
3730 Genetic Analyzer (Thermo Fisher Scientific, Inc., Waltham, MA,
USA) and the sequencing data was aligned against the corresponding
genomic sequence (ESR1, NM_000125) in the National Center for
Biotechnology Information (NCBI) database (www.ncbi.nlm.nih.gov). The identified somatic
mutations were confirmed by sequencing the paired, adjacent
non-cancerous tissues.
 | Table I.Primer sequences used for polymerase
chain reaction amplification of the estrogen receptor 1 gene. |
Table I.
Primer sequences used for polymerase
chain reaction amplification of the estrogen receptor 1 gene.
| Exon | Forward primer
(5′-3′) | Reverse primer
(5′-3′) | Annealing
temperature (°C) | Amplicon length
(bp) |
|---|
| 1–1 |
GAGCCTTCTGCCCTGCGG |
GGTCTGACCGTAGACCTG | 53 | 270 |
| 1–2 |
CCGCGGCCGCCGCCAACG |
GGCGCGGGCGCGGGTAC | 50 | 279 |
| 2 |
TAATGTTAATGGATTTAC |
TTCAACACACTATTACCT | 56 | 242 |
| 3 |
TAGATTCTGACTGGCTAA |
CTGGGAGAGATGTACCTA | 52 | 197 |
| 4–1 |
TGTATAAAAGTTTACACG |
GCACTGACCATCTGGTCG | 52 | 253 |
| 4–2 |
TGGCCTTGTCCCTGACG |
GTTCTTGAAAAGCTATTG | 58 | 228 |
| 5 |
TCATTTGAGTCAGCAGG |
GCTACAGCCAGGTCACTTA | 58 | 197 |
| 6 |
TCATGTCTTGTGGAAGA |
ATCTTGTGTTATCAACTC | 53 | 226 |
| 7 |
TCTCACTCTCTCTCTGC |
GTAGGAAGCCCACAGAT | 55 | 233 |
| 8 |
TGTCTTCCCACCTACAG |
GGAGCTCTCAGACCGTGG | 57 | 259 |
In silico analysis of the ESR1
mutations
Two online prediction programs, MutationTaster
(http://www.mutationtaster.org) (27) and Polymorphism Phenotyping v2
(PolyPhen-2; http://genetics.bwh.harvard.edu/pph2) (28), were used to predict the associations
between the identified ESR1 mutations and disease occurrence. These
programs assessed the identified ESR1 mutations as either ‘benign’
or ‘pathogenic’, according to the automatically predicted
score.
Evolutionary conservation
analysis
The evolutionary conservation of the mutated amino
acids of ESR1 was analyzed in a total of 18 vertebrate species
retrieved from the NCBI database, including Homo sapiens
(NP_000116), Pan troglodytes (XP_009450519), Mus
musculus (NP_001289460), Rattus norvegicus (NP_036821),
Ovis aries (NP_000116), Bos taurus (NP_001001443),
Gallus gallus (NP_990514), Sus scrofa (NP_999385),
Canis lupus familiaris (NP_001273887), Equus caballus
(NP_001075241), Tupaia chinensis (NP_001304001), Mustela
putorius furo (XP_004753629), Oryctolagus cuniculus
(XP_008261925), Pongo abelii (XP_002817538), Coturnix
japonica (NP_001310118), Alligator sinensis
(XP_014375965), Ceratotherium simum simum (NP_001266182) and
Xenopus tropicalis (NP_988866). The Molecular Evolutionary
Genetics Analysis 4.0 software (29)
was used for multiple sequence alignment.
Protein structure modeling
DeepView Swiss-PdbViewer 4.0 software (30) was used to predict the potential
protein structural changes for the identified ESR1 mutations. An
available 3D protein structure of human ESR1 (protein data bank
code, 2OCF) (31) was retrieved from
the SWISS-MODEL repository in the ExPasy web interface (http://www.expasy.org).
