Introduction
Salivary adenoid cystic carcinoma (SACC) is a rare
head and neck malignancy, which has an incidence of 0.5–2.5 per
100,000 people with a slightly higher occurrence in women (60%) in
the USA (1,2). The mean diagnostic age is ~57 years old
and the overall 5-, 10- and 15-year survival rates are ~90, 80 and
69%, respectively (2). A total of
70–80% of SACC cases arises from the parotid glands, including the
submandibular gland (10%) or the sublingual and minor salivary
glands (5%) (1). Salivary gland
malignancies are characterized by unpredictable growth expansion,
considerable perineural invasion (PNI) and high risk of metastasis,
which eventually leads to low survival outcomes (3). The lung is the most common metastatic
site, followed by bone, liver and other sites, which account for
80, 15 and 5% of SACC cases, respectively. Because of its low
frequency and unique clinical characteristics, the underlying
mechanisms of SACC metastasis and PNI remain unknown.
In addition to the extracellular matrix, cancer
cells are surrounded by fibroblasts, vascular endothelial cells and
immune cells (4). It has been
demonstrated that cancer-associated fibroblasts (CAFs) localized
within the tumor microenvironment can promote cancer malignant
transition by enhancing tumor growth, blood vessel formation,
inflammation and metastasis (5).
CAFs exert pro-tumorigenic effects and provide a distinctive tumor
microenvironment to promote tumor cell proliferation and invasion
(6). The crosstalk between cancer
cells and CAFs is critical for tumor initiation and progression,
and targeting CAFs may therefore represent a promising therapeutic
tool in the management of cancer (7).
Exosomes are cell-derived vesicles with a diameter
of 40–100 nm, which comprise a variety of DNA, RNA, lipids and
proteins that can be transferred from one cell to another via
membrane vesicle trafficking (8).
Previous studies demonstrated that intercellular communication
between tumor-derived exosomes and all types of cell within the
tumor microenvironment can influence cancer signaling pathways
involving epithelial-mesenchymal transition, cancer growth,
angiogenesis and metastasis (9,10). For
example, gastric cancer cells regulate liver microenvironment
through epidermal growth factor receptor-containing exosomes, which
can therefore lead to liver metastasis (9). Furthermore, bladder cancer cells can
trigger fibroblast differentiation into CAFs through
exosome-mediated transforming growth factor β delivery and SMAD
signaling activation (10). The role
of exosomes in the crosstalk between SACC cells and CAFs requires
therefore more attention.
The present study explored the mechanism underlying
metastasis and PNI features in SACC mainly by focusing on the
exosome-mediated communication between SACC cells and CAFs. The
downstream effect of this communication on nerve growth factor
(NGF) signaling was also evaluated.
Materials and methods
Cell culture
The human salivary adenoid cystic carcinoma SACC-83
(Shanghai Jingkang Biotechnology) and human periodontal ligament
fibroblast (HPLF) (Shanghai Zishi Biotechnology) cell lines were
tested negative for mycoplasma and were authenticated by short
tandem repeat markers. Cells were cultured in high-glucose
Dulbecco's modified Eagle's medium (DMEM; Invitrogen; Thermo Fisher
Scientific, Inc.) containing 10% fetal bovine serum (FBS; Gibco;
Thermo Fisher Scientific, Inc.) and 1% penicillin/streptomycin
(Thermo Fisher Scientific, Inc.), and maintained at 37°C in a
humidified incubator containing 5% CO2.
Exosome isolation and fluorescence
labeling
SACC-83 cells (5×106) were seeded and
cultured in T-75 cm2 flask with DMEM complete medium
until they reach 80% confluence and further grown for 24 h in
serum-free medium. Cell conditioned medium was then collected to
isolate exosomes using the Total Exosome Isolation Reagent kit
(Invitrogen; Thermo Fisher Scientific, Inc.; cat. no. 4478359).
Briefly, conditioned medium was centrifuged at 2,000 × g for 30 min
at 4°C to remove cells and debris, and supernatant was collected,
mixed with 5 ml Total Exosome Isolation Reagent and incubated at
4°C overnight. The sample was centrifuged at 10,000 × g for 1 h at
4°C and the subsequent exosome pellet was resuspended in 100 µl
PBS. The exosomes from five flasks of cells were pooled. The size
and particle concentration of exosomes were then determined with
the ZetaView nanoparticle tracking analysis (NTA; Particle Metrix
Inc.). Exosomes were quantified with a BCA Protein Assay kit
(Thermo Scientific) using an Absorbance Reader (ELX-800, BioTek
Instruments, Inc.), labeled with PKH67 Green Fluorescent Cell
Linker (Sigma-Aldrich; Merck KGaA; cat. no. PKH67GL) according to
the manufacturer's protocol.
Observation of exosome
internalization
HPLF cells (5×105) were incubated with
various amounts of PKH67-labeled exosomes (0, 25, 50 or 75 µg) and
for different incubation times (0, 3, 6 or 9 h). Cell nucleus was
stained with DAPI and cells were then fixed in 4% paraformaldehyde
for 10 min at room temperature (RT). Cells were visualized with
Zeiss LSM 510 confocal microscope (magnification, ×630, Oberkochen,
Germany).
Exosome visualization by transmission
electron microscopy (TEM)
Briefly, freshly isolated exosomes of which the
density was ~1.15 g/cm3 were suspended in 2%
paraformaldehyde over night at 4°C, mounted on formvar carbon
coated copper grids, fixed with 2% glutaraldehyde for 5 min and
washed in sterile distilled water (11). Samples were dried and examined with a
JEOL JEM-1010 transmission electron microscope (JEOL, Ltd.).
Western blotting
Exosomes or HPLF cell pellets were lysed in RIPA
buffer containing 50 mM Tris (pH 7.4), 150 mM NaCl, 1% Triton
X-100, 1% sodium deoxycholate and 0.1% SDS with protease inhibitor
cocktail (Roche Applied Science), and quantified with a BCA Protein
Assay kit (Thermo Scientific). Then, proteins (15 µg) isolated from
exosomes or cell lysate were separated by SDS-PAGE (10% gel) and
transferred onto polyvinylidene fluoride membranes (EMD Millipore).
Membranes were blocked with 5% skimmed milk in 1X Tris-buffered
saline (TBS) containing 0.1% Tween-20 at RT for 2 h, and incubated
with primary antibodies against CD63 (1:1,000; Abcam; cat. no.
ab59479), TSG101 (1:1,000; Abcam; cat. no. ab83), CD81 (1:1,000;
Thermo Fisher Scientific Inc.; cat. no. 10630D), glucose-regulated
protein 94 (GRP94; Cell Signaling Technology Inc.; cat. no. 20292),
NGF (Abcam; cat. no. ab6199, NGF neutralizing antibody), or β-actin
(Cell Signaling Technologies Inc.; cat. no. 4970) overnight at 4°C.
Membranes were washed and then incubated with horseradish
peroxidase-conjugated secondary antibodies (1:50,000; goat
anti-mouse; cat. no. 115-035-146, or goat anti-rabbit; cat. no.
