Introduction
Gastric cancer (GC) is a common cancer worldwide and
remains the third leading cause of cancer-related death, following
lung and liver cancer, accounting for 1.3 million cases of GC and
819,000 GC-associated cases of mortality in 2015 (1). The high incidence and mortality rates
of GC have been demonstrated throughout the developed world, and GC
adversely affects patients' health and quality of life (2). Although notable progress has been
achieved in treating patients with GC based on advances in
technology (3,4), the precise molecular mechanisms
underlying GC development and progression remain unknown, and
require further investigation.
MicroRNAs (miRNAs/miRs) can function as oncogenes or
tumor suppressor genes, and regulate gene expression through
translational repression or mRNA degradation by binding to the
3′-untranslated region (3′UTR) of their target mRNAs (5,6).
Previous studies have identified that miRNAs exhibit aberrant
expression and serve essential roles in GC (7–10).
Therefore, further investigation into the expression and function
of miRNAs may improve understanding of the molecular mechanisms
responsible for the pathogenesis of GC.
Previous studies have demonstrated that the
expression levels of miR-520a-3p are downregulated in non-small
cell lung cancer cells. Decreased levels of miR-520a-3p expression
were associated with poorer overall survival (11). Additionally, the upregulation of
miR-520a-3p significantly suppressed the proliferation, cell cycle
progression and metastatic activity by targeting mitogen-activated
protein kinase kinasekinase2, HOXD8 or PI3K/AKT/mTOR (12,13).
miR-520a-3p was also shown to inhibit proliferation and metastatic
activity in breast cancer through cyclinD1, and in colorectal
cancer via epidermal growth factor receptor (14,15). In
osteosarcoma, dexmedetomidine has been reported to upregulate
miR-520-3p, which directly targeted AKT1 to inhibit MG63 cell
proliferation and migration and promote apoptosis. In addition,
LINC01116 has been reported to target miR-520a-3p, which
affectedinterleukin-6 receptor, thereby promoting proliferation and
migration via the JAK-STAT signaling pathway (16,17).
However, the expression profile and biological significance of
miR-520a-3p in GC are unknown at present, to the best of our
knowledge.
The present study examined the expression of
miR-520a-3p in GC tissues and cells and demonstrated the functions
of miR-520a-3p overexpression in GC cell proliferation, invasion
and migration. Together, these data demonstrate that miR-520a-3p
may be a novel therapeutic target for the treatment of GC.
Materials and methods
Patients
GC specimens and adjacent tissues were received from
patients who were diagnosed with GC and underwent surgery at Ningbo
No. 2 Hospital (Zhejiang, China) between August 2016 and May 2017.
A total of 80 female patients, aged between 41–75 (55.3±11.4)
years, and had not received local or systemic therapy prior to
surgery at Ningbo No. 2 Hospital. The present study was approved by
The Research Ethics Committee of Ningbo No. 2 Hospital, and all
subjects provided the information written consent. The samples were
graded according to the 8th edition of the AJCC staging
classification system (18). The
relationship between miR-520a-3p levels and the clinical
characteristics of enrolled patients were analyzed.
Cell culture
All GC cell lines (SGC-7901, BGC-823, and MGC-803)
and a normal gastric epithelial cell line (GES-1) used in the
present study were obtained from the Institute of Biochemistry and
Cell Biology (Shanghai, China) and maintained in DMEM (HyClone; GE
Healthcare Life Sciences) medium (SGC-7901, BGC-823) or RPMI 1640
(HyClone; GE Healthcare Life Sciences) medium (MGC-803, GES-1)
containing 10% FBS (Shanghai Ex CellBiology, Inc.) at 37°C, in a
humidified incubator with 5% CO2 (Memmert GmbH).
Cell transfection
Cells (SGC-7901 and MGC803) were transfected with
miR-520a-3p mimics (100 pmol; forward, 5′-AAAGUGCUUCCCUUUGGACUGU-3′
and reverse, 5′-AGUCCAAAGGGAAGCACUUUUU-3′) or miR-520a-3p negative
control (NC; 100 pmol; forward, 5′-UUCUCCGAACGUGUCACGUTT-3′ and
reverse, 5′-ACGUGACACGUUCGGAGAATT-3′; both from Shanghai GenePharma
Co. Ltd.), or spindle and kinetochore associated 2 (SKA2) cDNA
plasmid (pLenti-SKA2-Puro) or NC (pLenti-C-Myc-DDK-P2A-Puro) (4 µg;
both from OriGene Technologies, Inc.) using Lipofectamine™ 2000
transfection reagent (Invitrogen; Thermo Fisher Scientific, Inc.)
following the manufacturer's protocol. Cells were incubated with
the transfection reagent for 24 h, following which RNA and protein
were extracted. No treatment or transfection was performed in the
MOCK group.
The SKA2 cDNA plasmid (without the 3′UTR) and
miR-520a-3p mimics were co-transfected into SGC-7901 cells, which
subsequently underwent western blot analyses, cell proliferation
assays, wound healing assay and cell invasion assays.
Western blotting
Total cell protein was extracted by 1XSDS
(Sigma-Aldrich; Merck KGaA) and quantified by a bicinchoninic acid
assay (Beyotime Institute of Biotechnology). Cellular proteins
(40–50 µg) were loaded onto a 12% polyacrylamide gel and resolved
by SDS-PAGE. After separation, the proteins were transferred onto
polyvinylidene difluoride membranes (EMD Millipore) for
immunoblotting. Membranes were blocked with 5% BSA (Beijing
Solarbio Science & Technology Co., Ltd.) for 2 h at room
temperature, the membranes were incubated with primary antibodies
overnight at 4°C. The primary antibodies used were Ki-67 (1:1,500;
Abcam; cat no. ab15580), matrix metalloproteinase (MMP)-2 (1:1,500;
Abcam; cat no. ab7033), MMP-9 (1:1,500; Abcam; cat no. ab137651),
SKA2 (1:1,500; Abcam; cat no. ab91551) and GAPDH (1:2,000; Cell
Signaling Technology, Inc.; cat. no. 5174). The membranes were
subsequently incubated for 1 h at room temperature with goat
anti-rabbit IgG-horseradish peroxidase (HRP) or goat anti-mouse
IgG-HRP (1:5,000; Wuhan Boster Biological Technology, Ltd.; cat.
nos. BA1054 and BA1050, respectively). The protein signals on the
membrane were detected using an ECL reagent (Absin Biotechnology
Co., Ltd.) and visualized using a chemiluminescence imaging system
(LI-COR Biosciences).
