Introduction
Bladder cancer is the sixth most common cause of
cancer mortality and its incidence has increased markedly in recent
decades (1,2) as well as an increasing prevalence of
established risk factors such as smoking, overweight, physical
inactivity, and changing reproductive patterns associated with
urbanization and economic development. Based on GLOBOCAN estimates,
about 14.1 million new cancer cases and 8.2 million deaths occurred
in 2012 worldwide. Over the years, the burden has shifted to less
developed countries, which currently account for about 57% of cases
and 65% of cancer deaths worldwide. Lung cancer is the leading
cause of cancer death among males in both more and less developed
countries, and has surpassed breast cancer as the leading cause of
cancer death among females in more developed countries; breast
cancer remains the leading cause of cancer death among females in
less developed countries. Other leading causes of cancer death in
more developed countries include colorectal cancer among males and
females and prostate cancer among males. In less developed
countries, liver and stomach cancer among males and cervical cancer
among females are also leading causes of cancer death. Although
incidence rates for all cancers combined are nearly twice as high
in more developed than in less developed countries in both males
and females, mortality rates are only 8 to 15% higher in more
developed countries. This disparity reflects regional differences
in the mix of cancers, which is affected by risk factors and
detection practices, and/or the availability of treatment. Risk
factors associated with the leading causes of cancer death include
tobacco use (lung, colorectal, stomach, and liver cancer. Current
prognostic factors, namely TNM stage and pathological grade,
insufficiently predict outcome at the individual level. The
different outcome for patients with the same stage and grade calls
for new prognostic molecular markers that may also serve as
therapeutic targets.
Several high-throughput studies have focused on
delineating genomic changes and gene expression in the various
stages of bladder-cancer development and progression. Recent
genomic analyses, part of The Cancer Genome Atlas, have identified
several new target genes not previously described in bladder
carcinogenesis (3–6).
Pleckstrin homology domain-containing S1
(PLEKHS1) is one of the most remarkable targets, because
after telomerase reverse transcriptase (TERT), it is the
second gene of the human genome showing frequent somatic non-coding
mutations within its promoter (7).
This gene, located on chromosome 10q25.3, encodes the PLEKHS1
protein with unknown function. These mutations are
single-nucleotide substitutions in the PLEKHS1 proximal
promoter (in intron 1 with a non-coding exon 1 from NM_024889.4):
substitutions that effect guanine at position Chr.10:
115511590 and cytosine at position Chr.10: 115511593 from the
GRCh37 (hg19) genomic coordinate (or c.-20+70G and c.-20+73C
from NM_024889.4). These mutations are close to the translation
start site of the PLEKHS1 gene (−3,447 and −3,444 bp
upstream of the translation start site) and are flanked by
stretches of 10 bp on both sides that are palindromic to each
other. The two PLEKHS1 promoter mutations could be
considered among the most common noncoding mutations in cancer. In
bladder cancer, almost 40% of tumors could be affected by promoter
mutations of this gene (8). However,
data on messenger ribonucleic acid (mRNA) expression levels in
bladder cancer are still lacking.
We analyzed the frequency of promoter mutations and
mRNA expression of PLEKHS1 in two large series of 335 bladder
tumors: 162 non-muscle-invasive bladder cancer (NMIBC) (71 and 91,
respectively) and 173 muscle-invasive bladder cancers (MIBC) (83
and 90, respectively). A retrospective review of patient outcomes
allowed for performing survival analyses to highlight the putative
prognostic value of this gene.
Patients and methods
Patients and samples
We analyzed samples from two series of patients with
urothelial carcinoma of the bladder. The first series consisted of
154 patients who had undergone transurethral bladder resection or a
radical cystectomy in our hospital between 2002 and 2007.
Immediately after surgery, tumor samples from each patient were
frozen in liquid nitrogen and stored at −80°C [for DNA
(deoxyribonucleic acid) and RNA extraction] and fixed in
formaldehyde. Specimens of normal bladder tissue from 20 patients
undergoing surgery unrelated to bladder tumors (transurethral
resection of the prostate, prostatic adenomectomy) were used as
sources of normal bladder tissues.
Each tumor was reviewed by two pathologists (DD and
MS) who were blinded to the clinical outcomes. Tumors were
re-staged according to the 2009 TNM classification of bladder
tumors (9) and were graded according
to the WHO 2004 tumor-grading scheme (10). Standard follow-up visits followed
current guidelines.
The second series was collected from 181 patients
treated surgically between 1988 and 2006 at Henri Mondor Hospital,
Institut Gustave Roussy (Villejuif, France) and Foch Hospital
(Suresnes, France). Pure SCC and pure adenocarcinoma were excluded.
Immediately after surgery, tumor samples from each patient were
frozen in liquid nitrogen and stored at −80°C (for DNA and RNA
extraction) and fixed in formaldehyde. For non muscle invasive
cases (stage Ta or T1), TURB material was used; for muscle invasive
cases, TURB or cystectomy material was used, without prior
neoadjuvant chemotherapy, as previously described in Rebouissou
et al (11).
All patients provided written informed consent.
These studies received approval from an institutional review board,
Centre d'Éthique Clinique de l'Hôpital Cochin, and was conducted
according to the principles outlined in the Declaration of
Helsinki.
DNA mutation analysis
PLEKHS1 has two mutational spots that are
well described. The assessment was performed by a screening with
high-resolution melt (HRM) analysis followed by Sanger sequencing
of samples with a mutated profile on HRM to validate the HRM data
and determine the nomenclature of mutations found. The nucleotide
sequences for the primers were for PLEKHS1-U
(5′-CTTCCAAGGCTGGGATGATCTA-3′) and PLEKHS1-L
(5′-AAGAAAGTGCCCATAACAGAAATACA-3′) (polymerase chain reaction (PCR)
product of 107 bp).