111-035-144; Jackson ImmunoResearch Inc.) for 1 h at RT. Reactive
bands were detected with Immobilon ECL Ultra Western HRP Substrate
(EMD Millipore). The relative intensities of the bands were
quantified by densitometry and normalized to β-actin signal levels.
Densitometric quantification was performed by Image-Pro Plus 6.0
software (Media Cybernetics, Inc. Rockville, MD, USA).
Cell invasion assay
SACC-83 cells (1×105) were suspended in
the supernatant collected from PBS or exosomes-treated HPLF cells
and placed into the upper chamber of Transwell cell culture insert
(diameter, 6.5 mm; pore size, 8.0 µm; Corning Inc.) of Matrigel (BD
Biosciences) pre-coated 24-well plates. DMEM containing 10% FBS was
added into the lower chamber. Following 24 h culture at 37°C,
SACC-83 cells that have migrated onto the lower surface of the
inserts were fixed in 4% paraformaldehyde for 30 min at RT, and
stained with 0.1% crystal violet dissolved in PBS for 20 min at RT.
Non-migrated cells at the bottom of the insert were removed and
cells adherent to the lower surface of the insert were counted in
six random fields using a light microscope at ×100 magnification
(Olympus IX5, Olympus America Inc.). The experiment was repeated at
least three times.
Cell culture wound closure assay
SACC-83 cells were grown to 100% confluence in
12-well plates and a scratch was generated with a sterile pipette
tip. Cells were then incubated with the supernatant collected from
PBS or exosomes-treated HPLF cells. Images were taken immediately
(0 h) at ×100 magnification following 24 h culture at 37°C using a
light microscope (Olympus IX5). Cell migration ability was
determined according to the percentage of area reduction or wound
closure by Image-Pro Plus 6.0 software.
RNA isolation reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was isolated from HPLF cells using
TRIzol® modified Ultrapure RNA kit (CW Biotech)
according to the manufacturer's instructions. The first strand of
complementary DNA was synthesized using Oligo(dT) primer by
SuperScript II reverse transcriptase (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol.
Subsequently, qPCRs were performed to analyze mRNA levels with SYBR
Green PCR Master mix (Roche Applied Science) using StepOnePlus™
Real-Time PCR system (Thermo Fisher Scientific, Inc.). The PCR
cycling conditions were 95°C for 30 sec followed by 40 cycles of
95°C for 5 sec and 60°C for 30 sec. The relative expression levels
were normalized to the housekeeping gene GAPDH and calculated using
the 2−∆∆Cq method (12).
The sequences of the primers used in the present study are listed
in Table I, and were synthesized by
Thermo Fisher Scientific, Inc.
 | Table I.Sequences of primers used for for
reverse transcription-quantitative polymerase chain reaction. |
Table I.
Sequences of primers used for for
reverse transcription-quantitative polymerase chain reaction.
| Genes | Primer
sequences |
|---|
| IL-6 |
|
Forward |
5′-TACATCCTCGACGCATCTC-3′ |
|
Reverse |
5′-AGCTCTGGCTGTTCCTCAC-3′ |
| IL-1β |
|
Forward |
5′-AAACAGATGAAGTGCTCCTTCCAGG-3′ |
|
Reverse |
5′-GTCGGAGATTCGTAGCTGGA-3′ |
| IL-8 |
|
Forward |
5′-GAGAGTGATTGAGAGTGGACCAC-3′ |
|
Reverse |
5′-CACAACCCTCTGCACCCAGTTT-3′ |
| TNF-α |
|
Forward |
5′-CCAGGCATCAGATCATCTTC-3′ |
|
Reverse |
5′-GGATGTTCGTCCTCCTCACAG-3′ |
| MCP-1 |
|
Forward |
5′-CAGCCAGATGCAATCAATGCC-3′ |
|
Reverse |
5′-TGGAATCCTGAACCCACTTCT-3′ |
| NGF |
|
Forward |
5′-ACCCGCAACATTACTGTGGACC-3′ |
|
Reverse |
5′-GACCTCGAAGTCCAGATCCTGA-3′ |
| NTRK1 |
|
Forward |
5′-CACTAACAGCACATCTGGAGACC-3′ |
|
Reverse |
5′-TGAGCACAAGGAGCAGCGTAGA-3′ |
| NTRK2 |
|
Forward |
5′-ACAGTCAGCTCAAGCCAGACAC-3′ |
|
Reverse |
5′-GTCCTGCTCAGGACAGAGGTTA-3′ |
| NTF4 |
|
Forward |
5′-GCAAGGCTGATAACGCTGAGGA-3′ |
|
Reverse |
5′-CCTGGGCATCAGCGGTCAATG-3′ |
| CXCR4 |
|
Forward |
5′-CTCCTCTTTGTCATCACGCTTCC-3′ |
|
Reverse |
5′-GGATGAGGACACTGCTGTAGAG-3′ |
| CXCL12 |
|
Forward |
5′-CTCAACACTCCAAACTGTGCCC-3′ |
|
Reverse |
5′-CTCCAGGTACTCCTGAATCCAC-3′ |
| BDNF |
|
Forward |
5′-CATCCGAGGACAAGGTGGCTTG-3′ |
|
Reverse |
5′-GCCGAACTTTCTGGTCCTCATC-3′ |
| iNOS |
|
Forward |
5′-GCTCTACACCTCCAATGTGACC-3′ |
|
Reverse |
5′-CTGCCGAGATTTGAGCCTCATG-3′ |
| MMP2 |
|
Forward |
5′-ATAACCTGGATGCCGTCGT-3′ |
|
Reverse |
5′-AGGCACCCTTGAAGAAGTAGC-3′ |
| MMP9 |
|
Forward |
5′-GAACCAATCTCACCGACAGG-3′ |
|
Reverse |
5′-GCCACCCGAGTGTAACCATA-3′ |
| GAPDH |
|
Forward |
5′-TTGCCATCAATGACCCCTTCA-3′ |
|
Reverse |
5′-CGCCCCACTTGATTTTGGA-3′ |
RNA sequencing and data analysis
HPLF cells were treated with PBS or SACC-83-derived
exosomes. Total RNA was isolated using TRIzol® modified
Ultrapure RNA kit (CW Biotech) and used for RNA sequencing
analysis. The complementary DNA library was constructed and
sequenced by BGI Genomics Services using BGISEQ-500 platform.
Bioinformatics workflow including data filtering, mapping
transcript prediction, differential gene expression analysis, gene
ontology (GO; http://geneontology.org) and Kyoto
Encyclopedia of Genes and Genomes (KEGG; http://www.genome.jp/kegg) pathway analysis were
performed by procedures established by BGI Genomics Services. The
in-depth bioinformatics analysis of NTRK1 expression in the
Oncomine database was carried out at https://www.oncomine.org/.
Cytokine secretion measurement
Human β-NGF secreted by HPLF cells in culture
supernatants was quantified using ELISA (cat. no. DY256-05; R&D
Systems, Minneapolis, MN, USA), according to the manufacturer's
instructions.