RT-qPCR
The expression levels of miR-520a-3p and SKA2 were
determined using RT-qPCR. Total RNA was extracted using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) according to the manufacturer's protocol. The RNA was reverse
transcribed to cDNA using a TaqMan reverse transcription kit
(Thermo Fisher Scientific, Inc.) with the following RT protocol; 15
min at 37°C and 5 sec at 85°C. cDNA was amplified using SYBR-Green
PCR Master mix (Roche Diagnostics) on a LightCycler 480 system. The
thermocycling conditions were as follows: 95°C for 10 min; and
followed by 45 cycles, 95°C for 10 sec and 60°C for 60 sec. Fold
changes were calculated by relative quantification
(2−∆∆Cq) using endogenous U6 and GAPDH as references for
miR-520a-3p and SKA2 expression, respectively (19). The primers used for miR-520a-3p were:
Forward, 5′-ACACTCCAGCTGGGAAAGTGCTTCCC-3′ and reverse,
5′-CTCAACTGGTGTCGTGGA-3′. For U6, the primers used were: Forward,
5′-CTCGCTTCGGCAGCACA-3′ and reverse, 5′-AACGCTTCACGAATTTGCGT-3′.
For SKA2, the primers used were: Forward,
5′-CTGAAACTATGCTAAGTGGGGGAG-3′ and reverse,
5′-TTCCAAACATCCTGACACTCAAAAG-3′. For GAPDH, the primers used were:
Forward, 5′-AAGCCTGCCGGTGACTAAC-3′ and reverse,
5′-GCATCACCCGGAGGAGAAAT-3′.
Cell proliferation assay
miR-520a-3p mimics, SKA2 cDNA and negative
control-transfected GC cells (SGC-7901 and MGC803) were seeded in
96-well plates (1×104 cells/well). A total of 20 µl Cell
Titer 96® AQueous One Solution (Promega Corporation) was
added to each well to determine cell viability at 4, 24, 48, 72 and
96 h after seeding. Cells were incubated at 37°C, in a humidified
incubator with 5% CO2 for 3 h. Subsequently the
absorbance of each well was measured at 490 nm on a
spectrophotometer (Beckman Coulter, Inc.).
Wound healing assay
Cells were cultured in 6-well plates
(1×105 cells/well) for 24 h. Once the cellular density
had reached ~100%, a vertical line was scraped in the culture using
a 1-ml pipette tip. The cells were rinsed with PBS and incubated
with fresh serum-free medium at 37°C. The widths of the gap at 0
and 24 h were compared to assess the distance of migration using
fluorescence microscopy.
Cell invasion assays
Cell invasion assays were performed using a Matrigel
invasion chamber (24-well plates; 8-µm pore size; Corning Inc.)
according to the manufacturer's protocol. A total of
5×104 cells were seeded in the upper chambers of the
wells in 100 µl FBS-free medium, and the lower chambers contained
DMEM supplemented with 20% FBS. The cells were incubated for 24 h
at 37°C, after which the cells on the filter surface were stained
with 0.1% crystal violet for 30 min at room temperature and images
were captured with an inverted fluorescence microscope (Nikon
Corporation), representative images were captured at ×100
magnification. The absorbance was measured at 590 nm.
Bioinformatics
Potential miR-520a-3p targets were predicted and
analyzed using the following two publicly available databases:
TargetScanv7.2 (http://www.targetscan.org), and miRanda (http://www.microrna.org/microrna/getGeneForm.do).
Luciferase assay
The predicted miR-520a-3p binding sites on the 3′UTR
of wild-type (WT) SKA2 (MirTarget-SKA2-3U-WT), together with a
corresponding mutant (mut) miR-520a-3p binding sites on the 3′UTR
of SAK2 (MirTarget-SKA2-3U-Mut), were synthesized and transfected
into the pGL3 vector (Promega Corporation). SGC-7901 cells were
seeded in a 24-well plates (5×104 cells/well), the WT or
Mut 3′UTR vectors, and miR-520a-3p mimics or NC were co-transfected
using Lipofectamine™ 2000. The relative luciferase activity was
measured after 48 h using a Dual-Luciferase Reporter assay system
according to the manufacturer's protocols (Promega Corporation).
Luciferase activity was normalized to Renilla luciferase
activity.
Statistical analysis
All experiments were repeated three times and data
are presented as the mean ± standard deviation. One-way ANOVA and
Fisher's least significant difference tests were used to calculate
P-values between multiple groups. Data in Table I were analyzed using χ2
tests of four-fold table. Statistical analyses were performed using
SPSS 15.0 software (SPSS, Inc.). P<0.05 was considered to
indicate a statistically significant difference.
 | Table I.Association between miR-520a-3p
expression and clinicopathological factors in 80 primary gastric
cancer tissues. |
Table I.
Association between miR-520a-3p
expression and clinicopathological factors in 80 primary gastric
cancer tissues.