Real-time RT-qPCR
The theoretical basis, primers and PCR consumables,
RNA extraction, complementary deoxyribonucleic acid (cDNA)
synthesis, and PCR-reaction conditions were previously described in
detail (12). One endogenous RNA
control gene was chosen, namely TBP (GenBank accession no.
NM_003194), which encodes the TATA box-binding protein. Because
PLEKHS1 was expressed in tumor samples but not normal
bladder tissue, values were normalized so that a Ct value of 35 was
set to 1. Taking into account the optimal cut-off for
PLEKHS1, mRNA values ≥100 were considered
overexpression.
Primers were chosen with the assistance of the Oligo
6.0 computer program (National Biosciences, Plymouth, MN). The
nucleotide primer sequences were PLEKHS1-U
(5′-AAGATGTTTAAATGCCACCCTGATG-3′) and PLEKHS1-L
(5′-CCAGTCTTTAATCTTCTCCCTGTCGT-3′) (PCR product of 99 bp).
Experiments were performed in duplicate for each data point.
Statistical analysis
The clinicopathologic features of NMIBC and MIBC
were tested for their association with tumor recurrence and
survival by Student's t-test for continuous variables or
χ2 test for qualitative variables. The distribution of
mRNA levels was described with medians (range). Relationships
between clinical and histological variables and mRNA levels of
PLEKHS1 were tested by the non-parametric Mann-Whitney U
test and Kruskal-Wallis H-test. Overall survival (OS) was
calculated from the date of surgery until death or the last
follow-up. Recurrence-free survival (RFS) was defined as the time
from the date of surgery to the first local relapse or first
metastasis. For NMIBC, progression-free survival (PFS) was defined
as the time from the date of surgery to progression to
muscle-invasive disease. Survival curves were derived from
Kaplan-Meier estimates. The log-rank test was used to compare
survival between subgroups. The prognostic impact of mRNA levels,
adjusted for the other prognostic factors, was assessed by Cox
proportional-hazards regression analysis, estimating hazard ratios
and 95% confidence intervals. The variables significant in
univariate analysis (P<0.10) were included in multivariate
analysis. Differences were considered significant at P<0.05.
Results
Clinicopathologic characteristics of
the cohorts
Complete clinical, histological and survival data
were obtained from medical records for these 154 patients [129 men
and 25 women; median age 70 years (range, 31–91)]. Pathological
staging showed NMIBC in 71 patients (25 low-grade pTa, 17
high-grade pTa, 29 high-grade pT1) and high-grade MIBC in 83
patients. For NMIBC, the median follow-up was 57.4 months (range,
1–158 months; mean follow-up, 61 months). For MIBC, the median
follow-up was 12.5 months (range, 1–152 months; mean follow-up, 29
months). In the MIBC cohort, 12 patients (14.5%) received
neoadjuvant chemotherapy before cystectomy and 25 (30.1%) received
adjuvant chemotherapy taking into account pathological
characteristics of the tumor and renal function. Clinical,
histological and survival characteristics for the first series are
presented in Tables I and II.
 | Table I.Clinical, pathological and survival
characteristics of the 71 NMIBC of the first series. |
Table I.
Clinical, pathological and survival
characteristics of the 71 NMIBC of the first series.
|
|
|
| Recurrence | Muscle-invasive
progression |
|---|
|
|
|
|
|
|
|---|
| Characteristic | Whole population, n
(%) | No recurrence, n
(%) | n (%) | P-valuea | n (%) | P-valueb |
|---|
| Total population | 71 (100.0) | 25 (35.2) | 36 (50.7) |
| 10 (14.1) |
|
| Age (years) |
|
|
|
|
| 0.08 |
| ≥60 | 56 (78.9) | 19 (76.0) | 27 (75.0) | 0.93 | 10 (100.0) |
|
|
<60 | 15 (21.1) | 6 (24.0) | 9 (25.0) |
| 0 (0.0) |
|
| Sex |
|
|
|
|
|
|
| Male | 63 (88.7) | 22 (88.0) | 32 (88.9) | 0.91 | 9 (90.0) | 0.89 |
|
Female | 8 (11.3) | 3 (12.0) | 4 (11.1) |
| 1 (10.0) |
|
| Smoking status |
|
|
|
|
|
|
|
Non-smoker | 33 (46.5) | 13 (52.0) | 15 (41.7) | 0.43 | 5 (50.0) | 0.81 |
|
Smoker | 38 (53.5) | 12 (48.0) | 21 (58.3) |
| 5 (50.0) |
|
| History of
NMIBC |
|
|
|
|
|
|
| No | 39 (54.9) | 22 (88.0) | 13 (36.1) | <0.0001 | 4 (40.0) | 0.31 |
|
Yes | 32 (45.1) | 3 (12.0) | 23 (63.9) |
| 6 (60.0) |
|
| Associated
pTis |
|
|
|
|
|
|
| No | 69 (97.2) | 25 (100.0) | 36 (100.0) | 0.99 | 8 (80.0) | 0.0004 |
|
Yes | 2 (2.8) | 0 (0.0) | 0 (0.0) |
| 2 (20.0) |
|
| Grade |
|
|
|
|
|
|
|
Low | 25 (35.2) | 10 (40.0) | 14 (38.9) | 0.93 | 1 (10.0) | 0.07 |
|
High | 46 (64.8) | 15 (60.0) | 22 (61.1) |
| 9 (90.0) |
|
| Tumor stage |
|
|
|
|
|
|
| Ta | 42 (59.2) | 15 (60.0) | 24 (66.7) | 0.59 | 3 (30.0) | 0.043 |
| T1 | 29 (40.8) | 10 (40.0) | 12 (33.3) |
| 7 (70.0) |
|
 | Table II.Clinical, pathological and survival
characteristics of the 83 muscle-invasive bladder cancer of the
first series. |
Table II.