Immunohistochemistry (IHC)
analyses
Human SACC tissues were obtained by the Changhai
Hospital. The SACC patients were treated in the Hospital between
January 2016 and May 2018, and diagnosed with a surgical biopsy by
histopathology. The mean age was 56 years (range, 50–65 years). All
patients provided written informed consent, and the use of human
samples was approved by the Shanghai Changhai Hospital Ethics
Committee. Tissues were fixed in 10% formalin for 24 h at RT and
embedded in paraffin. Tissue sections (5 µm-thick) were cut for IHC
staining. The antigen retrieval was performed using a pressure
cooker at 121°C. Endogenous peroxidase activity was blocked by 3%
hydrogen peroxide at RT for 20 min. Normal goat serum (10%; CW
Biotech) was used to block nonspecific binding at RT for 1 h.
Sections were incubated with NTRK1 rabbit monoclonal primary
antibody (Abcam; cat. no. ab76291; 1:100; clone, EP1058Y) at 4°C
overnight. The subsequent procedures were performed using a SP
Rabbit HRP kit (DAB) (cat. no. CW2035, CW Biotech) in accordance
with the manufacturer's protocol. The tissue sections were
counterstained by immersing sides in hematoxylin for 2 min at RT.
Antibody staining was then observed under a light microscope at
×100, ×200 and ×400 magnification (Olympus IX5).
Statistical analysis
Two-tailed Student's t-test was used to compare two
groups. Comparison among multiple groups was performed using
one-way analysis of variance followed by Tukey's post hoc test of
significance between individual groups. Data are presented as the
means ± standard error of the mean. Statistical analyses were
performed with GraphPad Prism (v7.0; GraphPad Software, Inc.).
P<0.05 was considered to indicate a statistically significant
difference. Each experiment was repeated at least three times.
Results
SACC-derived exosomes are efficiently
internalized into fibroblasts
Exosomes were isolated from conditioned medium of
SACC-83 cells. As presented in Fig.
1A, exosomes were characterized by the three specific exosome
markers CD63, CD81 and TSG101 (13);
however, GRP94 was absent. Furthermore, the size and concentration
(~8.2×1010 particles/ml) of exosomes were determined by
NTA (Fig. 1B) and TEM (Fig. 1C). The results demonstrated that
exosome diameter ranged between 100 and 150 nm. To examine the
transfer ability of exosomes to HPLF cells, SACC-83-derived
exosomes were labeled with PKH67 and incubated with HPLF cells for
3, 6 or 9 h (Fig. 1D). The results
from confocal microscopy analysis demonstrated that green
fluorescent exosomes were internalized into HPLF cells in a time-
and concentration-dependent manner (Fig.
1D and E, respectively). These results indicated that
SACC-83-derived exosomes may be transferred into HPLF cells.
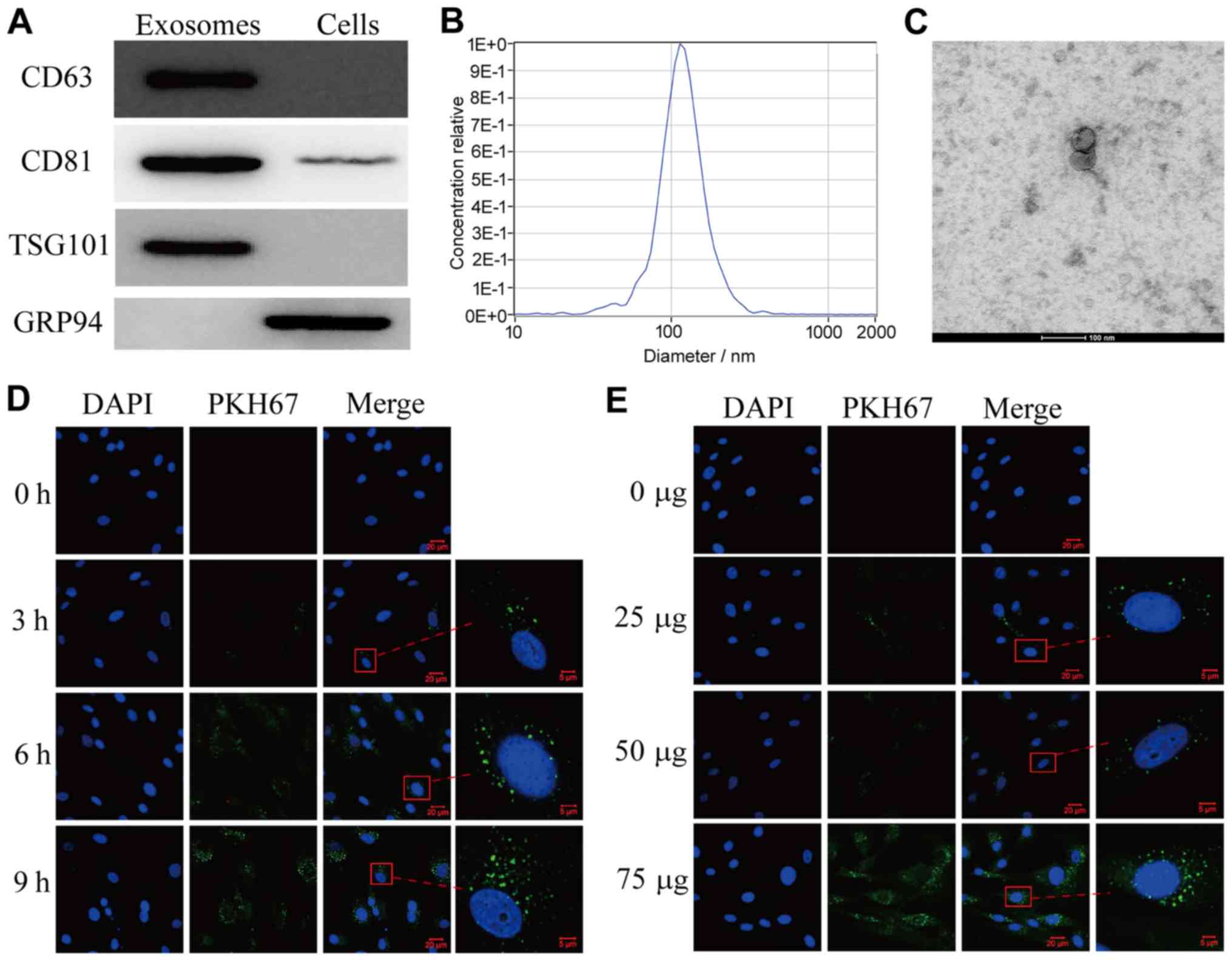 | Figure 1.Exosomes purified from SACC-83 cells
are delivered into HPLF cells. (A) Western blotting analysis
showing the presence of CD63, CD81, TSG101 and the absence of GRP94
in SACC-83 cell-derived exosomes. (B) Nanoparticle tracking
analysis and (C) transmission electron microscopy images showing
exosomes isolated from SACC-83 cells. Magnification, ×630; scale
bar, 20 µm. (D) Confocal microscopy images showing the
internalization of PKH67-labeled exosomes in HPLF cells at
different incubation times. Scale bar, 5 µm. (E) Confocal
microscopy images showing the delivery of 0, 25, 50 or 75 µg of
exosomes into HPLF cells after 6 h. Scale bar, 5 µm. Magnified
views are shown on the right. GRP94, glucose-regulated protein 94;
HPLF, human periodontal ligament fibroblast; SACC, salivary adenoid
cystic carcinoma. |
Tumor-derived exosomes stimulate HPLF
cells to facilitate SACC-83 cell migration and enhance the
expression of proinflammatory and perineural mediators in HPLF
cells
Exosomes secreted from cells can transfer biological
information to other recipient cells and may therefore regulate the
crosstalk between cancer cells and tumor microenvironment (14). The present study investigated whether
the administration of SACC-83-derived exosomes to HPLF cells would
result in HPLF cell change of phenotype, and subsequently influence
the metastasis of SACC-83 cells. The results demonstrated that
SACC-83 exhibited a higher invasive ability following stimulation
with the supernatant from exosome-educated HPLF cells compared with
control (Fig. 2A). Furthermore, the
expression levels of matrix metalloproteinases 2 and 9 (MMP2 and
MMP9), which are responsible for promoting cancer invasion
(15), were significantly increased
compared with control (Fig. 2B).