|
|
| miR-520a-3p
expression level |
|
|---|
|
|
|
|
|
|---|
| Characteristic | Patients, n | Low, n (%) | High, n (%) | P-value |
|---|
| Age, years |
|
|
| 0.990 |
|
≤50 | 38 | 28 (73.7) | 10 (26.3) |
|
|
>50 | 42 | 31 (73.8) | 11 (26.2) |
|
| Tumor diameter,
cm |
|
|
| 0.067 |
| ≤3 | 24 | 21 (87.5) | 3 (12.5) |
|
|
>3 | 56 | 38 (67.9) | 18 (32.1) |
|
| Tumor TNM
staging |
|
|
| 0.016a |
|
I+II | 43 | 27 (62.8) | 16 (37.2) |
|
|
III+IV | 37 | 32 (86.5) | 5 (13.5) |
|
| Tumor histological
overall NHS grade |
|
|
| 0.305 |
| 1 | 7 | 4 (57.1) | 3 (42.9) |
|
| 2 | 41 | 33 (80.5) | 8 (19.5) |
|
| 3 | 32 | 22 (68.8) | 10 (31.3) |
|
| Invasion |
|
|
| 0.001b |
|
T1+T2 | 27 | 14 (51.9) | 13 (48.1) |
|
|
T3+T4 | 53 | 45 (84.9) | 8 (15.1) |
|
| Pathological
type |
|
|
| 0.412 |
|
Adenocarcinoma | 69 | 52 (75.4) | 17 (24.6) |
|
|
Non-adenocarcinoma | 11 | 7 (63.6) | 4 (36.4) |
|
Results
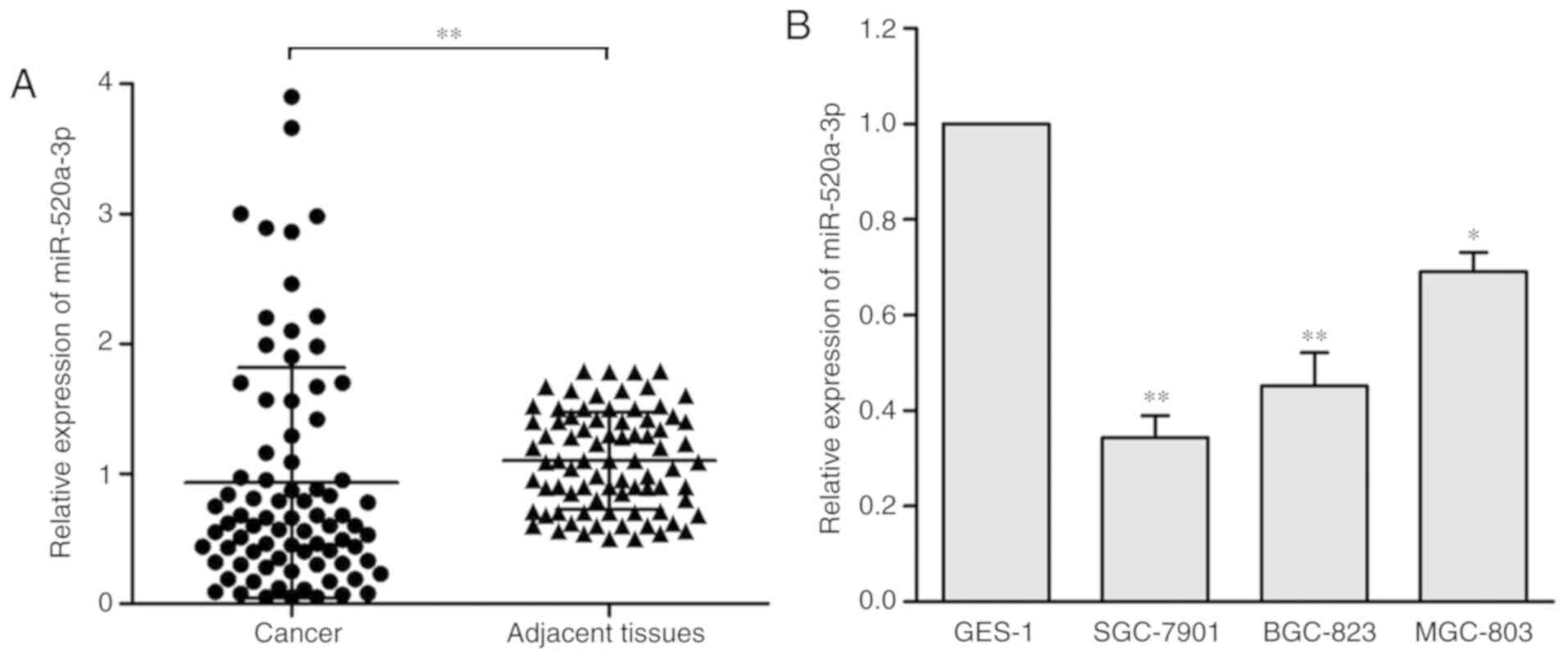
miR-520a-3p expression levels are
downregulated in GC and are associated with clinical stage and
tumor invasion
miR-520a-3p expression levels in 80 GC samples and
adjacent tissue samples were measured by RT-qPCR, and the levels
were normalized to U6. The results demonstrated that miR-520a-3p
expression levels were significantly decreased in GC tissues
compared with the adjacent normal tissues (P<0.01; Fig. 1A). Furthermore, the expression levels
of miR-520a-3p in three human GC cell lines (SGC-7901, BGC-823 and
MGC-803) were evaluated and compared with the normal gastric
epithelial cell line (GES-1). miR-520a-3p expression was
significantly upregulated in the GES-1 compared with the other
three cell lines (Fig. 1B).
To investigate the clinical significance of
miR-520a-3p in patients with GC, miR-520a-3p expression in a cohort
of 80 specimens was analyzed (Table
I). The miR-520a-3p levels were significantly associated with
clinical stage (P<0.05) and tumor invasion (P<0.001) in GC
tissues; however, they were not associated with patient age, tumor
size, histological grade or pathological type (P>0.05).