Clinical, pathological and survival
characteristics of the 83 muscle-invasive bladder cancer of the
first series.
|
|
| Disease-free
survival | Overall
survival |
|---|
|
|
|
|
|
|---|
| Characteristic | Whole population, n
(%) | Number of events
(%)a |
P-valuec | Number of events
(%)b |
P-valuec |
|---|
| Total
population | 83 (100.0) | 48 (57.8) |
| 46 (55.4) |
|
| Age (years) |
|
|
|
|
|
|
≥60 | 61 (73.5) | 40 (65.5) | 0.017 | 39 (63.9) | 0.009 |
|
<60 | 22 (26.5) | 8 (36.4) |
| 7 (31.8) |
|
| Sex |
|
|
|
|
|
|
Male | 66 (79.5) | 36 (54.5) | 0.23 | 38 (57.6) | 0.44 |
|
Female | 17 (20.5) | 12 (70.6) |
| 8 (47.1) |
|
| Smoking status |
|
|
|
|
|
|
Non-smoker | 34 (41.0) | 18 (52.9) | 0.45 | 12 (35.3) | 0.002 |
|
Smoker | 49 (59.0) | 30 (61.2) |
| 34 (69.4) |
|
| History of
NMIBC |
|
|
|
|
|
| No | 59 (71.1) | 30 (50.8) | 0.043 | 31 (52.5) | 0.41 |
|
Yes | 24 (28.9) | 18 (75.0) |
| 15 (62.5) |
|
| Associated
pTis |
|
|
|
|
|
| No | 73 (88.0) | 43 (58.9) | 0.59 | 40 (54.8) | 0.76 |
|
Yes | 10 (12.0) | 5 (50.0) |
| 6 (60.0) |
|
| Tumor stage |
|
|
|
|
|
| T2 | 34 (41.0) | 17 (50.0) | 0.10 | 13 (38.2) | 0.009 |
|
≥T3 | 49 (59.0) | 31 (63.3) |
| 33 (67.3) |
|
| Lymph node
status |
|
|
|
|
|
| N− | 58 (69.9) | 27 (46.6) | 0.002 | 25 (43.1) | 0.0006 |
| N+ | 25 (30.1) | 21 (84.0) |
| 21 (84.0) |
|
The second series consisted of an independent cohort
of 181 patients with bladder cancer [150 men and 31 women; median
age 67 years (range, 30–95)]. Pathological staging showed NMIBC in
91 patients (33 low-grade pTa, 17 high-grade pTa, 41 high-grade
pT1) and high-grade MIBC in 90 patients. In this second cohort,
molecular subgroups (basal phenotype, called MC7, vs non-basal
phenotype) were previously determined by using the CIT (Carte
d'Identité des Tumeurs) classification (11) we identified an MIBC subgroup
accounting for 23.5% of MIBC, associated with shorter survival and
displaying a basal-like phenotype, as shown by the expression of
epithelial basal cell markers. Basal-like tumors presented an
activation of the epidermal growth factor receptor (EGFR). None
patient of the second cohort received neoadjuvant chemotherapy. As
in the first cohort, patients received adjuvant chemotherapy taking
into account pathological characteristics of the tumor if they were
eligible. Clinical and histological variables did not differ
between the two series, except for history of NMIBC (Table III).
 | Table III.Clinical and pathological
characteristics of NMIBC and MIBC the two series. |
Table III.
Clinical and pathological
characteristics of NMIBC and MIBC the two series.
| A, NMIBC |
|---|
|
|---|
| Characteristic | Whole population, n
(%) | First series, n
(%) | Second series, n
(%) |
P-valueh |
|---|
| Total
population | 162 (100.0) | 71 (100.0) | 91 (100.0) |
|
| Age
(years)a |
|
|
|
|
|
≥60 | 108 (72.0) | 56 (78.9) | 52 (65.8) | 0.08 |
|
<60 | 42 (28.0) | 15 (21.1) | 27 (34.2) |
|
| Sex |
|
|
|
|
|
Male | 138 (85.2) | 63 (88.7) | 75 (82.4) | 0.26 |
|
Female | 24 (14.8) | 8 (11.3) | 16 (17.6) |
|
| History of
NMIBCb |
|
|
|
|
| No | 108 (71.5) | 39 (54.9) | 69 (86.3) | <0.0001 |
|
Yes | 43 (28.5) | 32 (45.1) | 11 (13.7) |
|
| Associated
pTisc |
|
|
|
|
| No | 134 (93.7) | 69 (97.2) | 65 (90.3) | 0.09 |
|
Yes | 9 (6.3) | 2 (2.8) | 7 (9.7) |
|
| Grade |
|
|
|
|
|
Low | 58 (35.8) | 25 (35.2) | 33 (36.3) | 0.89 |
|
High | 104 (64.2) | 46 (64.8) | 58 (63.7) |
|
| Tumor stage |
|
|
|
|
| Ta | 92 (56.8) | 42 (59.2) | 50 (54.9) | 0.59 |
| T1 | 70 (43.2) | 29 (40.8) | 41 (45.1) |
|
|
| B, MIBC |
|
|
Characteristic | Whole
population, n (%) | First series, n
(%) | Second series, n
(%) |
P-valueh |
|
| Total
population | 173 (100.0) | 83 (100.0) | 90 (100.0) |
|
| Age
(years)d |
|
|
|
|
|
≥60 | 124 (73.8) | 61 (73.5) | 63 (74.1) | 0.93 |
|
<60 | 44 (26.6) | 22 (26.5) | 22 (25.9) |
|
| Sex |
|
|
|
|
|
Male | 141 (81.5) | 66 (79.5) | 75 (83.3) | 0.52 |
|
Female | 32 (18.5) | 17 (20.5) | 15 (16.7) |
|
| History of
NMIBCe |
|
|
|
|
| No | 135 (84.4) | 73 (88.0) | 62 (80.5) | 0.20 |
|
Yes | 25 (15.6) | 10 (12.0) | 15 (19.5) |
|
| Associated
pTisf |
|
|
|
|
| No | 135 (84.4) | 73 (88.0) | 62 (80.5) | 0.20 |
|
Yes | 25 (15.6) | 10 (12.0) | 15 (19.5) |
|
| Tumor stage |
|
|
|
|
| T2 | 66 (38.2) | 34 (41.0) | 32 (35.6) | 0.46 |
|
≥T3 | 107 (61.8) | 49 (59.0) | 58 (64.4) |
|
| Lymph node
statusg |
|
|
|
|
|
N− | 93 (63.7) | 58 (69.9) | 35 (55.6) | 0.07 |
|
N+ | 53 (36.3) | 25 (30.1) | 28 (44.4) |
|
DNA mutations of PLEKHS1
DNA mutation analysis involved 103 available tumor
DNA samples (44 NMIBC and 59 MIBC) from the first series and 18
normal bladder samples, then the 181 tumor samples (91 NMIBC and 90
MIBC) from the second series.