Consistent with the invasion assay results, SACC-83 cells migration
was significantly increased following stimulation with the culture
medium collected from exosome-educated HPLF cells (Fig. 2C). To determine the expression of
common proinflammatory factors in exosome-educated HPLF cells,
RT-qPCR was performed to detect the mRNA levels of monocyte
chemoattractant protein 1 (MCP-1), inducible nitric oxide synthase
(iNOS), interleukin (IL)-6 1β and 8 and tumor necrosis factor α
(TNF-α). As presented in Fig. 2D,
the expression levels of MCP-1, iNOS, IL-6, IL-1β, IL-8 and TNF-α
were significantly increased in exosomes-treated HPLF cells
compared with PBS-treated HPLF cells. Furthermore, the expression
levels of factors involved in tumor metastasis and PNI, including
C-X-C chemokine receptor type 4 (CXCR4), C-X-C motif chemokine
ligand 12 (CXCL12), neurotrophic receptor tyrosine kinases (NTRK) 1
and 2, neurotrophin 4 (NTF4), NGF and brain-derived neurotrophic
factor (BDNF) were examined. The results demonstrated that the
expression levels of CXCR4, NTRK2, NGF and BDNF were significantly
increased in exosome-educated HPLF cells compared to those in
PBS-treated HPLF cells (Fig.
2E).
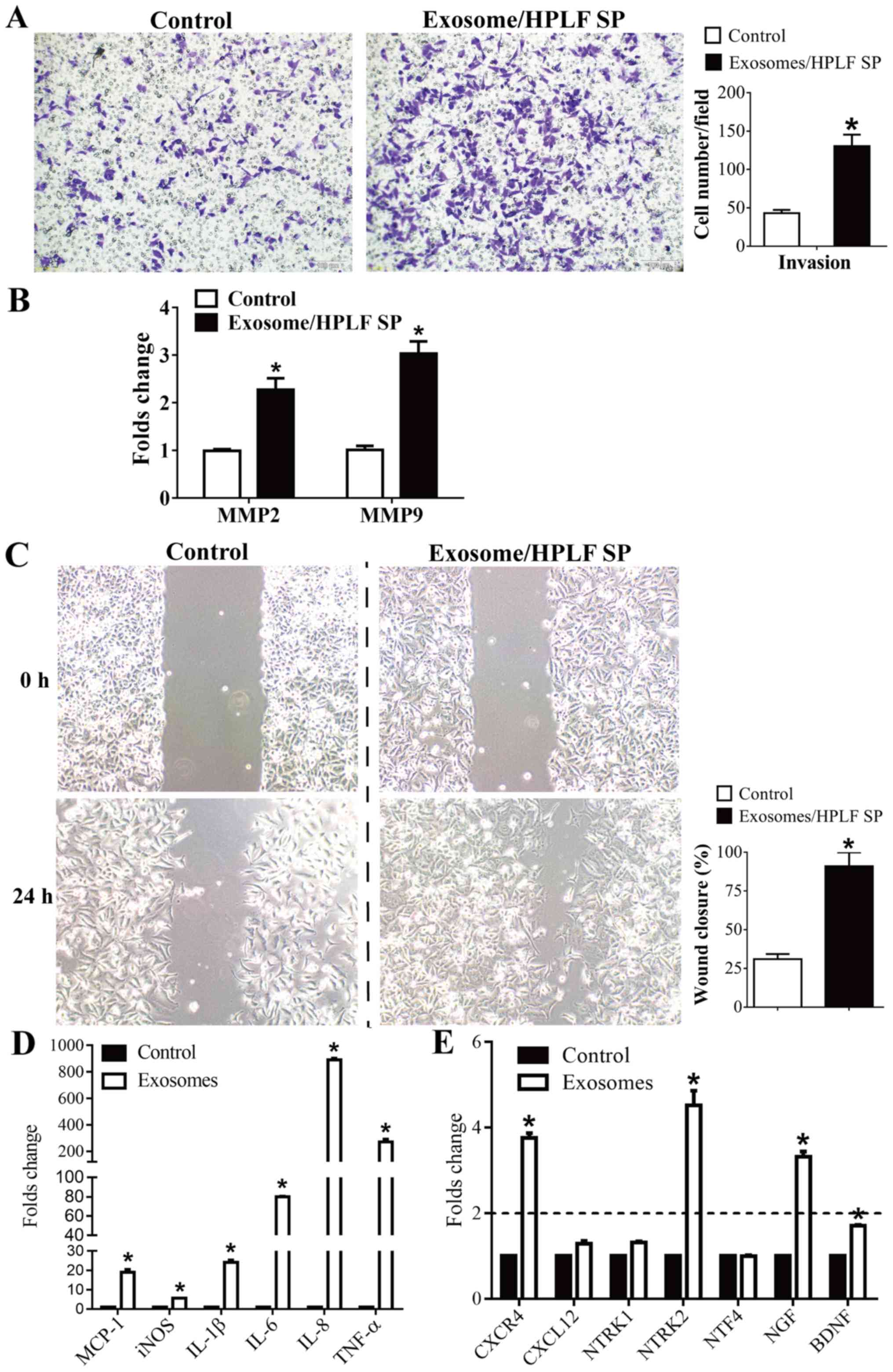 | Figure 2.Exosomes stimulated HPLF cells to
facilitate SACC-83 cell invasion and enhance the expression of
proinflammatory and perineural mediators in HPLF cells. (A)
Transwell assay was used to assess the invasive ability of SACC-83
cells treated with control or the culture SP from exosome-educated
HPLF cells (Exosome/HPLF SP). (B) RT-qPCR for analysis of MMP2 and
MMP9 gene expression levels in SACC-83 cells. (C) SACC-83 cell
migration was determined using wound-healing assay. (D) RT-qPCR for
analysis of MCP-1, iNOS, IL-1β, IL-6, IL-8 and TNF-α gene
expression levels in HPLF cells. (E) RT-qPCR for analysis of CXCR4,
CXCL12, NTRK1, NTRK2, NTF4, NGF and BDNF gene expression levels in
HPLF cells. Data represent the mean ± standard error of the mean of
at least three independent experiments. *P<0.05 vs. control
group. BDNF, Brain-derived neurotrophic factor; CXCL12, C-X-C motif
chemokine ligand 12; CXCR4, C-X-C chemokine receptor type 4; GAPDH,
glyceraldehyde 3-phosphate dehydrogenase; HPLF, human periodontal
ligament fibroblast; IL-1β, interleukin 1β; IL-6, interleukin 6;
iNOS, inducible nitroc oxide synthase; MCP-1, monocyte
chemoattractant protein 1; MMP2, matrix metalloproteinase 2; MMP9,
matrix metalloproteinase 9; NGF, nerve growth factor; NTF4,
neurotrophin 4; NTRK1, neurotrophic receptor tyrosine kinase 1;
NTRK2, neurotrophic receptor tyrosine kinase 2; TNF-α, tumor
necrosis factor α; RT-qPCR, reverse transcription-quantitative
polymerase chain reaction; SACC, salivary adenoid cystic carcinoma;
SP, supernatant. |
Exosomes mediate the pro-tumorigenic
effects of HPLF cells and enhance NGF expression in HPLF cells
In order to identify how the culture supernatant
from exosome-treated HPLF cells could enhance SACC-83 cell invasion
ability, an unbiased RNA sequencing analysis was performed to
determine the transcriptome profile of PBS or exosome-treated HPLF
cells. The average RNA sequencing depth was 21.89 million reads,
with an average genome mapping rate of 93.67%, and the total number
of genes identified was 17,648. Furthermore, 943 differentially
expressed genes with >2-fold expression change were identified.