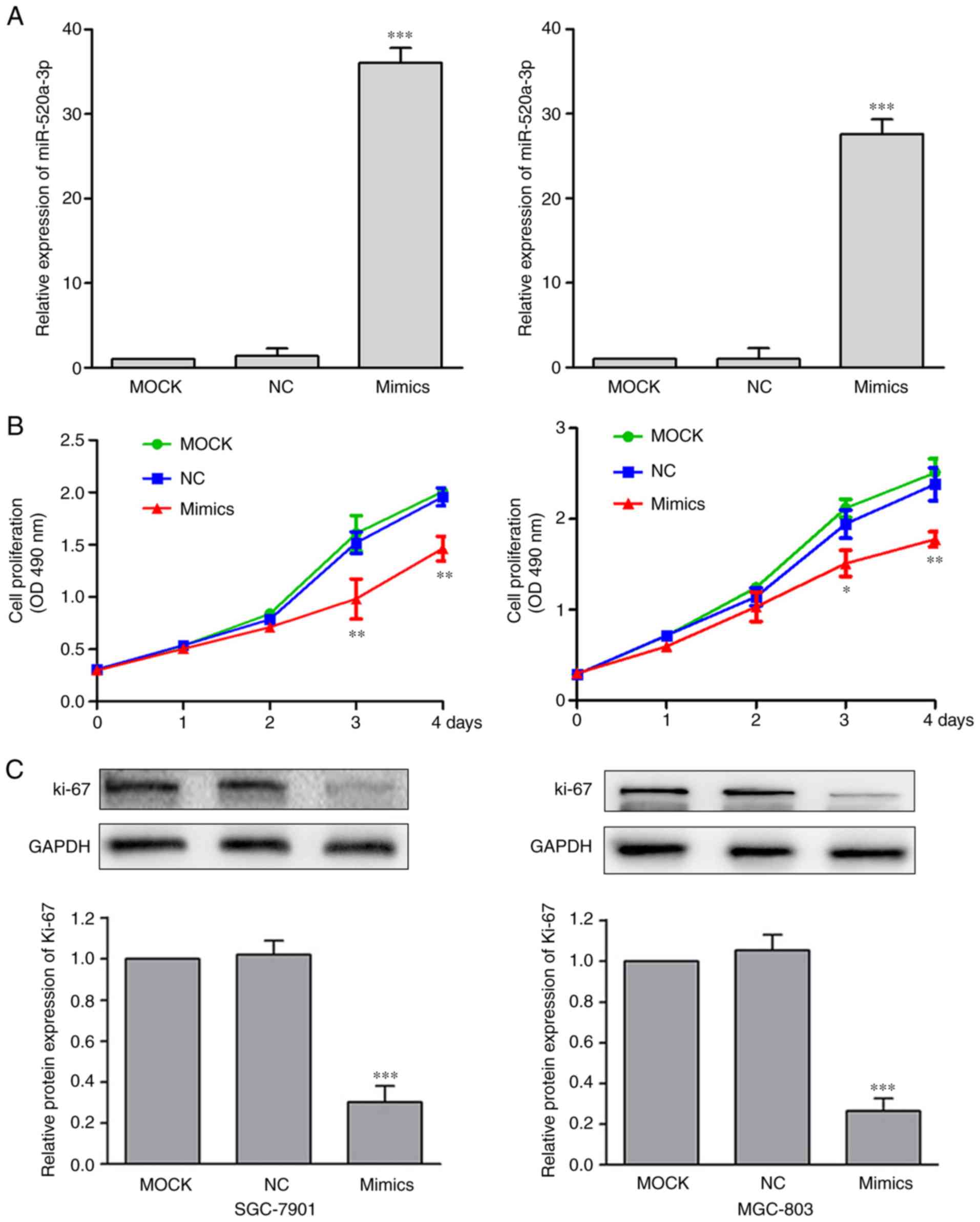
miR-520a-3p suppresses the
proliferation of GC cells
To investigate the functional effects of miR-520a-3p
expression in GC, SGC-7901 and MGC-803 cells were transfected with
MOCK, miR-NC or miR-520a-3p mimics. Treatment with miR-520a-3p
mimics significantly increased the expression levels of miR-520a-3p
(P<0.001), compared with the MOCK group (Fig. 2A). Following the upregulation of
miR-520a-3p expression, the effect on cell proliferation was
examined using cell proliferation assay. Compared with the control
cells, cells transfected with miR-520a-3p mimics demonstrated
significantly reduced levels of proliferation (Fig. 2B). Recent studies have demonstrated
that Ki-67 is closely associated with cell proliferation (20,21).
Investigating the levels of Ki-67 showed that miR-520a-3p mimics
significantly decreased Ki-67 expression in GC cells (P<0.01;
Fig. 2C).
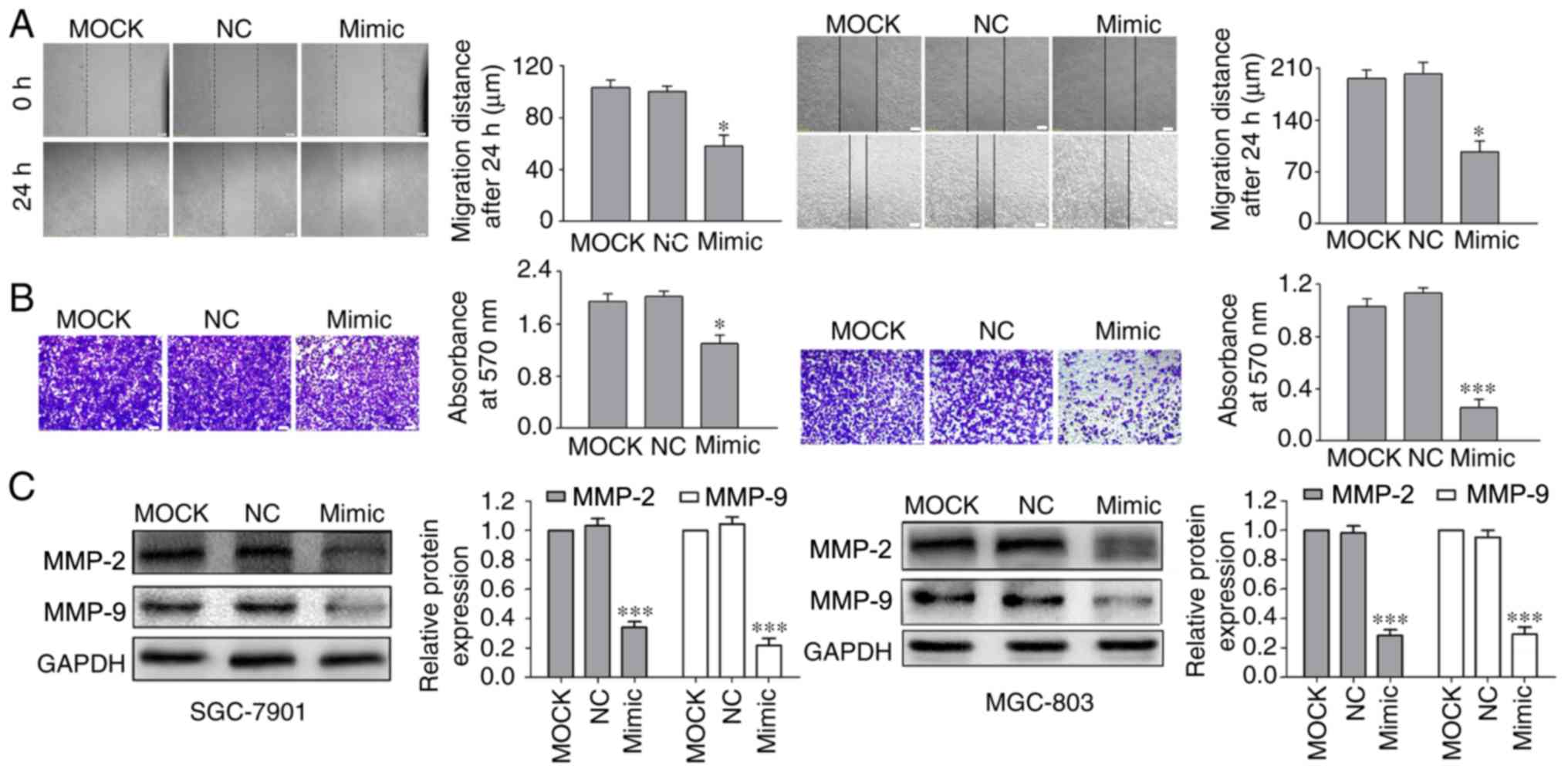
miR-520a-3p decreases the migration
and invasion of GC cells
To examine how miR-520a-3p affects the migration and
invasion of GC cells, the effects on cell migration and invasion
were investigated in vitro using scratch wound healing and
Transwell assays, respectively. The data demonstrated that
transfection of cells with miR-520a-3p mimics significantly
inhibited GC cell migration and invasion (Fig. 3A and B) compared with MOCK and NC
groups.