In the first series, 11/44 NMIBC (25.0%) and 19/59
MIBC (32.2%) samples had a promoter-mutated profile (Table IV). The PLEKHS1 promoter
exhibited recurrent mutations at two genomic positions: c.-20+70
(G>A and G>C) and c.-20+73 (C>A, C>T and C>G). NMIBC
and MIBC samples did not differ by type of mutation (P=0.43).
 | Table IV.Frequency of mutations in the
PLEKHS1 promoter in the two series. |
Table IV.
Frequency of mutations in the
PLEKHS1 promoter in the two series.
|
| First series
(n=103) | Second series
(n=181) |
|---|
|
|
|
|
|---|
| PLEKHS1
profile | Mutated n (%) | Not mutated n
(%) |
P-valuea | Mutated n (%) | Not mutated n
(%) |
P-valuea |
|---|
| All tumors | 30 (29.1) | 73 (70.9) |
| 64 (35.4) | 117 (64.4) |
|
| NMIBC | 11 (25.0) | 33 (75.0) | 0.43 | 30 (33.0) | 61 (67.0) | 0.50 |
| MIBC | 19 (32.2) | 40 (67.8) |
| 34 (37.8) | 56 (62.2) |
|
In the second series, 30/91 NMIBC (33.0%) and 34/90
MIBC (37.8%) samples had a promoter-mutated profile (Table IV). The DNA-mutated profile was not
significantly associated with clinical parameters for patients with
NMIBC (Table V) or MIBC (Table VI). Regarding molecular subgroups in
MIBC, a DNA-mutated profile was significantly more frequent with
the basal phenotype (MC7) than the non-basal phenotype (61.5 vs.
27.1%, P=0.0025). PLEKHS1 DNA mutation was not associated with
prognosis, in terms of recurrence or progression with NMIBC and in
terms of RFS or OS with MIBC (data not shown).
 | Table V.Association of clinicopathological
variables and PLEKHS1 DNA mutated profile with NMIBC from
the second series. |
Table V.
Association of clinicopathological
variables and PLEKHS1 DNA mutated profile with NMIBC from
the second series.
| Characteristic | Total population, n
(%) | PLEKHS1
mutated, n (%) | Not mutated, n
(%) |
P-valuea |
|---|
| Total | 91 (100.0) | 30 (33.0) | 61 (67.0) |
|
| Age
(years)b |
|
|
|
|
|
≥60 | 52 (65.8) | 17 (32.7) | 35 (67.3) | 0.95 |
|
<60 | 27 (34.2) | 9 (33.3) | 18 (66.7) |
|
| Sex |
|
|
|
|
|
Male | 75 (82.4) | 26 (35.1) | 48 (64.9) | 0.74 |
|
Female | 16 (17.6) | 4 (26.7) | 11 (73.3) |
|
| History of
NMIBCc |
|
|
|
|
| No | 69 (86.3) | 26 (37.7) | 43 (62.3) | 0.36 |
|
Yes | 11 (13.7) | 2 (18.2) | 9 (81.8) |
|
| Associated
pTisd |
|
|
|
|
| No | 65 (90.3) | 21 (32.3) | 44 (67.7) | 0.37 |
|
Yes | 7 (9.7) | 4 (57.1) | 3 (42.9) |
|
| Grade |
|
|
|
|
|
Low | 33 (36.3) | 7 (21.2) | 26 (78.8) | 0.098 |
|
High | 58 (63.7) | 21 (38.2) | 34 (61.8) |
|
| Tumor stage |
|
|
|
|
| Ta | 50 (54.9) | 14 (28.0) | 36 (72.0) | 0.27 |
| T1 | 41 (45.1) | 16 (39.0) | 25 (61.0) |
|
|
Phenotypee |
|
|
|
|
|
Basal | 1 (1.3) | 0 (0.0) | 1 (100.0) | 0.76 |
|
Non-basal | 78 (98.7) | 26 (33.3) | 52 (66.7) |
|
 | Table VI.Association of clinicopathological
variables and PLEKHS1 DNA mutated profile with MIBC from the
second series. |
Table VI.