Results
ESR1 mutations
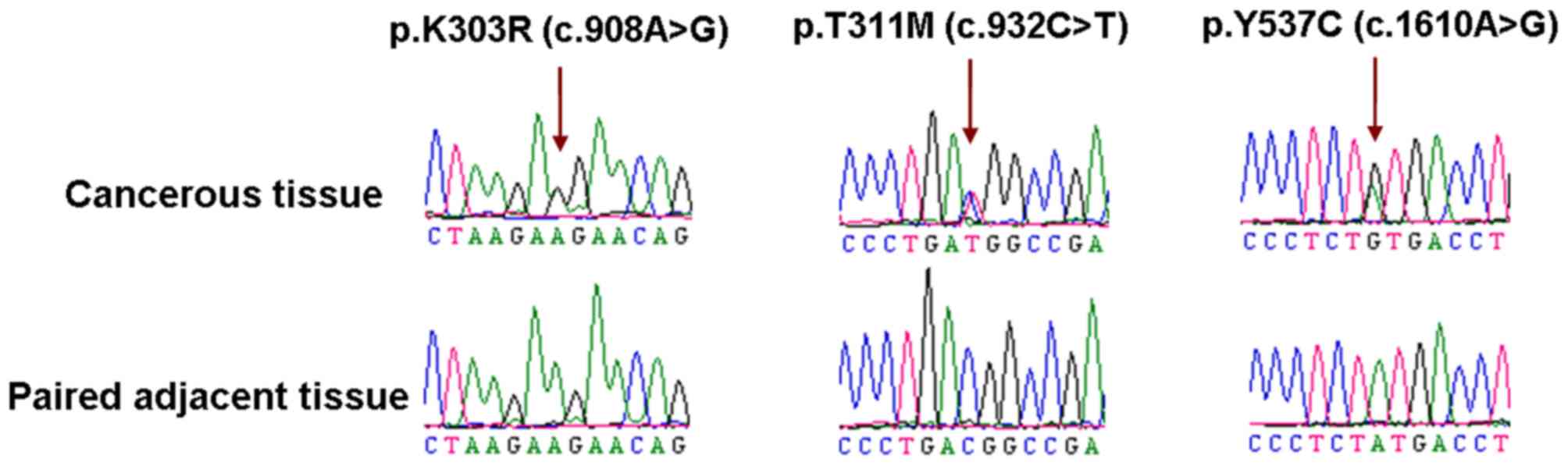
A total of three heterozygous missense ESR1
mutations, p.K303R (c.908A>G), p.T311M (c.932C>T) and p.Y537C
(c.1610A>G), were identified from 207 cervical squamous cell
carcinoma samples (3/207, 1.4%), while no mutations were detected
in the adenosquamous carcinoma and adenocarcinoma samples. The
mutations were absent in the paired non-cancerous tissues and were
therefore considered to be somatic (Fig.
1). The K303R and T311M mutations are located in the
‘hingeregion’ and the Y537C mutation is located in the
‘ligand-binding domain’ (9,10). Of the three individuals with ESR1
mutations, two were further diagnosed with uterine fibroid and one
with ovarian endometriosis.
In silico analysis of the ESR1
mutations
Two publicly available bioinformatics programs,
MutationTaster and PolyPhen-2, were used to predict the potential
functional significance of the ESR1 mutations. The predictions by
MutationTaster for the three ESR1 mutations (p.K303R, p.T311M and
p.Y537C) were ‘disease causing’ and ‘protein features (might be)
affected’, while PolyPhen-2 predicted these mutations to be
‘probably damaging’ (p.T311M and p.Y537C) or ‘possibly damaging’
(p.K303R), with a prediction score of >0.90. Furthermore, the
Y537C (c.1610A>G) and K303R (c.908A>G) mutations were not
identified in the 1,000Genomes (https://www.ncbi.nlm.nih.gov/variation/tools/1000genomes/)
(32) or the Exome Aggregation
Consortium (EXAC; http://exac.broadinstitute.org/) (33) databases, while the p.T311M
(c.932C>T) mutation was identified in the general population
with an extremely low frequency (1/121,362) in the EXAC
database.