Interactions between cell-surface proteins and the
extracellular matrix (ECM) serve a critical role in tumor migration
and invasion (22). As MMPs,
particularly MMP-2 and MMP-9, digest ECM components, they are
closely associated with the metastatic capabilities of cancerous
cells (23). Transfection of
miR-520a-3p mimics led to significantly decreased protein
expression levels of MMP-2 and MMP-9 in GC cells compared with the
MOCK and NC transfections (Fig.
3C).
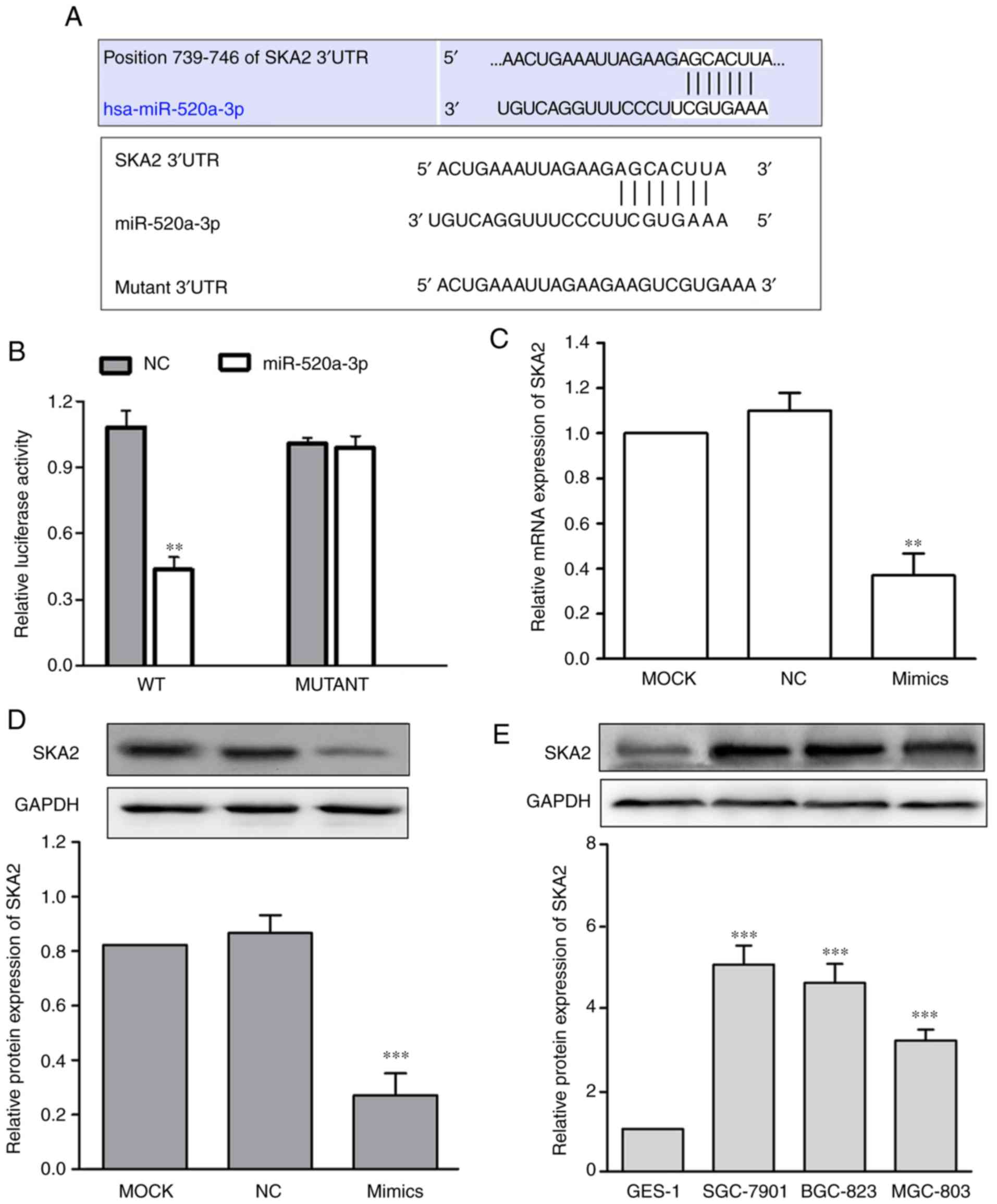
miR-520a-3p directlytargets SKA2 in
the SGC-7901GC cell line
To investigate the molecular mechanism underlying
the effects of miR-520a-3p on GC cellular functions, SKA2 mRNA was
identified as a potential miR-520a-3p target using bioinformatics
(Fig. 4A). Subsequently, the WT or
Mut target region sequence of the SKA2 3′UTR were cloned and
inserted into a luciferase reporter vector (Fig. 4A). The luciferase reporter assays
revealed that miR-520a-3p significantly decreased the luciferase
activity in the reporter gene containing the WT 3′UTR (P<0.01);
however, no difference was observed with the Mut 3′UTR (P>0.05;
Fig. 4B). To demonstrate whether
miR-520a-3p affected SKA2 expression in GC, expression of SKA2 in
SGC-7901 cells transfected with miR-520a-3p mimics was measured.
The data demonstrated that transfection with miR-520a-3p mimics
significantly decreased SKA2 mRNA and protein expression levels,
compared with the MOCK group (P<0.01; Fig. 4C and D). Furthermore, the protein
expression of SKA2 in SGC-7901, BGC-823, and MGC-803 was
significantly in all 3 GC cell lines compared with GES-1
(P<0.001; Fig. 4E).