Association of clinicopathological
variables and PLEKHS1 DNA mutated profile with MIBC from the
second series.
| Characteristic | Total population, n
(%) | PLEKHS1
mutated, n (%) | Not mutated, n
(%) |
P-valuea |
|---|
| Total | 90 (100) | 34 (64.8) | 56 (35.2) |
|
| Ageb |
|
|
|
|
|
≥60 | 63 (74.1) | 26 (41.3) | 37 (58.7) | 0.24 |
|
>60 | 22 (25.9) | 6 (27.3) | 16 (72.7) |
|
| Sex |
|
|
|
|
|
Male | 75 (83.3) | 29 (38.7) | 46 (61.3) | 0.70 |
|
Female | 15 (16.7) | 5 (33.3) | 10 (66.7) |
|
| History of
NMIBCc |
|
|
|
|
| No | 71 (85.5) | 26 (36.6) | 45 (63.4) | 0.38 |
|
Yes | 12 (14.5) | 6 (50.0) | 6 (50.0) |
|
| Associated
pTisd |
|
|
|
|
| No | 62 (80.5) | 25 (40.3) | 37 (59.7) | 0.62 |
|
Yes | 15 (19.5) | 5 (33.3) | 10 (66.7) |
|
| Tumor stage |
|
|
|
|
| T2 | 32 (35.6) | 16 (50.0) | 16 (50.0) | 0.076 |
|
≥T3 | 58 (64.4) | 18 (31.0) | 40 (69.0) |
|
| Lymph node
statuse |
|
|
|
|
| N− | 35 (55.6) | 14 (40.0) | 21 (60.0) | 0.52 |
| N+ | 28 (44.4) | 9 (32.1) | 19 (67.9) |
|
|
Phenotypef |
|
|
|
|
|
Basal | 26 (30.6) | 16 (61.5) | 10 (38.5) | 0.0025 |
|
Non-basal | 59 (69.4) | 16 (27.1) | 43 (72.9) |
|
mRNA expression of PLEKHS1
The mRNA expression of PLEKHS1 was assessed
in the first series (n=154). The median mRNA PLEKHS1 level
was 19.7 [range, 0.0–972.6] with NMIBC and 25.7 [0.0–2288.1] with
MIBC, vs. 0.0 [0.0–21.4] in normal bladder samples. PLEKHS1
was overexpressed in NMIBC and MIBC versus normal bladder tissue
(P=0.00031 and P=0.000025), with no difference in level between
NMIBC and MIBC (P=0.51). Overall, 22.5 and 27.7% of NMIBC and MIBC
tumors showed PLEKHS1 overexpression (vs. 0% in normal
bladder samples, P<0.01). PLEKHS1 mRNA level was not
associated with DNA mutations in NMIBC (P=0.39) or MIBC
(P=0.84).
Prognostic value of PLEKHS1 mRNA
overexpression
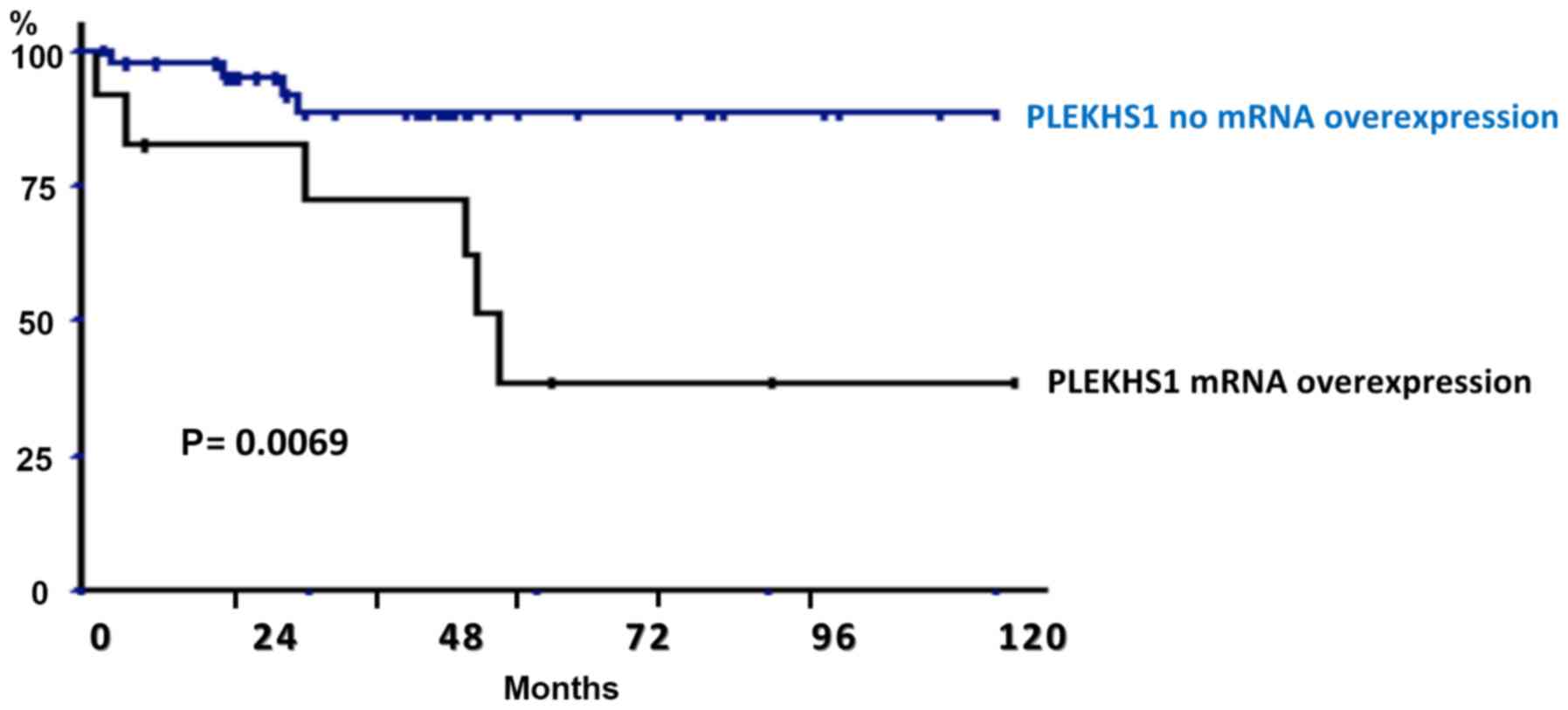
PLEKHS1 mRNA overexpression was not
significantly associated with clinical variables in NMIBC (Table VII) or MIBC (Table VIII). With NMIBC, PLEKHS1
overexpression was associated with worse PFS on univariate analysis
(P=0.0069) (Fig. 1). Five- and
10-year PFS rates were 72.4 and 38.8% with PLEKHS1
overexpression versus 88.9 and 88.7% without PLEKHS1
overexpression. Other clinical variables affecting PFS on
univariate analysis with P<0.10 on log-rank test included T
stage and grade and were retained for multivariate analysis. On
multivariate analysis, PLEKHS1 overexpression remained an
independent prognostic factor of PFS (P=0.034) (Table IX).