Evolutionary conservation analysis and
protein structural modeling
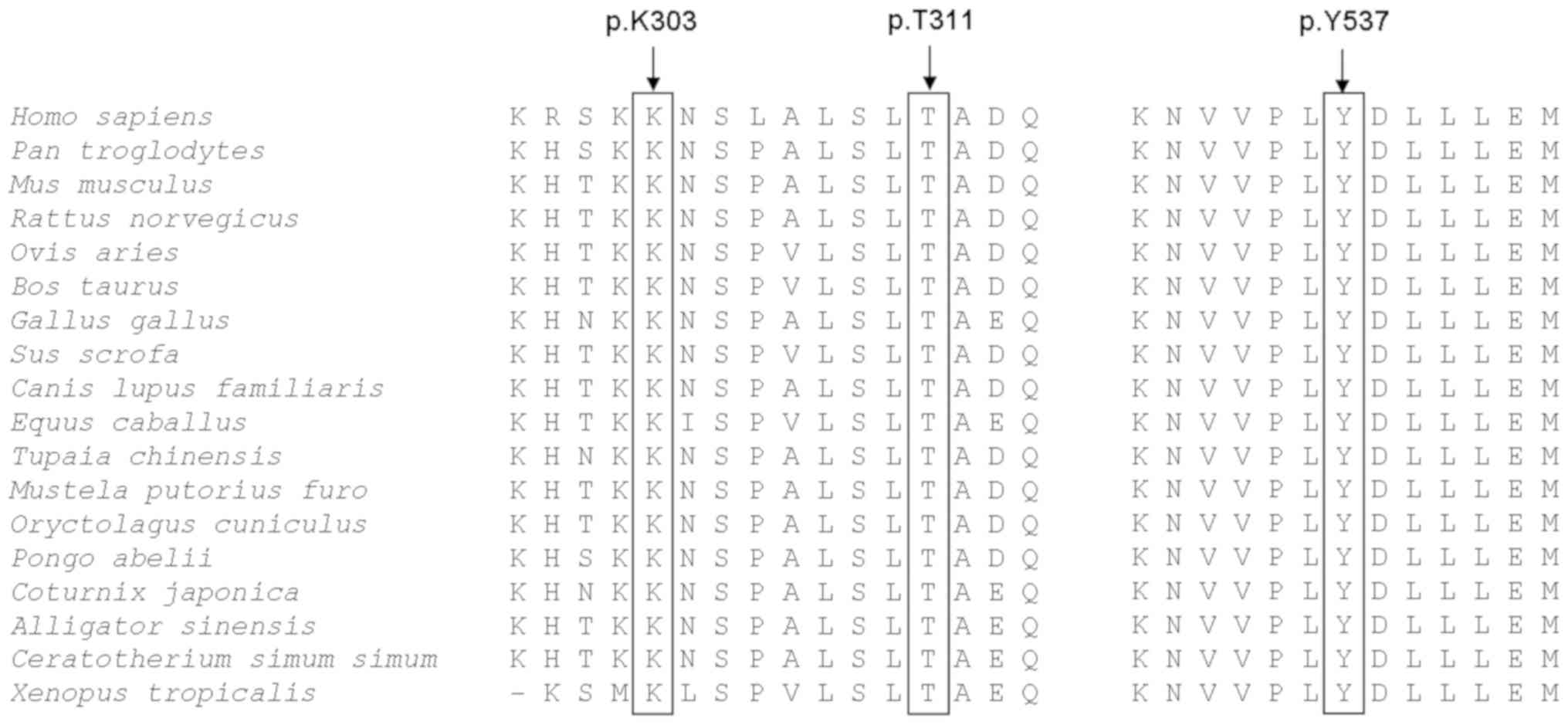
The results of evolutionary conservation analysis
demonstrated that the three ESR1 mutations were associated with
highly conserved amino acid changes among 18 vertebrate species,
ranging from Homo sapiens to Xenopus tropicalis
(Fig. 2). The protein structural
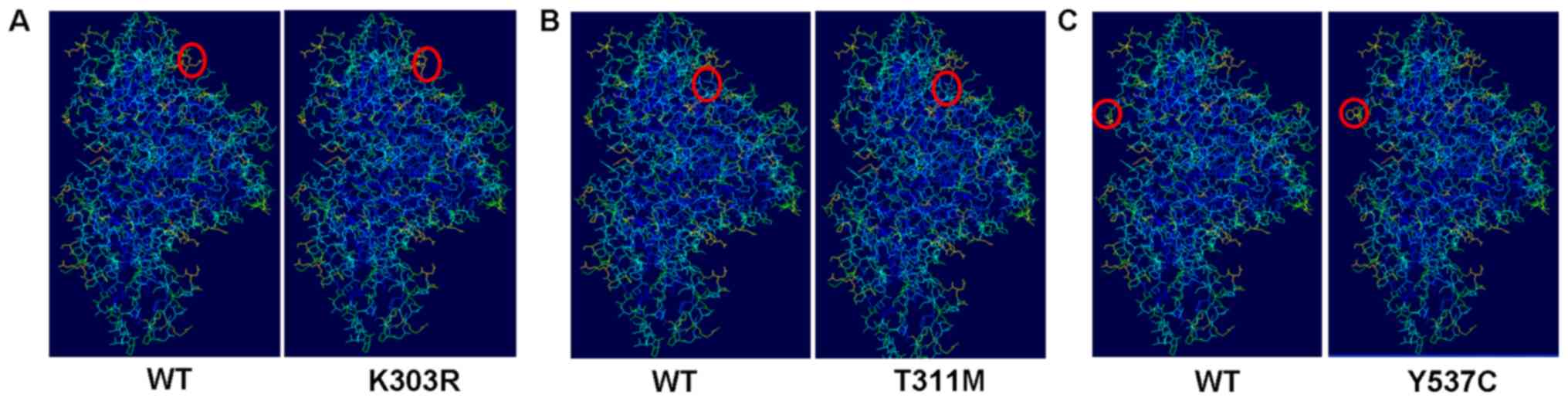
prediction results suggested that the three ESR1 mutations may
induce the structural changes in the side chain of ESR1 protein
(Fig. 3); results that were
consistent with the prediction results by MutationTaster.