GO term analysis of these altered genes exhibited enrichment in the
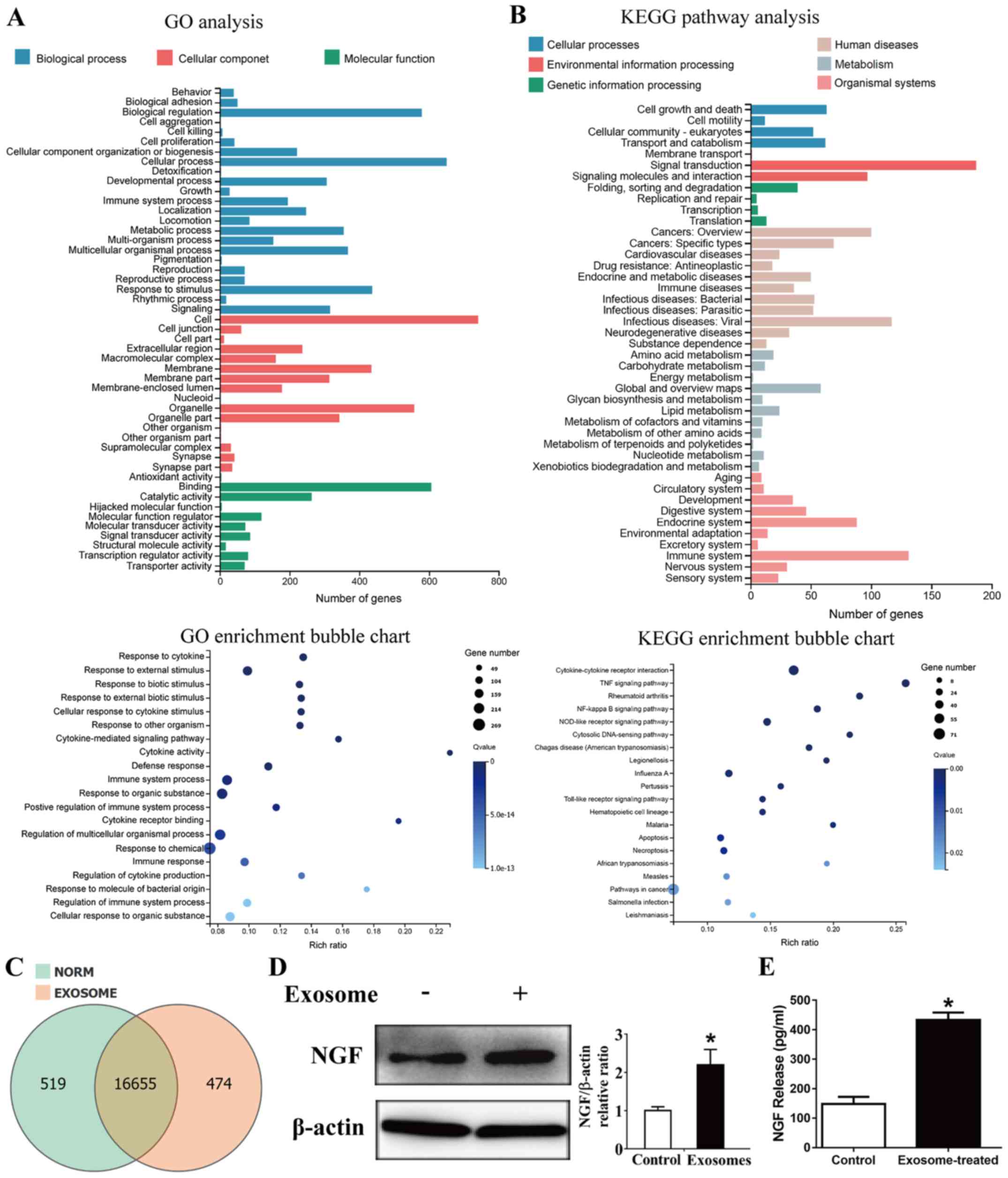
‘response to chemical’ and ‘immune system process’ (Fig. 3A). The results from KEGG pathway
analysis reported enrichment in ‘cytokine-cytokine receptor
interaction’ (Fig. 3B). There were
16,655 genes expression in both PBS or exosome-treated HPLF cells,
whereas 519 genes were expressed only in PBS-treated HPLF cells,
and 474 genes were expressed only in exosome-treated HPLF cells
(Fig. 3C). The expression of NGF,
which was proposed as a potential prognosis and pathogenic
biomarker of SACC (16–18), was then examined. As presented in
Fig. 3D and E, NGF protein
expression was significantly increased in exosome-educated HPLF
cells.