 | Table VII.Association of clincopathological
variables and PLEKHS1 mRNA level with NMIBC from the first
series. |
Table VII.
Association of clincopathological
variables and PLEKHS1 mRNA level with NMIBC from the first
series.
| Characteristic | Total population, n
(%) | PLEKHS1 mRNA
no overexpression, n (%) | PLEKHS1 mRNA
overexpression, n (%) |
P-valuea |
|---|
| Total | 71 (100) | 55 (77.5) | 16 (22.5) |
|
| Age |
|
|
|
|
|
≥60 | 56 (78.9) | 43 (76.8) | 13 (23.2) | 0.79 |
|
<60 | 15 (21.1) | 12 (80.0) | 3 (20.0) |
|
| Sex |
|
|
|
|
|
Male | 63 (88.7) | 48 (76.2) | 15 (23.8) | 0.79 |
|
Female | 8 (11.3) | 7 (87.5) | 1 (12.5) |
|
| Smoking status |
|
|
|
|
|
Non-smoker | 33 (46.5) | 25 (75.8) | 8 (24.2) | 0.75 |
|
Smoker | 38 (53.5) | 30 (78.9) | 8 (21.1) |
|
| History of
NMIBC |
|
|
|
|
| No | 39 (54.9) | 31 (79.5) | 8 (20.5) | 0.65 |
|
Yes | 32 (45.1) | 24 (75.0) | 8 (25.0) |
|
| Associated
pTis |
|
|
|
|
| No | 69 (97.2) | 54 (78.3) | 15 (21.7) | 0.35 |
|
Yes | 2 (2.8) | 1 (50.0) | 1 (50.0) |
|
| Grade |
|
|
|
|
|
Low | 25 (35.2) | 21 (84.0) | 4 (16.0) | 0.33 |
|
High | 46 (64.8) | 34 (73.9) | 12 (26.1) |
|
| Tumor stage |
|
|
|
|
|
pTa | 42 (59.2) | 35 (83.3) | 7 (16.7) | 0.15 |
|
pT1 | 29 (40.8) | 20 (69.0) | 9 (31.0) |
|
 | Table VIII.Association of clinicopathological
variables and PLEKHS1 mRNA level with MIBC from the first
series. |
Table VIII.
Association of clinicopathological
variables and PLEKHS1 mRNA level with MIBC from the first
series.
| Characteristic | Total population, n
(%) | PLEKHS1 mRNA
no overexpression, n (%) | PLEKHS1 mRNA
overexpression, n (%) |
P-valuea |
|---|
| Total | 83 (100) | 60 (72.3) | 23 (27.7) |
|
| Age |
|
|
|
|
|
≥60 | 61 (73.5) | 44 (72.1) | 17 (27.9) | 0.96 |
|
<60 | 22 (26.5) | 16 (72.7) | 6 (27.3) |
|
| Sex |
|
|
|
|
|
Male | 66 (79.5) | 45 (68.2) | 21 (31.8) | 0.10 |
|
Female | 17 (20.5) | 15 (88.2) | 2 (11.8) |
|
| Smoking status |
|
|
|
|
|
Non-smoker | 34 (41.0) | 23 (67.6) | 11 (32.4) | 0.43 |
|
Smoker | 49 (59.0) | 37 (75.5) | 12 (24.5) |
|
| History of
NMIBC |
|
|
|
|
| No | 59 (71.1) | 39 (66.1) | 20 (33.9) | 0.05 |
|
Yes | 24 (28.9) | 21 (87.5) | 3 (12.5) |
|
| Associated
pTis |
|
|
|
|
| No | 73 (88.0) | 52 (71.2) | 21 (28.7) | 0.56 |
|
Yes | 10 (12.0) | 8 (80.0) | 2 (20.0) |
|
| Grade |
|
|
|
|
|
Low | 34 (41.0) | 25 (73.5) | 9 (26.5) | 0.83 |
|
High | 49 (59.0) | 35 (71.4) | 14 (28.6) |
|
| Tumor stage |
|
|
|
|
|
pTa | 58 (69.9) | 39 (68.4) | 19 (32.8) | 0.12 |
|
pT1 | 25 (30.1) | 21 (84.0) | 4 (16.0) |
|
 | Table IX.Cox proportional-hazards regression
analysis of factors affecting progression-free survival with
non-muscle-invasive bladder cancer. |
Table IX.