Discussion
Previous studies have identified prevalent ESR1
mutations in breast cancer (9,10);
however, it remains largely unknown whether ESR1 mutations exist in
other types of cancer, including cervical cancer.
In the present study, a total of 260 samples from
Chinese patients with distinct subtypes of cervical cancer were
tested for the presence of ESR1 mutations. In total, three missense
somatic mutations in ESR1, p.K303R (c.908A>G), p.T311M
(c.932C>T) and p.Y537C (c.1610A>G), were identified from 207
squamous cell carcinomas (3/207, 1.4%), which were not present in
other cervical cancer subtypes. Evolutionary conservation analysis
demonstrated that the three ESR1 mutations were associated with
highly conserved amino acid, which may describe the potential to
cause protein structural changes. In silico analysis
suggested that these mutations may be pathogenic. Furthermore, the
three ESR1 mutations have previously been identified in other types
of cancer. The p.K303R (c.908A>G) mutation has been observed in
206/6,556 samples of breast cancer (24–37). The
p.T311M (c.932C>T) mutation has been identified in 2/2,218
colorectal cancer samples (38) and
1/2,105 liver cancer samples (38).
In addition, the p.Y537C (c.1610A>G) mutation has been detected
in 10/6,556 breast cancer samples (11,39–41).
Similarly, a previous comprehensive study identified ESR1 somatic
mutations p.K206R (c.617A>G) and p.L372L (c.1116C>G) in 2/306
(0.7%) cervical squamous cell carcinoma samples (http://cancer.sanger.ac.uk/cosmic). However, a
previous genomic analysis of cervical cancer failed to detect any
ESR1 mutations in 79 squamous cell carcinoma samples (17). Considering the low frequency of ESR1
mutation in cervical squamous cell carcinoma, it is suggested that
the small sample size analyzed in this previous study may have
caused the inconsistency in the results (17). In combination, both previous studies
and the present study suggest that ESR1 mutations may participate
in the carcinogenesis of cervical cancer, albeit at a low
frequency.
A number of previous functional assays for the
identified ESR1 mutations, including p.K303R (42) and p.Y537C (9,43),
demonstrated that these mutations are associated with acquired
endocrine resistance in hormonal therapy in breast cancer (7,8,40,41).
Therefore, it is proposed that the ESR1 mutations identified in
cervical cancer in the present study may further cause acquired
endocrine resistance to hormonal therapy in breast cancer.
Antihormonal agents have recently been used to
improve effects of chemo- and radiotherapy in cervical cancer
(44). However, due to the potential
acquired endocrine resistance in cervical cancer samples with ESR1
mutations, antihormonal agents should be used with caution during
chemo-and radiotherapy.
The three ESR1 mutations were not detected in the 27
adenosquamous carcinoma or the 26 adenocarcinoma samples of the
present study, which is consistent with a prior genomic analysis of
cervical cancer with distinct subtypes, in which no ESR1 mutations
were detected in either 24 adenocarcinoma or 7 adenosquamous
carcinoma samples (17). In summary,
these results suggest that the ESR1 mutations may not be positively
involved in the pathogenesis of cervical adenocarcinoma and
adenosquamous carcinoma. However, the absence of ESR1 mutations in
patients with adenosquamous carcinoma and adenocarcinoma may be due
to the small sample sizes analyzed in the present study.