Neutralizing NGF reduces SACC-83 cell
invasion stimulated by the supernatant from exosome-treated HPLF
cells
In order to determine whether the enhanced SACC-83
cell migratory ability was caused by NGF released from
exosome-treated HPLF cells, the functional NGF neutralizing
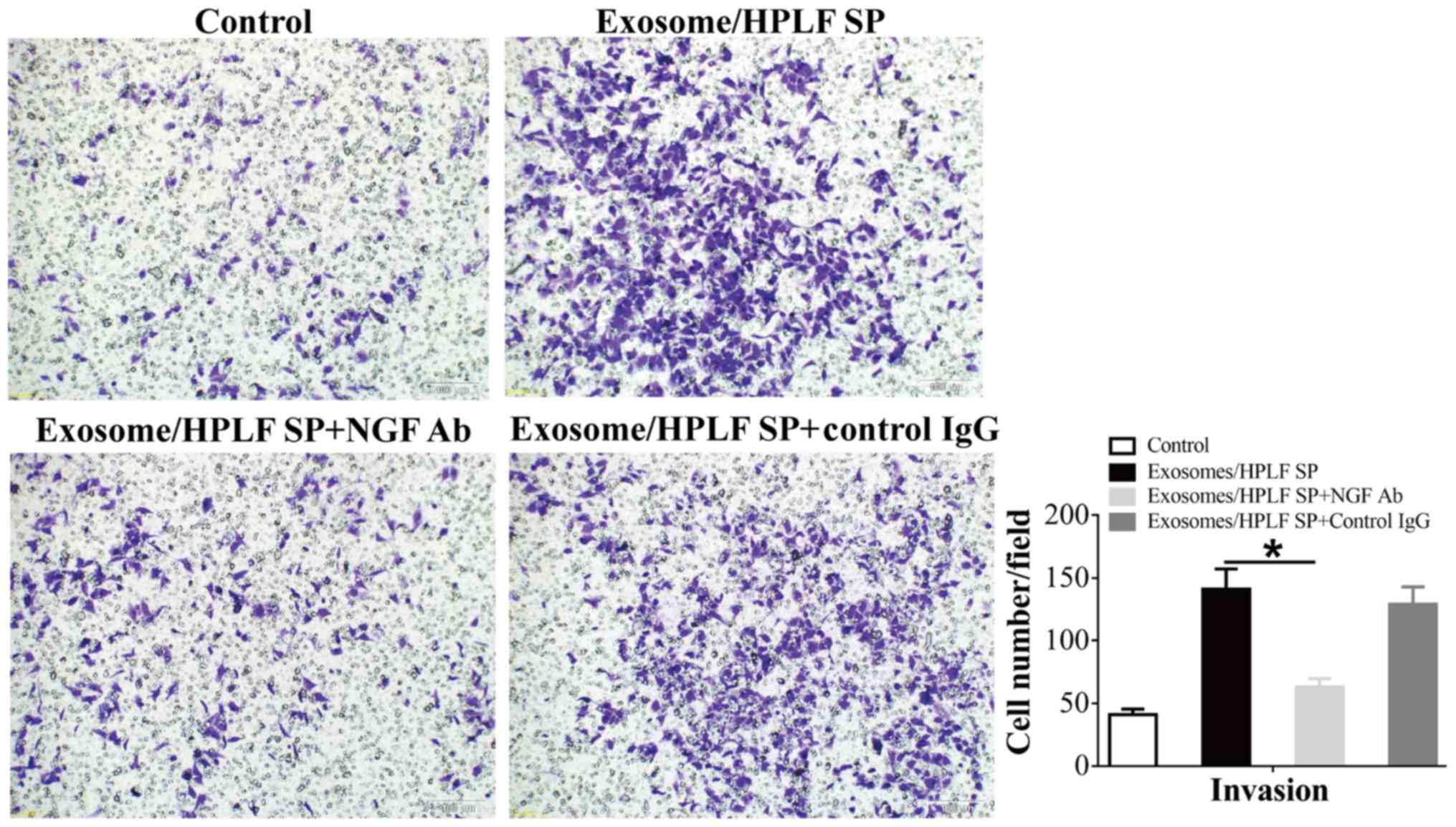
antibody was used in the invasion assay. As presented in Fig. 4, NGF blockage significantly reduced
the enhanced invasive ability of SACC-83 cells following treatment
with supernatant from exosome-educated HPLF cells.
Upregulation of NTRK1 expression in
patients with SACC
To illustrate the roles of NGF signaling in SACC
progression, in-depth bioinformatics analysis of the Oncomine
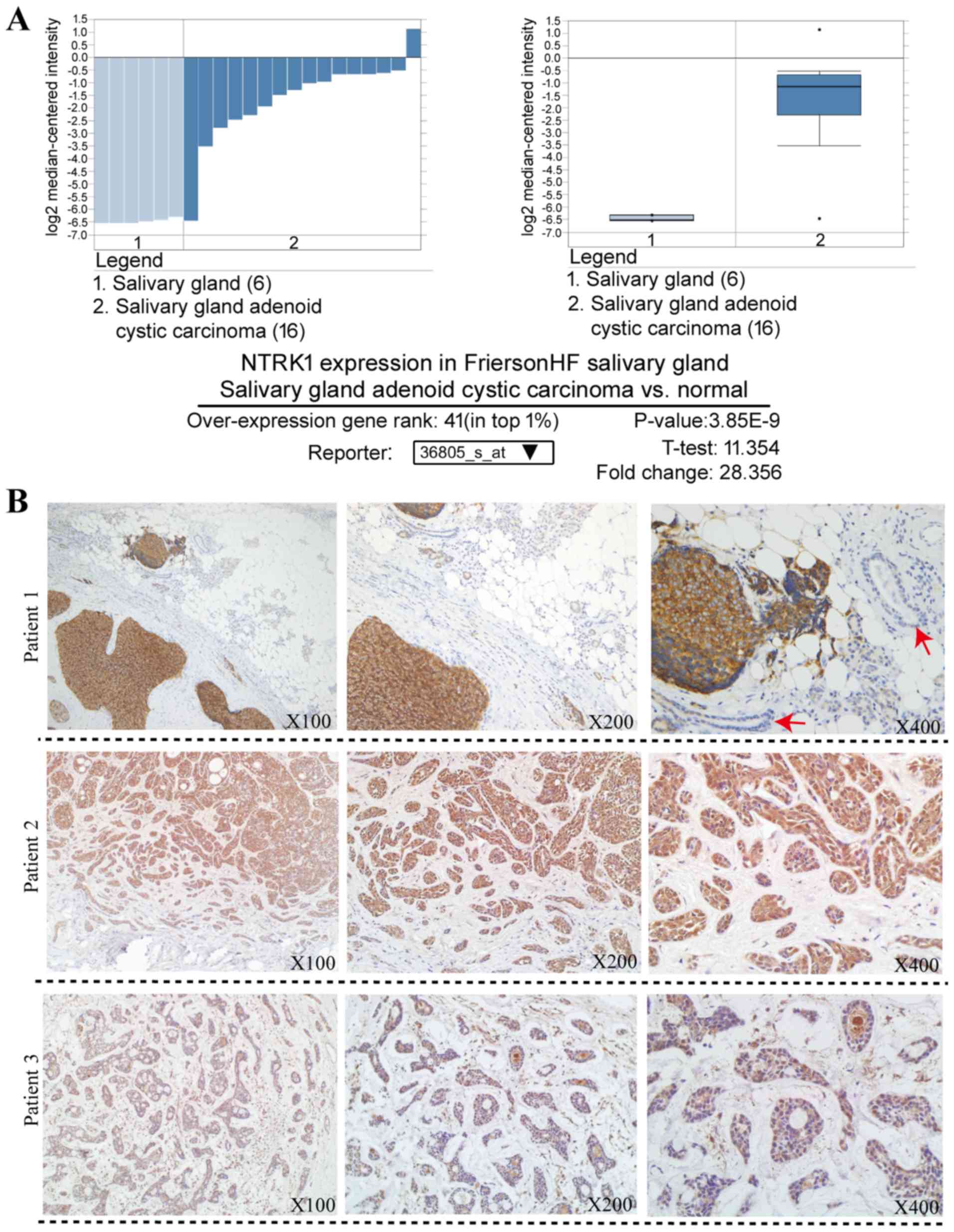
database was carried out. The results demonstrated that NTRK1
overexpression ranked in the top 1% of increased gene expression in
salivary gland adenoid cystic carcinoma vs. normal (Fig. 5A), and that NTRK1 mRNA level was
increased by a 28.356-fold in SACC tissue compared to normal tissue
from the Oncomine database (19)
(Fig. 5A). Furthermore, NTRK1
expression was examined by IHC in tumor tissues from patients with
SACC. As presented in Fig. 5B, the
cancerous tissues staining revealed strongly positive NTRK1
expression, whereas the non-cancerous tissues staining (red arrow)
revealed negative NTRK1 expression. The protein expression of NTRK1
was markedly upregulated in the cancerous tissues from three
patients with SACC.