Cox proportional-hazards regression
analysis of factors affecting progression-free survival with
non-muscle-invasive bladder cancer.
|
| Progression-free
survival |
|---|
|
|
|
|---|
| Prognostic
factor | HR | 95% CI | P-value |
|---|
| T stage | 3.92 | (0.79–19.36) | 0.093 |
| Grade | 2.19 | (0.20–24.58) | 0.52 |
| PLEKHS1
overexpression | 4.01 | (1.11–14.50) | 0.034 |
Overall, 21/28 patients (75%) who received Bacillus
Calmette-Guerin (BCG) therapy showed recurrent NMIBC or progression
to an invasive tumor during follow-up, including 15 (53.6%) within
the first 2 years. The median PLEKHS1 mRNA level was higher
but not significantly with BCG-refractory tumor or early recurrence
(<24 months) than for BCG responders (no recurrence or >24
months): 40.8 [range, 0.0–375.5] vs. 15.2 [0.6–381.6] (P=0.80).
With MIBC, PLEKHS1 overexpression was not
associated with RFS or OS (P=0.33 and P=0.057, respectively).
Discussion
Mutations of PLEKHS1 are non-coding and are
located in the gene promoter and therefore cannot be detected by
whole-exome analyses. This is also the case for non-coding
mutations located on the upstream regulatory sequences for genes
such as TERT, WD repeat domain 74 (WDR74) and
succinate dehydrogenase complex subunit D (SDHD) (8). Point mutations in the TERT
promoter are the best known and occur in a subset of tumors with
clinical interest. In bladder urothelial carcinomas, almost two
thirds of tumors show somatic test mutations (13).
To our knowledge, this is the first study to assess
PLEKHS1 DNA mutations and mRNA expression in bladder cancer. This
assessment is of outmost importance because the association of gene
expression and the prognostic impact of promoter mutations in the
cancer remain unknown. Whole-genome sequencing of non-coding
somatic mutations, rather than these exome studies, might provide
answers to these deliberations.
We investigated DNA mutations and mRNA expression in
a large series of 154 bladder cancer cases. The results of DNA
analysis were then validated in an independent second series
(n=181). In these two series, tumors were half non-muscle invasive
and half muscle invasive. Characteristics of both populations are
consistent with urothelial carcinoma presentation. Moreover, we
observed the classical prognostic factors (TNM stage and
grade).
We showed a high frequency of promoter DNA mutations
for PLEKHS1, detected in about one-third of bladder tumors
(NMIBC and MIBC). Our results are consistent with the princeps
study of Weinhold et al describing PLEKHS1 DNA
mutations in 8/20 samples (40%) of bladder cancer (8). This high rate of DNA mutations supports
the concept of an essential involvement of this gene in bladder
carcinogenesis, possibly similar to human TERT. It is also
of interest as a biomarker to detect circulating tumor DNA that can
be used for a variety of clinical and research purposes for bladder
cancer.
In our study, PLEKHS1 mutations were
particularly frequent with the basal MIBC phenotype, which is known
to have poor prognosis but is associated with improved survival
after neoadjuvant chemotherapy (14). Further research is needed to assess
the role of PLEKHS1 in the chemosensitivity of MIBC.
PLEKHS1 mRNA was overexpressed in
approximatively one-quarter of bladder tumors, regardless of stage
and grade. As well, the expression was particularly high in some
tumor samples, but normal bladder tissue showed very low
levels.
Although the rates of mutations appear to be almost
the same as the proportion of mRNA overexpression, we did not find
an association between DNA alterations and mRNA expression. This is
an unexpected result suggesting that mutations in the
PLEKHS1 promoter probably do not affect transcription and
are not responsible for the increased mRNA expression in our
bladder tumor series. Our results disagree with those of Weinhold
et al., who showed PLEKHS1 mutations inversely
correlated with mRNA expression level in a small series of 20
bladder tumors (8). In contrast,
somatic mutations in the TERT promoter increased the expression of
telomerase (13).
In NMIBC, PLEKHS1 overexpression was
associated with poor prognosis and increased risk of progression to
muscle-invasive disease. The overexpression remained an independent
prognostic factor on multivariate analysis, which is quite
remarkable. Indeed, except for TNM stage and pathological grade, we
have no molecular markers to distinguish tumors that would be able
to progress in muscle-invasive disease during follow-up. This is
especially crucial for pT1 high-grade tumors, which have been shown
to progress in 30 to 50% of cases during the first 5 years. Early
identification of patients at risk of disease progression may lead
to more aggressive therapeutic strategies, such as early
cystectomy. One of the main issues is the potential response of
these tumors to BCG therapy. In our series, even though
PLEKHS1 mRNA expression was three times higher with early
recurrent or refractory tumor than other tumors, PLEKHS1 was
not a significant predictive factor of response to BCG therapy.
However, the small number of patients who received BCG may explain
the lack of statistical power in our study.
One limitation of our study is that a longer
follow-up or a larger cohort of patients may be needed to show
significance, especially for pT1 tumors and response to BCG. To
confirm these data, prospective clinical studies associated with a
molecular evaluation of tumors are needed. We tried to assess
protein expression by western blot analysis and
immunohistochemistry, but the two primary anti-PLEKHS1 antibodies
used (i.e., HPA037583, Sigma and H00079949-T01, Abnova) showed no
specific protein bands on western blot analysis or on
immunostaining (data not shown), which suggests no qualitative
antibody available for PLEKHS1. Functional analyses should probably
be done in parallel to investigate the role of PLEKHS1 in
carcinogenesis. Recently, Grossmann et al. identified an
interaction between PIK3R3 (p55c regulatory subunit of PI3 kinase)
and PLEKHS1 (15), but the
significance of this protein interaction remains uncertain. In our
series, even if PLEKHS1 mutations were significantly more
frequent with the basal than non-basal MIBC phenotype, we did not
identify any other associated gene or pathway alterations.