Overexpression of SKA2 reverses the
miR-520a-3p-mediated inhibition of SGC-7901 cell proliferation,
migration and invasion
To determine whether SKA2 overexpression reversed
the inhibitory effects of miR-520a-3p mimics on SGC-7901 cells,
SKA2 cDNA without the 3′UTR and miR-520a-3p mimics were
co-transfected into SGC-7901 cells (P<0.01; Fig. 5A). Western blotting and cell
viability assays showed that the decrease in SKA2 expression and
cell viability following miR-520-3p overexpression was reversed
when SKA2 was co-transfected with miR-520a-3p mimics (Fig. 5B and C). Migration and invasion
assays demonstrated that SKA2 co-transfection significantly
reversed the miR-520a-3p-mediated decrease in cell migration and
invasion in SGC-7901 cells (Fig. 5D and
E).
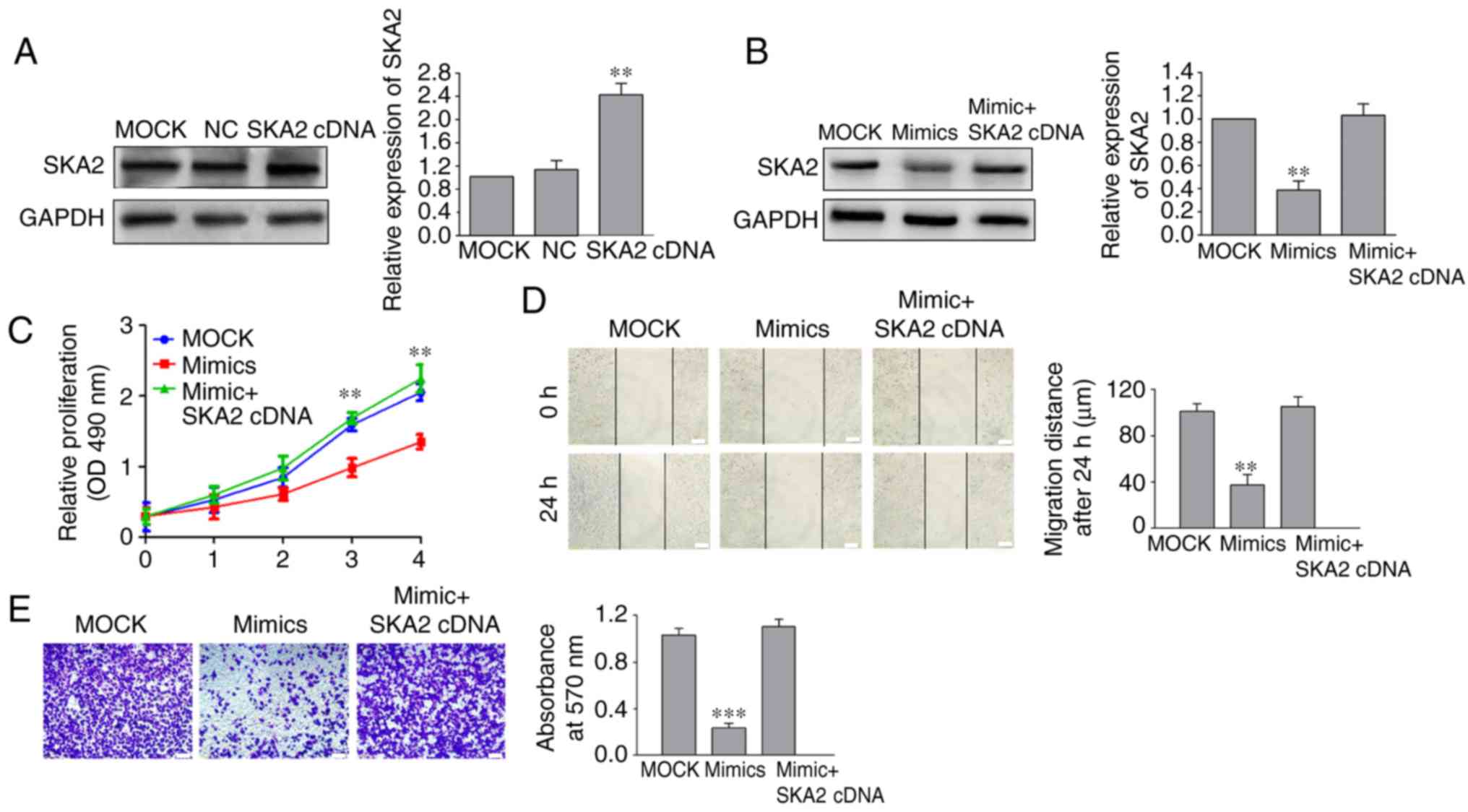 | Figure 5.SKA2 regulates miR-520a-3p-mediated
inhibition of SGC-7901 cells proliferation, migration and invasion.
(A) After cells were transfected with a SKA2 cDNA plasmid, the
expression of SKA2 was notably increased in SGC-790 cells.
Following co-transfection of miR-520a-3p mimics and SKA2 cDNA into
SGC-7901 cells, (B) expression of SKA2 and (C) cell viability were
examined by western blotting and cell proliferation assay,
respectively. (D) Migration distance and (E) number of invasive
cells in GC cells following treatment with miR-520a-3p mimics and
SKA2 cDNA were detected by wound healing and Transwell assays,
respectively. **P<0.01, ***P<0.001 vs. the MOCK group. ×100
magnification. Scale bar, 100 µm. SKA2, spindle
kinetochore-associated protein 2; miR, microRNA; GC, gastric
cancer; MOCK, mock transfected cells; NC, negative control; OD,
optical density. |
Discussion
miR-520a-3p serves an tumor suppressive role in
certain types of cancer (11–17).
However, its functions and expression in GC have not been studied,
to the best of our knowledge.
The results demonstrated that miR-520a-3p was
downregulated both in human GC tissues and GC cell lines compared
with adjacent tissues and a normal gastric cell line. In addition,
the expression of miR-520a-3p was associated with clinical stage
and tumor invasion in patients with GC. Subsequent experimentation
revealed that the upregulation of miR-520a-3p inhibited the
proliferation, migration and invasion of GC cells (SGC-7901 and
MGC-803), potentially by directly inhibiting SKA2. Furthermore,
miR-520a-3p significantly inhibited the level of SKA2, and
overexpression of SKA2 rescued the miR-520a-3p-mediated inhibition
of SGC-7901 cell proliferation, migration and invasion. Therefore,
the present study further confirmed the several biological
functions attributed to miR-520a-3p, including the reduction in GC
cell viability, and inhibition of cell migration and invasion
following transfection with miR-520a-3p mimics, as well as the
molecular mechanism underlying the miR-520a-3p-mediated inhibitory
effect (11–17). SKA2 is located on chromosome 17 of
the human genome and is a conserved protein involved in the
kinetochore complex (24). SKA2,
together with its cofactors SKA1 and SKA3, constitute the SKA
complex, which maintains the metaphase plate and/or spindle
checkpoint silencing (25,26). Recently, SKA2 was also identified to
exert a function in tumorigenesis. Aberrant expression patterns of
SKA2 have been reported in lung cancer (27–29),
breast cancer (30–32), osteosarcoma (33,34) and
kidney cancer (35). Nevertheless,
the potential roles of SKA2 in GC are unknown. The results, to the
best of our knowledge, are the first to demonstrate that SKA2 was
significantly upregulated in gastric cell lines.