Discussion
SACC is a rare type of head and neck cancer
characterized by unpredictable growth, extensive PNI and high risk
of metastasis. PNI corresponds to the neoplastic invasion of nerves
along with increased recurrence rates and reduced survival
(2,3). Although the clinical significance of
PNI is widely recognized, its underlying mechanisms remain unclear.
Factors involved in nerve microenvironment, including chemokines,
neurotrophic factors and cellular adhesion molecules, have recently
gained more attention (20).
Cancer-associated fibroblasts are localized within the tumor
microenvironment and produce certain mediators, including growth
factors, chemokines and extracellular matrix that enhance cancer
cell metastasis (21). CAFs can
therefore provide a unique tumor microenvironment that stimulates
cancer development and progression (22). Perineurium originates from the
differentiation of fibroblasts (21)
and any change in composition and malfunction of the perineurium
can lead to tumor invasion (20).
The role of fibroblast in tumor metastasis and PNI requires
therefore more attention.
In the present study, the crosstalk between human
SACC cells and fibroblasts was investigated. Cell-derived exosomes
act as a bridge between cells to deliver molecules and link
cellular functions. It has been demonstrated that pancreatic
fibroblast-derived exosomes promote the proliferation and survival
of cancer cells (23). A recent
study reported that CAFs account for oral squamous cell carcinoma
progression and metastasis through exosome-derived paracrine
miR-34a-5p (24). This study
therefore investigated the role of tumor-derived exosome-educated
HPLF cells in SACC-83 cell metastasis. The results demonstrated
that supernatant from exosome-educated HPLF cells markedly promoted
SACC-83 invasion, which suggested that exosomes may be considered
as key mediators of the tumorigenic effect of fibroblasts on SACC
pathogenesis.
To comprehensively understand the underlying
mechanisms by which tumor-derived exosomes influenced fibroblast
functions and subsequently SACC progression, RNA sequencing
analysis was performed in order to analyze the gene expression
changes in exosome-treated HPLF cells. A total of 943
differentially expressed genes (upregulated or downregulated) with
>2-fold expression change were identified, and these genes
exhibited enriched cytokine-cytokine receptor interaction. Chronic
inflammation, which has been recognized as one of the major tumor
hallmarks, can influence almost all stages of tumorigenesis
(25). The present study
demonstrated that the expression levels of the proinflammatory
cytokines MCP-1, IL-1β, IL-6, IL-8 and TNF-α were significantly
increased in exosome-stimulated HPLF cells. These inflammatory
factors, including IL-1β, have been reported to serve a crucial
role in the proliferation, migration and invasion of malignant
tumor cells (26,27). The results from the present study
suggested that SACC-83 cell-derived exosomes may educate HPLF cells
to create an inflammatory microenvironment that could facilitate
tumor progression.
Chemokines and their receptors can directly affect
cancer cell proliferation and movement or indirectly modulate tumor
growth by stimulating the secretion of growth, chemotactic and
angiogenic factors from stromal cells into the cancer
microenvironment (28). For example,
nerve-derived C-C motif chemokine ligand 2 facilitates prostate
cancer cell invasion and PNI via C-C chemokine receptor type
2-mediated signaling (29).
Furthermore, CXCL12 can facilitate squamous cell/carcinoma cell
survival and migration via its receptor CXCR4 (30). The results from the present study
demonstrated that expression levels of CXCR4, NTRK2, NGF and BDNF
were significantly increased in exosome-stimulated HPLF cells. It
has been demonstrated that NGF and its receptor NTRK1 are
associated with metastasis and PNI in several types of cancer. For
example, expression levels of NGF and its high and low affinity
receptors NTRK1 and p75NTR, respectively, were markedly elevated in
pancreatic cancer cells and surrounding nerves, which was
positively correlated with the presence of PNI (31). Furthermore, in oral tongue tumor
tissues, stronger NGF and NTRK1 staining was observed in tissues
with PNI compared with tissues of the same stage but without PNI
(32). The present study
demonstrated that NGF protein level was significantly increased in
exosome-educated HPLF cells. In particular, mRNA levels of NTRK1 in
HPLF cells did not change following exosome treatment. Furthermore,
NGF blockage significantly reduced the enhanced invasive ability of
SACC-83 cells following treatment with culture medium from
exosome-educated HPLF cells. Cancer cell-derived exosomes can be
internalized into the recipient cells and regulate their biological
function and signaling (33).
Similarly, the results from this study indicated that exosomes may
serve a crucial role in tumor microenvironment and enhance the
crosstalk between SACC-83 cells and HPLF cells. Furthermore,
results from the Oncomine database analysis demonstrated that NTRK1
overexpression ranked in the top 1% of increased gene expression in
SACC tissues compared with normal tissues. Consistent with this
finding, higher expression level of NTRK1 was observed in the
cancerous tissues compared with in non-cancerous tissues from
patients with SACC.
In conclusion, the present study demonstrated that
SACC-83 cell-derived exosomes stimulated HPLF cells to produce NGF,
which may enhance SACC-83 cell invasion. The results also indicated
that tumor-derived exosomes may lead to HPLF cell activation and
downstream inflammation. This mechanism may illustrate the
crosstalk between SACC and fibroblasts and explain the occurrence
of metastasis and PNI in SACC. Since NGF-NTRK1 signaling results in
poor prognosis in patients with SACC (18), targeting tumor-derived exosomes and
CAFs by blocking NGF-NTRK1 pathway may potentially serve as a novel
therapy that could be used in synergy with conventional therapies
in order to improve patient survival.
Acknowledgements
Not applicable.
Funding
This study was supported by the National Natural
Science Foundation of China (grant nos. 81772577 and 81602497).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
ZX and JZ designed the study and wrote the
manuscript. ZX and XZ performed the experiments. All authors read
and approved the final manuscript.
Ethics approval and consent to
participate
All procedures performed in studies involving human
tissues were in accordance with the ethical standards of the
Institutional Committee and with the Declaration of Helsinki. All
patients provided written informed consent prior to the study
start. Use of human samples in the present study was approved by
Shanghai Changhai Hospital Ethics Committee.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Venteicher AS, Walcott BP, Sheth SA,
Snuderl M, Patel AP, Curry WT and Nahed BV: Clinical features of
brain metastasis from salivary gland tumors. J Clin Neurosci.