Therefore, despite its prognostic value, PLEKHS1 cannot be
considered a therapeutic target at this time.
Our results support the involvement of
PLEKHS1 in bladder carcinogenesis. DNA mutations were
observed in almost one-third of bladder cancers and approximately
one quarter of NMIBC and MIBC tumors showed mRNA overexpression. In
MIBC, PLEKHS1 mutations seem frequent with the basal
phenotype. In NMIBC, PLEKHS1 overexpression may be an
independent prognostic factor of progression to muscle-invasive
disease. Our study has many biases since it is a retrospective
clinical study, small cohort and can only be considered as a
preliminary study. The clinical interest remains to be demonstrated
by a prospective study centered on high grade NMIBC. Further
studies are needed to dissect the mechanisms of action and the
specific role of PLEKHS1 in bladder carcinogenesis and to
develop therapeutic inhibitors.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
GP and CLG contributed to the acquisition and
interpretation of data, and manuscript writing. SV, AS and FL
contributed to the acquisition of data. FR, YA, NBD, MZ and BT
aquired the data. DD contributed to acquisition and interpretation
of data. IB contributed to the conception and design of the study,
and interpretation of data. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The present study was approved by the local ethics
and research committee of Mondor and Foch Hospitals, Centre
d'Éthique Clinique de l'Hôpital Cochin (descriptive retrospective
study). All patients provided written informed consent.
Patient consent for publication
All patients provided informed consent.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lebret T, Hervé JM, Yonneau L, Barré P,
Lugagne PM, Butreau M, Molinié V and Botto H: Study of survival
after cystectomy for bladder cancer. Report of 504 cases. Prog
Urol. 10:553–560. 2000.(In French). PubMed/NCBI
|
|
3
|
Salvi S, Calistri D, Gurioli G, Carretta
E, Serra L, Gunelli R, Zoli W and Casadio V: Copy number analysis
of 24 oncogenes: MDM4 identified as a putative marker for low
recurrence risk in non muscle invasive bladder cancer. Int J Mol
Sci. 15:12458–12468. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Cancer Genome Atlas Research Network, .
Comprehensive molecular characterization of urothelial bladder
carcinoma. Nature. 507:315–322. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Williams SV, Hurst CD and Knowles MA:
Oncogenic FGFR3 gene fusions in bladder cancer. Hum Mol Genet.
22:795–803. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Massari F, Bria E, Ciccarese C, Munari E,
Modena A, Zambonin V, Sperduti I, Artibani W, Cheng L, Martignoni
G, et al: Prognostic value of beta-tubulin-3 and c-Myc in muscle
invasive urothelial carcinoma of the bladder. PLoS One.
10:e01279082015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Pignot G, le Goux C and Bieche I: Recent
advances in bladder urothelial carcinogenesis. Bull Cancer.
102:1020–1035. 2015.(In French). View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Weinhold N, Jacobsen A, Schultz N, Sander
C and Lee W: Genome-wide analysis of noncoding regulatory mutations
in cancer. Nat Genet. 46:1160–1165. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sobin L, Gospodarowicz M and Wittekind C:
TNM Classification of Malignant Tumors. John Wiley and Sons;
Hoboken, NJ, USA: pp. 262–265. 2009
|
|
10
|
Molinié V: Classification of bladder
tumors in 2006. Prog Urol FMC. pp16. 7–10. 2006.
|
|
11
|
Rebouissou S, Bernard-Pierrot I, de
Reyniès A, Lepage ML, Krucker C, Chapeaublanc E, Hérault A, Kamoun
A, Caillault A, Letouzé E, et al: EGFR as a potential therapeutic
target for a subset of muscle-invasive bladder cancers presenting a
basal-like phenotype. Sci Transl Med. 6:244ra912014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Pignot G, Bieche I, Vacher S, Güet C,
Vieillefond A, Debré B, Lidereau R and Amsellem-Ouazana D:
Large-scale real-time reverse transcription-PCR approach of
angiogenic pathways in human transitional cell carcinoma of the
bladder: Identification of VEGFA as a major independent prognostic
marker. Eur Urol. 56:678–688. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Killela PJ, Reitman ZJ, Jiao Y, Bettegowda
C, Agrawal N, Diaz LA Jr, Friedman AH, Friedman H, Gallia GL,
Giovanella BC, et al: TERT promoter mutations occur frequently in
gliomas and a subset of tumors derived from cells with low rates of
self-renewal. Proc Natl Acad Sci USA. 110:6021–6026. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
McConkey DJ, Choi W, Shen Y, Lee IL,
Porten S, Matin SF, Kamat AM, Corn P, Millikan RE, Dinney C, et al:
A prognostic gene expression signature in the molecular
classification of chemotherapy-naïve urothelial cancer is
predictive of clinical outcomes from neoadjuvant chemotherapy: A
phase 2 trial of dose-dense methotrexate, vinblastine, doxorubicin,
and cisplatin with bevacizumab in urothelial cancer. Eur Urol.
69:855–862. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Grossman HB, Natale RB, Tangen CM,
Speights VO, Vogelzang NJ, Trump DL, deVere White RW, Sarosdy MF,
Wood DP Jr, Raghavan D and Crawford ED: Neoadjuvant chemotherapy
plus cystectomy compared with cystectomy alone for locally advanced
bladder cancer. N Engl J Med. 349:859–866. 2003. View Article : Google Scholar : PubMed/NCBI
|