In conclusion, the current study identified three
potentially pathogenic ESR1 mutations in cervical squamous cell
carcinoma samples from Chinese patients, which were not observed in
other subtypes. These results, together with numerous previous
studies, suggested that ESR1 mutations may be involved in the
carcinogenesis of squamous cell carcinoma, but not in other
subtypes of cervical cancer.
Acknowledgements
Not applicable.
Funding
The present study was supported by grants from the
Natural Science Foundation of China (grant no. 81160079) and the
Natural Science Foundation of Jiangxi Province (grant nos.
20143ACG70016 and 20161ACB21021).
Availability of data and materials
All the data generated or analyzed during the
present study are available from the corresponding author on
reasonable request.
Authors' contributions
XMY, HH, LXX and YTC performed the experiments. ZMW
and LQW analyzed the data. XYC and JL collected samples and
clinical data. XMY prepared the manuscript. OPH designed the study
and revised the manuscript.
Ethics approval and consent to
participate
The experimental protocol was established according
to the ethical guidelines of the Helsinki Declaration and was
approved by the Human Ethics Committee of Nanchang University.
Written informed consent was obtained from all participants or
their guardian.
Patients consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2018. CA Cancer J Clin. 68:7–30. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jemal A, Simard EP, Dorell C, Noone AM,
Markowitz LE, Kohler B, Eheman C, Saraiya M, Bandi P, Saslow D, et
al: Annual report to the nation on the status of cancer, 1975–2009,
featuring the burden and trends in human papillomavirus
(HPV)-associated cancers and HPV vaccination coverage levels. J
Natl Cancer Inst. 105:175–201. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Pedersen K, Burger EA, Nygård M,
Kristiansen IS and Kim JJ: Adapting cervical cancer screening for
women vaccinated against human papillomavirus infections: The value
of stratifying guidelines. Eur J Cancer. 91:68–75. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Philp L, Jembere N, Wang L, Gao J, Maguire
B and Kupets R: Pap tests in the diagnosis of cervical cancer: Help
or hinder? Gynecol Oncol. 150:61–66. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bogani G, Leone Roberti Maggiore U,
Signorelli M, Martinelli F, Ditto A, Sabatucci I, Mosca L, Lorusso
D and Raspagliesi F: The role of human papillomavirus vaccines in
cervical cancer: Prevention and treatment. Crit Rev Oncol Hematol.
122:92–97. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lytwyn A, Elit L and Sellors JW: Human
papillomavirus DNA versus papanicolaou screening tests for cervical
cancer. N Engl J Med. 358:6412008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yaşar P, Ayaz G, User SD, Güpür G and
Muyan M: Molecular mechanism of estrogen-estrogen receptor
signaling. Reprod Med Biol. 16:4–20. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bado I, Gugala Z, Fuqua SAW and Zhang XH:
Estrogen receptors in breast and bone: From virtue of remodeling to
vileness of metastasis. Oncogene. 36:4527–4537. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Robinson DR, Wu YM, Vats P, Su F, Lonigro
RJ, Cao X, Kalyana-Sundaram S, Wang R, Ning Y, Hodges L, et al:
Activating ESR1 mutations in hormone-resistant metastatic breast
cancer. Nat Genet. 45:1446–1451. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Toy W, Shen Y, Won H, Green B, Sakr RA,
Will M, Li Z, Gala K, Fanning S, King TA, et al: ESR1
ligand-binding domain mutations in hormone-resistant breast cancer.
Nat Genet. 45:1439–1445. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bartels S, Christgen M, Luft A, Persing S,
Jödecke K, Lehmann U and Kreipe H: Estrogen receptor (ESR1)
mutation in bone metastases from breast cancer. Mod Pathol.
31:56–61. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yanagawa T, Kagara N, Miyake T, Tanei T,
Naoi Y, Shimoda M, Shimazu K, Kim SJ and Noguchi S: Detection of
ESR1 mutations in plasma and tumors from metastatic breast cancer
patients using next-generation sequencing. Breast Cancer Res Treat.