As a potential target gene of miR-520a-3p predicted
by miRanda and TargetScan software, the present study analyzed the
relationship between SKA2 and miR-520a-3p in SGC-7901 cells. The
results demonstrated that miR-520a-3p mimics significantly
attenuated the mRNA and protein expression of SKA2. Furthermore,
downregulation of SKA2 by miR-520a-3p mimics was reversed by SKA2
cDNA plasmid. These results suggested that miR-520a-3p directly
repressed the expression of SKA2 in GC by binding to its 3′UTR.
A previous study reported that miR-520d-3p inhibited
GC cell growth and metastatic activity by targeting EphA2 (36). miR-520d-3p and miR-520a-3p are
members of the same microRNA family and share a large amount of
homology. As they have different sequences and binding sites of
target genes, they play a role in tumorigenesis through different
regulatory mechanisms. In the present study, the potential value of
miRNA-520a-3p and SKA2 as therapeutic targets in cancer diagnosis
and targeted therapy was demonstrated.
In summary, miR-520a-3p was downregulated in GC
tissues and cell lines. miR-520a-3p also serves a role as a tumor
suppressor gene in GC cell lines, decreasing cell proliferation,
invasion and migration by targeting SKA2. The novel interaction
between miR-520a-3p and SKA2 may contribute to the development of
improved targeted therapies for patients with GC. However, due to
the complex signaling pathways regulated by microRNAs and the
limited sample size, further investigation into the function and
regulatory mechanisms of miR-520a-3p in vivo is required. In
particular, the expression levels of miR-520a-3p and SKA2 should be
determined in a range of cancer types, both in vitro and
in vivo, to determine whether the mechanisms underlying
miR-520a-3p-mediated oncogenic activity in GC cell lines are
replicated elsewhere.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
HS and FR collected and analyzed the data, and wrote
and revised the manuscript. XF conceived and designed the
experiments. HJ and YC conducted the experiments. All authors read
and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by The Research
Ethics Committee of Ningbo No. 2 Hospital (Zhejiang, China), and
all patients provided written informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Global Burden of Disease Cancer
Collaboration, ; Fitzmaurice C, Allen C, Barber RM, Barregard L,
Bhutta ZA, Brenner H, Dicker DJ, Chimed-Orchir O, Dandona R, et al:
Global, regional and national cancer incidence, mortality, years of
life lost, years lived with disability, and disability-adjusted
life-years for 32 cancer groups, 1990 to 2015: A systematic
analysis for the global burden of disease study. JAMA Oncol.
3:524–548. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Smyth EC and Cunningham D: Gastric cancer
in 2012: Defining treatment standards and novel insights into
disease biology. Nat Rev Clin Oncol. 10:73–74. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhang J, Song Y, Zhang C, Zhi X, Fu H, Ma
Y, Chen Y, Pan F, Wang K, Ni J, et al: Circulating miR-16-5p and
miR-19b-3p as two novel potential biomarkers to indicate
progression of gastric cancer. Theranostics. 5:733–745. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Jiang X and Wang Z: MiR-16 targets SALL4
to repress the proliferation and migration of gastric cancer. Oncol
Lett. 16:3005–3012. 2018.PubMed/NCBI
|
|
5
|
Lan H, Lu H, Wang X and Jin H: MicroRNAs
as potential biomarkers in cancer: Opportunities and challenges.
Biomed Res Int. 2015:1250942015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Valencia-Sanchez MA, Liu J, Hannon GJ and
Parker R: Control of translation and mRNA degradation by miRNAs and
siRNAs. Genes Dev. 20:515–524. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Luo X, Wang GH, Bian ZL, Li XW, Zhu BY,
Jin CJ and Ju SQ: Long non-coding RNA CCAL/miR-149/FOXM1 axis
promotes metastasis in gastric cancer. Cell Death Dis. 9:9932018.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Liu J, Wei Y, Li S, Li Y, Liu H, Liu J and
Zhu X: MicroRNA-744 promotes cell apoptosis via targeting B cell
lymphoma-2 in gastric cancer cell line SGC-7901. Exp Ther Med.
16:3611–3616. 2018.PubMed/NCBI
|
|
9
|
Yu H, Zhang J, Wen Q, Dai Y, Zhang W, Li F
and Li J: MicroRNA-6852 suppresses cell proliferation and invasion
via targeting forkhead box J1 in gastric cancer. Exp Ther Med.
16:3249–3255. 2018.PubMed/NCBI
|
|
10
|
Peng X, Kang Q, Wan R and Wang Z:
MiR-26a/HOXC9 dysregulation promotes metastasis and stem cell-like
phenotype of gastric cancer. Cell Physiol Biochem. 49:1659–1649.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yu J, Tan Q, Deng B, Fang C, Qi D and Wang
R: The microRNA-520a-3p inhibits proliferation, apoptosis and
metastasis by targeting MAP3K2 in non-small cell lung cancer. Am J
Cancer Res. 5:802–811. 2015.PubMed/NCBI
|
|
12
|
Liu Y, Miao L, Ni R, Zhang H, Li L, Wang
X, Li X and Wang J: MicroRNA-520a-3p inhibits proliferation and
cancer stem cell phenotype by targeting HOXD8 in non-small cell
lung cancer. Oncol Rep. 36:3529–3535. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lv X, Li CY, Han P and Xu XY:
MicroRNA-520a-3p inhibits cell growth and metastasis of non-small
cell lung cancer through PI3K/AKT/mTOR signaling pathway. Eur Rev
Med Pharmacol Sci. 22:2321–2327. 2018.PubMed/NCBI
|
|
14
|
Li J, Wei J, Mei Z, Yin Y, Li Y, Lu M and
Jin S: Suppressing role of miR-520a-3p in breast cancer through
CCND1 and CD44. Am J Transl Res. 9:146–154. 2017.PubMed/NCBI
|
|
15
|
Zhang R, Liu R, Liu C, Niu Y, Zhang J, Guo
B, Zhang CY, Li J, Yang J and Chen X: A novel role for MiR-520a-3p
in regulating EGFR expression in colorectal cancer. Cell Physiol
Biochem. 42:1559–1574. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang X, Xu Y, Chen X and Xiao J:
Dexmedetomidine inhibits osteosarcoma cell proliferation and
migration, and promotes apoptosis by regulating MiR-520a-3p. Oncol
Res. 26:495–502. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhang B, Yu L, Han N, Hu Z, Wang S, Ding L
and Jiang J: LINC01116 targets miR-520a-3p and affects IL6R to
promote the proliferation and migration of osteosarcoma cells
through the Jak-stat signaling pathway. Biomed Pharmacother.