20:1533–1537. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ellington CL, Goodman M, Kono SA, Grist W,
Wadsworth T, Chen AY, Owonikoko T, Ramalingam S, Shin DM, Khuri FR,
et al: Adenoid cystic carcinoma of the head and neck: Incidence and
survival trends based on 1973–2007 Surveillance, Epidemiology and
End Results data. Cancer. 118:4444–4451. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ho AS, Kannan K, Roy DM, Morris LG, Ganly
I, Katabi N, Ramaswami D, Walsh LA, Eng S, Huse JT, et al: The
mutational landscape of adenoid cystic carcinoma. Nat Genet.
45:791–798. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Shiga K, Hara M, Nagasaki T, Sato T,
Takahashi H and Takeyama H: Cancer-associated fibroblasts: Their
characteristics and their roles in tumor growth. Cancers.
7:2443–2458. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Madar S, Goldstein I and Rotter V: ‘Cancer
associated fibroblasts’-more than meets the eye. Trends Mol Med.
19:447–453. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Liotta LA and Kohn EC: The
microenvironment of the tumour-host interface. Nature. 411:375–379.
2001. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Xing F, Saidou J and Watabe K: Cancer
associated fibroblasts (CAFs) in tumor microenvironment. Front
Biosci (Landmark Ed). 15:166–179. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Che Y, Geng B, Xu Y, Miao X, Chen L, Mu X,
Pan J, Zhang C, Zhao T, Wang C, et al: Helicobacter
pylori-induced exosomal MET educates tumour-associated
macrophages to promote gastric cancer progression. J Cell Mol Med.
22:5708–5719. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhang H, Deng T, Liu R, Bai M, Zhou L,
Wang X, Li S, Wang X, Yang H, Li J, et al: Exosome-delivered EGFR
regulates liver microenvironment to promote gastric cancer liver
metastasis. Nat Commu. 8:150162017. View Article : Google Scholar
|
|
10
|
Ringuette Goulet C, Bernard G, Tremblay S,
Chabaud S, Bolduc S and Pouliot F: Exosomes induce fibroblast
differentiation into cancer-associated fibroblasts through TGFβ
signaling. Mol Cancer Res. 16:1196–1204. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Théry C, Amigorena S, Raposo G and Clayton
A: Isolation and characterization of exosomes from cell culture
supernatants and biological fluids. Curr Protoc Cell Biol Chapter.
3:Unit 3.22. 2006.
|
|
12
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Andreu Z and Yáñez-Mó M: Tetraspanins in
extracellular vesicle formation and function. Front Immunol.
5:4422014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ruivo CF, Adem B, Silva M and Melo SA: The
biology of cancer exosomes: Insights and new perspectives. Cancer
Res. 77:6480–6488. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Deryugina EI and Quigley JP: Matrix
metalloproteinases and tumor metastasis. Cancer Metastasis Rev.
25:9–34. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hao L, Xiao-Lin N, Qi C, Yi-Ping Y,
Jia-Quan L and Yan-Ning L: Nerve growth factor and vascular
endothelial growth factor: Retrospective analysis of 63 patients
with salivary adenoid cystic carcinoma. Int J Oral Sci. 2:35–44.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang L, Sun M, Jiang Y, Yang L, Lei D, Lu
C, Zhao Y, Zhang P, Yang Y and Li J: Nerve growth factor and
tyrosine kinase A in human salivary adenoid cystic carcinoma:
Expression patterns and effects on in vitro invasive behavior. J
Oral Maxillofac Surg. 64:636–641. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kobayashi K, Ando M, Saito Y, Kondo K,
Omura G, Shinozaki-Ushiku A, Fukayama M, Asakage T and Yamasoba T:
Nerve growth factor signals as possible pathogenic biomarkers for
perineural invasion in adenoid cystic carcinoma. Otolaryngol Head
Neck Surg. 153:218–224. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Frierson HF Jr, El-Naggar AK, Welsh JB,
Sapinoso LM, Su AI, Cheng J, Saku T, Moskaluk CA and Hampton GM:
Large scale molecular analysis identifies genes with altered
expression in salivary adenoid cystic carcinoma. Am J Pathol.
161:1315–1323. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Bakst RL and Wong RJ: Mechanisms of
perineural invasion. J Neurol Surg B Skull Base. 77:96–106. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kalluri R and Zeisberg M: Fibroblasts in
cancer. Nat Rev Cancer. 6:392–401. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bhowmick NA, Neilson EG and Moses HL:
Stromal fibroblasts in cancer initiation and progression. Nature.
432:332–337. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Richards KE, Zeleniak AE, Fishel ML, Wu J,
Littlepage LE and Hill R: Cancer-associated fibroblast exosomes
regulate survival and proliferation of pancreatic cancer cells.
Oncogene. 36:1770–1778. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Li YY, Tao YW, Gao S, Li P, Zheng JM,
Zhang SE, Liang J and Zhang Y: Cancer-associated fibroblasts
contribute to oral cancer cells proliferation and metastasis via
exosome-mediated paracrine miR-34a-5p. EBioMedicine. 36:209–220.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Colotta F, Allavena P, Sica A, Garlanda C
and Mantovani A: Cancer-related inflammation, the seventh hallmark
of cancer: Links to genetic instability. Carcinogenesis.
30:1073–1081. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Voronov E, Shouval DS, Krelin Y, Cagnano
E, Benharroch D, Iwakura Y, Dinarello CA and Apte RN: IL-1 is
required for tumor invasiveness and angiogenesis. Proc Natl Acad
Sci USA. 100:2645–2650. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Apte RN, Dotan S, Elkabets M, White MR,
Reich E, Carmi Y, Song X, Dvozkin T, Krelin Y and Voronov E: The
involvement of IL-1 in tumorigenesis, tumor invasiveness,
metastasis and tumor-host interactions. Cancer Metastasis Rev.
25:387–408. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chow MT and Luster AD: Chemokines in
cancer. Cancer Immunol Res. 2:1125–1131. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
He S, He S, Chen CH, Deborde S, Bakst RL,
Chernichenko N, McNamara WF, Lee SY, Barajas F, Yu Z, et al: The
chemokine (CCL2-CCR2) signaling axis mediates perineural invasion.
Mol Cancer Res. 13:380–390. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Muller A, Sonkoly E, Eulert C, Gerber PA,
Kubitza R, Schirlau K, Franken-Kunkel P, Poremba C, Snyderman C,
Klotz LO, et al: Chemokine receptors in head and neck cancer:
Association with metastatic spread and regulation during
chemotherapy. Int J Cancer. 118:2147–2157. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhu Z, Friess H, diMola FF, Zimmermann A,
Graber HU, Korc M and Buchler MW: Nerve growth factor expression
correlates with perineural invasion and pain in human pancreatic
cancer. J Clin Oncol. 17:2419–2428. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kolokythas A, Cox DP, Dekker N and Schmidt
BL: Nerve growth factor and tyrosine kinase A receptor in oral
squamous cell carcinoma: Is there an association with perineural
invasion? J Oral Maxillofac Surg. 68:1290–1295. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zheng P, Chen L, Yuan X, Luo Q, Liu Y, Xie
G, Ma Y and Shen L: Exosomal transfer of tumor-associated
macrophage-derived miR-21 confers cisplatin resistance in gastric
cancer cells. J Exp Clin Cancer Res. 36:532017. View Article : Google Scholar : PubMed/NCBI
|