163:231–240. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lefebvre C, Bachelot T, Filleron T,
Pedrero M, Campone M, Soria JC, Massard C, Lévy C, Arnedos M,
Lacroix-Triki M, et al: Mutational profile of metastatic breast
cancers: A retrospective analysis. PLoS Med. 13:e10022012016.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Clarke R, Tyson JJ and Dixon JM: Endocrine
resistance in breast cancer-An overview and update. Mol Cell
Endocrinol. 418:220–234. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Backes FJ, Walker CJ, Goodfellow PJ, Hade
EM, Agarwal G, Mutch D, Cohn DE and Suarez AA: Estrogen
receptor-alpha as a predictive biomarker in endometrioid
endometrial cancer. Gynecol Oncol. 141:312–317. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Cancer Genome Atlas Network, ; Muzny DM,
Bainbridge MN, Chang K, Dinh HH, Drummond JA, Fowler G, Kovar CL,
Lewis LR, Morgan MB, et al: Comprehensive molecular
characterization of human colon and rectal cancer. Nature.
487:330–337. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ojesina AI, Lichtenstein L, Freeman SS,
Pedamallu CS, Imaz-Rosshandler I, Pugh TJ, Cherniack AD, Ambrogio
L, Cibulskis K, Bertelsen B, et al: Landscape of genomic
alterations in cervical carcinomas. Nature. 506:371–375. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Muller E, Brault B, Holmes A, Legros A,
Jeannot E, Campitelli M, Rousselin A, Goardon N, Frébourg T,
Krieger S, et al: Genetic profiles of cervical tumors by
high-throughput sequencing for personalized medical care. Cancer
Med. 4:1484–1493. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sjöblom T, Jones S, Wood LD, Parsons DW,
Lin J, Barber TD, Mandelker D, Leary RJ, Ptak J, Silliman N, et al:
The consensus coding sequences of human breast and colorectal
cancers. Science. 314:268–274. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Banerji S, Cibulskis K, Rangel-Escareno C,
Brown KK, Carter SL, Frederick AM, Lawrence MS, Sivachenko AY,
Sougnez C, Zou L, et al: Sequence analysis of mutations and
translocations across breast cancer subtypes. Nature. 486:405–409.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Cancer Genome Atlas Research Network;
Albert Einstein College of Medicine; Analytical Biological
Services; Barretos Cancer Hospital; Baylor College of Medicine;
Beckman Research Institute of City of Hope; Buck Institute for
Research on Aging; Canada's Michael Smith Genome Sciences Centre;
Harvard Medical School; Helen F. Graham Cancer Center &
Research Institute at Christiana Care Health Services, ; et al
Integrated genomic and molecular characterization of cervical
cancer. Nature. 543:378–384. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
den Boon JA, Pyeon D, Wang SS, Horswill M,
Schiffman M, Sherman M, Zuna RE, Wang Z, Hewitt SM, Pearson R, et
al: Molecular transitions from papillomavirus infection to cervical
precancer and cancer: Role of stromal estrogen receptor signaling.
Proc Natl Acad Sci USA. 112:E3255–E3264. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Bronowicka-Kłys DE, Lianeri M and
Jagodziński PP: The role and impact of estrogens and xenoestrogen
on the development of cervical cancer. Biomed Pharmacother.
84:1945–1953. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Leung YK, Lee MT, Lam HM, Tarapore P and
Ho SM: Estrogen receptor-beta and breast cancer: Translating
biology into clinical practice. Steroids. 77:727–737. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Vazquez Rodriguez G, Abrahamsson A, Jensen
LD and Dabrosin C: Estradiol promotes breast cancer cell migration
via recruitment and activation of neutrophils. Cancer Immunol Res.