107:270–282. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
In H, Solsky I, Palis B, Langdon-Embry M,
Ajani J and Sano T: Validation of the 8th edition of the AJCC TNM
staging system for gastric cancer using the national cancer
database. Ann Surg Oncol. 24:3683–3691. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Pizon M, Schott DS, Pachmann U and
Pachmann K: B7-H3 on circulating epithelial tumor cells correlates
with the proliferation marker, Ki-67, and may be associated with
the aggressiveness of tumors in breast cancer patients. Int J
Oncol. 53:2289–2299. 2018.PubMed/NCBI
|
|
21
|
Krtinic D, Zivadinovic R, Jovic Z, Pesic
S, Mihailovic D, Ristic L, Cvetanovic A, Todorovska I, Zivkovic N,
Rankovic GN, et al: Significance of the Ki-67 proliferation index
in the assessment of the therapeutic response to cisplatin-based
chemotherapy in patients with advanced cervical cancer. Eur Rev Med
Pharmacol Sci. 22:5149–5155. 2018.PubMed/NCBI
|
|
22
|
Komemi O, Epstein Shochet G, Pomeranz M,
Fishman A, Pasmanik-Chor M, Drucker L, Tartakover Matalon S and
Lishner M: Placenta-conditioned extracellular matrix (ECM)
activates breast cancer cell survival mechanisms: A key for future
distant metastases. Int J Cancer Sep. 144:1633–1644. 2019.
View Article : Google Scholar
|
|
23
|
Shay G, Lynch CC and Fingleton B: Moving
targets: Emerging roles for MMPs in cancer progression and
metastasis. Matrix Biol. 44-46:200–206. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhang QH, Qi ST, Wang ZB, Yang CR, Wei YC,
Chen L, Ouyang YC, Hou Y, Schatten H and Sun QY: Localization and
function of the Ska complex during mouse oocyte meiotic maturation.
Cell Cycle. 11:909–916. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Gaitanos TN, Santamaria A, Jeyaprakash AA,
Wang B, Conti E and Nigg EA: Stable kinetochore-microtubule
interactions depend on the Ska complex and its new component
Ska3/C13Orf3. EMBOJ. 28:1442–1452. 2009. View Article : Google Scholar
|
|
26
|
Jeyaprakash AA, Santamaria A, Jayachandran
U, Chan YW, Benda C, Nigg EA and Conti E: Structural and functional
organization of the ska complex, a key component of the
kinetochore-microtubule interface. Mol Cell. 46:274–286. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Cao G, Huang B, Liu Z, Zhang J, Xu H, Xia
W, Li J, Li S, Chen L, Ding H, et al: Intronic miR-301 feedback
regulates its host gene, ska2, in A549 cells by targeting MEOX2 to
affect ERK/CREB pathways. Biochem Biophys Res Commun. 396:978–982.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang Y, Zhang Y, Zhang C, Weng H, Li Y,
Cai W, Xie M, Long Y, Ai Q, Liu Z, et al: The gene pair PRR11 and
SKA2 shares a NF-Y-regulated bidirectional promoter and contributes
to lung cancer development. Biochim Biophys Acta. 1849:1133–1144.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang Y, Weng H, Zhang Y, Long Y, Li Y, Niu
Y, Song F and Bu Y: The PRR11-SKA2 bidirectional transcription unit
is negatively regulated by p53 through NF-Y in lung cancer cells.
Int J Mol Sci. 18(pii): E5342017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Shi W, Gerster K, Alajez NM, Tsang J,
Waldron L, Pintilie M, Hui AB, Sykes J, P'ng C, Miller N, et al:
MicroRNA-301 mediates proliferation and invasion in human breast
cancer. Cancer Res. 71:2926–2937. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ren Z, Yang T, Ding J, Liu W, Meng X,
Zhang P, Liu K and Wang P: MiR-520d-3p antitumor activity in human
breast cancer via post-transcriptional regulation of spindle and
kinetochore associated 2 expression. Am J Transl Res. 10:1097–1108.
2018.PubMed/NCBI
|
|
32
|
Wu H, Wang Y, Chen T, Li Y, Wang H, Zhang
L, Chen S, Wang W and Chen C, Yang Q and Chen C: The N-terminal
polypeptide derived from vMIP-II exerts its antitumor activity in
human breast cancer by regulating lncRNA SPRY4-IT1. Biosci Rep.
38(pii): BSR201804112018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lin CC, Chao PY, Shen CY, Shu JJ, Yen SK,
Huang CY and Liu JY: Novel target genes responsive to apoptotic
activity by Ocimum gratissimum in human osteosarcoma cells. Am J
Chin Med. 42:743–767. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Chang IC, Chiang TI, Lo C, Lai YH, Yue CH,
Liu JY, Hsu LS and Lee CJ: Anemone altaica induces apoptosis in
human osteosarcoma cells. Am J Chin Med. 43:1031–1042. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhuang H, Meng X, Li Y, Wang X, Huang S,
Liu K, Hehir M, Fang R, Jiang L, Zhou JX, et al: Cyclic AMP
responsive element-binding protein promotes renal cell carcinoma
proliferation probably via the expression of spindle and
kinetochore-associated protein 2. Oncotarget. 7:16325–16337. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Li R, Yuan W, Mei W, Yang K and Chen Z:
MicroRNA 520d-3p inhibits gastric cancer cell proliferation,
migration, and invasion by downregulating EphA2 expression. Mol
Cell Biochem. 396:295–305. 2014. View Article : Google Scholar : PubMed/NCBI
|