5:234–247. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zou Y, Liu FY, Wu J, Wan L, Fang SF, Zhang
ZY, Luo Y, Chen MH, Huang MZ, He M and Huang OP: Mutational
analysis of the RAS/RAF/MEK/ERK signaling pathway in 260 Han
Chinese patients with cervical carcinoma. Oncol Lett. 14:2427–2431.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Schwarz JM, Cooper DN, Schuelke M and
Seelow D: MutationTaster2: Mutation prediction for the
deep-sequencing age. Nat Methods. 11:361–362. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Adzhubei IA, Schmidt S, Peshkin L,
Ramensky VE, Gerasimova A, Bork P, Kondrashov AS and Sunyaev SR: A
method and server for predicting damaging missense mutations. Nat
Methods. 7:248–249. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Tamura K, Dudley J, Nei M and Kumar S:
MEGA4: Molecular evolutionary genetics analysis (MEGA) software
version 4.0. Mol Biol Evol. 24:1596–1599. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Johansson MU, Zoete V, Michielin O and
Guex N: Defining and searching for structural motifs using
DeepView/Swiss-PdbViewer. BMC Bioinformatics. 13:1732012.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Koide A, Abbatiello S, Rothgery L and
Koide S: Probing protein conformational changes in living cells by
using designer binding proteins: Application to the estrogen
receptor. Proc Natl Acad Sci USA. 99:1253–1258. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
1000 Genomes Project Consortium, ;
Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker
RE, Kang HM, Marth GT and McVean GA: An integrated map of genetic
variation from 1,092 human genomes. Nature. 491:56–65. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lek M, Karczewski KJ, Minikel EV, Samocha
KE, Banks E, Fennell T, O'Donnell-Luria AH, Ware JS, Hill AJ,
Cummings BB, et al: Analysis of protein-coding genetic variation in
60,706 humans. Nature. 536:285–291. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Fuqua SA, Wiltschke C, Zhang QX, Borg A,
Castles CG, Friedrichs WE, Hopp T, Hilsenbeck S, Mohsin S,
O'Connell P and Allred DC: A hypersensitive estrogen receptor-alpha
mutation in premalignant breast lesions. Cancer Res. 60:4026–4029.
2000.PubMed/NCBI
|
|
35
|
Conway K, Parrish E, Edmiston SN, Tolbert
D, Tse CK, Geradts J, Livasy CA, Singh H, Newman B and Millikan RC:
The estrogen receptor-alpha A908G (K303R) mutation occurs at a low
frequency in invasive breast tumors: Results from a
population-based study. Breast Cancer Res. 7:R871–R880. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Herynk MH, Parra I, Cui Y, Beyer A, Wu MF,
Hilsenbeck SG and Fuqua SA: Association between the estrogen
receptor alpha A908G mutation and outcomes in invasive breast
cancer. Clin Cancer Res. 13:3235–3243. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Abbasi S, Rasouli M, Nouri M and Kalbasi
S: Association of estrogen receptor-α A908G (K303R) mutation with
breast cancer risk. Int J Clin Exp Med. 6:39–49. 2013.PubMed/NCBI
|
|
38
|
Lim B, Mun J, Kim JH, Kim CW, Roh SA, Cho
DH, Kim YS, Kim SY and Kim JC: Genome-wide mutation profiles of
colorectal tumors and associated liver metastases at the exome and
transcriptome levels. Oncotarget. 6:22179–22190. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wang P, Bahreini A, Gyanchandani R, Lucas
PC, Hartmaier RJ, Watters RJ, Jonnalagadda AR, Trejo Bittar HE,
Berg A, Hamilton RL, et al: Sensitive detection of mono- and
polyclonal ESR1 mutations in primary tumors, metastatic lesions,
and cell-free DNA of breast cancer patients. Clin Cancer Res.
22:1130–1137. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Niu J, Andres G, Kramer K, Kundranda MN,
Alvarez RH, Klimant E, Parikh AR, Tan B, Staren ED and Markman M:
Incidence and clinical significance of ESR1 mutations in heavily
pretreated metastatic breast cancer patients. Onco Targets Ther.
8:3323–3328. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Takeshita T, Yamamoto Y, Yamamoto-Ibusuki
M, Inao T, Sueta A, Fujiwara S, Omoto Y and Iwase H: Droplet
digital polymerase chain reaction assay for screening of ESR1
mutations in 325 breast cancer specimens. Transl Res.
166:540–553.e2. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Cui Y, Zhang M, Pestell R, Curran EM,
Welshons WV and Fuqua SA: Phosphorylation of estrogen receptor
alpha blocks its acetylation and regulates estrogen sensitivity.
Cancer Res. 64:9199–9208. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Toy W, Weir H, Razavi P, Lawson M,
Goeppert AU, Mazzola AM, Smith A, Wilson J, Morrow C, Wong WL, et
al: Activating ESR1 mutations differentially affect the efficacy of
ER antagonists. Cancer Discov. 7:277–287. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Segovia-Mendoza M, Jurado R, Mir R, Medina
LA, Prado-Garcia H and Garcia-Lopez P: Antihormonal agents as a
strategy to improve the effect of chemo-radiation in cervical
cancer: In vitro and in vivo study. BMC Cancer. 15:212015.
View Article : Google Scholar : PubMed/NCBI
|