Introduction
Human esophageal cancer is a malignancy that is
associated with a high mortality rate worldwide; ~>90% of all
cases of this disease present as esophageal squamous cell carcinoma
(ESCC) (1,2). Despite advancements in therapeutic
techniques, including chemotherapeutic, radiotherapeutic and
surgical treatments, the prognosis of ESCC remains poor; its 5-year
survival rate is only 10–15% (3).
Therefore, further research on the molecular mechanisms underlying
the development and progression of ESCC and the development of
effective treatments for ESCC is required. Cisplatin is a
first-line chemotherapeutic drug that is commonly used to treat
several types of cancer, including ESCC (4). However, cancer cells appear to be
chemoresistant to cisplatin during long-term treatment (4). Thus, the inhibition of cancer
chemoresistance to cisplatin may improve cisplatin chemotherapy in
various types of cancer.
MicroRNAs (miRNAs/miRs) are small non-coding RNAs
that regulate gene expression at the post-transcriptional level;
they function in various processes, such as cell proliferation,
apoptosis and differentiation (5).
miRNAs control gene expression by degrading their target mRNAs or
inhibiting their translation into functional proteins (5). Accumulated evidence has illustrated
that miRNAs play diverse roles in the regulation of cancer
initiation and progression of ESCC (6,7). Certain
studies have demonstrated that miR-544 serves as an oncogene in
osteosarcoma, liver cancer, gastric cancer and colorectal cancer
(8–11). However, miR-544 acts as a tumor
suppressor and inhibits tumor progression in breast cancer and
glioma (12,13); this contrast in function may be
tissue-specific. To date, the function and mechanism of miR-544 in
the progression of ESCC have not been well illustrated.
The present study for the first time determined
whether miR-544 was deregulated in ESCC tissues and cells.
Furthermore, the effects of miR-544 overexpression on ESCC cellular
proliferation and chemoresistance to cisplatin were determined. In
addition, the correlation between miR-544 and E2F5 expression in
ESCC was demonstrated.
Materials and methods
Tissue specimens
A total of 30 tumor tissues and paired adjacent
normal tissues were obtained from 30 patients with ESCC (22 males
and 8 females; age range, 49–77 years) who underwent surgery at the
Department of Oncology at Linyi Central Hospital, China between
June 2016 and June 2017. The normal paired tissues were obtained
from the distal resection margins (>2 cm away from the tumor
tissue). The histological grade and clinical stage of the tumors
were defined based on the seventh edition of the American Joint
Committee on Cancer system for esophageal cancer (14). The histological differentiation of
the tumors were graded as follows: Well, n=8; moderate, n=13; and
poor, n=9 Tumor-Node-Metastasis stage was also assigned: I, n=5;
II, n=18; III, n=7. All specimens were stored at −80°C until
analysis. No patients received radiotherapy or chemotherapy prior
to surgery. The present study was approved by the Research Ethics
Committee of Linyi Central Hospital. Written informed consent was
obtained from all patients prior to enrollment.
Cell culture and cell
transfection
The EC9706, KYSE450 and TE-1 human ESCC cell lines
and the human immortalized normal esophageal epithelial cell line
HET-1A were purchased from the Type Culture Collection of the
Chinese Academy of Sciences. The cells were incubated in RPMI-1640
medium (HyClone; GE Healthcare Life Sciences) containing 10% fetal
bovine serum (Gibco; Thermo Fisher Scientific, Inc.) at 37°C in a
humidified incubator containing 5% CO2. The target
sequences of miR-544 mimic were as follows:
5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACGAACTTT-3′. The
target sequences of NC were as follows:
5′-CAGUACUUUUGUGUAGUACAA-3′. The transfection efficiency was
confirmed by RT-qPCR.
Full-length E2F5 from the human cDNA library was
inserted into pcDNA3.1 vector (Invitrogen; Thermo Fisher
Scientific, Inc.) to generate the pcDNA3.1/E2F5 overexpression
vector. The pcDNA3.1 empty vector was the used as a NC. The KYSE450
and TE-1 cells were transfected with miR-NC (50 nM) or miR-544
mimic (50 nM) and/or pcDNA3.1/ E2F5 vector (100 nM) or pcDNA3.1
(100 nM) using Lipofectamine® 2000 reagent (Invitrogen;
Thermo Fisher Scientific, Inc.) according to the manufacturer's
protocol. Transfection efficiency was confirmed by using western
blot analysis. Subsequent experiments were performed 24–48 h after
transfection.
Western blot analysis
Total protein was extracted from ESCC and HET-1A
cells and tissues using lysis buffer (Beyotime Institute of
Biotechnology), and protein concentrations were measured with a
Bicinchoninic Acid Protein Assay kit (Beyotime Institute of
Biotechnology). A total of 30 µl protein extract was separated via
SDS-PAGE (10% gel) and transferred onto a PVDF membrane. The
membranes were blocked with 5% non-fat milk for 60 min at room
temperature, followed by incubation with primary antibodies against
E2F5 (1:500; cat. no. ab44996; Abcam), cleaved caspase-3 (1:500;
cat. no. ab2302; Abcam), cleaved poly (ADP-ribose) polymerase
(1:500; cleaved-PARP, cat. no. AF1567; Beyotime Institute of
Biotechnology) and β-actin (1:500; cat. no. ab8227; Abcam)
overnight at 4°C, then treated with a Horseradish
peroxidase-labeled goat anti-rabbit secondary antibody (1:1,000;
cat. no. ab150077; Abcam) for 2 h at room temperature. The protein
bands were visualized using an enhanced chemiluminescence system
(Beyotime Institute of Biotechnology). β-actin was acted as a
loading control for normalization.
Total mRNA extraction and reverse
transcription-quantitative (RT-q)PCR. According to the protocol of
mRNA extraction, the cells were dissolved in TRIzol®
reagent (Invitrogen; Thermo Fisher Scientific, Inc.) to extract
total mRNA. Total RNA was reverse transcribed with oligodT primers
using the miRcute Plus miRNA First-strand cDNA Synthesis kit
(Tiangen Biotech Co., Ltd.) at 37°C for 60 min and 85°C for 1 min.
qPCR was performed using SYBR Premix Ex Taq (Takara Biotechnology
Co., Ltd.). The PCR thermocycling conditions were: 94°C for 2 min
followed by 30 cycles of 94°C for 30 sec, 61°C for 30 sec, and 72°C
for 30 sec. The relative expression levels of miR-544 and E2F5 were
calculated as the inverse log of ΔΔCq and normalized to the
reference (15). The sequence of the
primers used for amplification were: E2F5 forward,
5′-CGGCTAGCTTCTGGATTTCAACTTTTCTTC-3′; E2F5 reverse,
5′-GCGTCGACGGAAAGTGGAATGTCAGAAGTC-3′; miR-544 forward,
5′-GCCCGATTCTGCATTTTTAGC-3′; miR-544 reverse
5′-CGGGCTAAGACGTAAAAACG-3′; β-actin forward
5′-GATCATTGCTCCTCCTGAGC-3′; β-actin reverse,
5′-ACTCCTGCTTGCTGATCCAC-3′; U6 forward,
5′-TGCGGGTGCTCGCTTCGCAGC-3′; and U6 reverse
5′-CCAGTGCAGGGTCCGAGGT-3′.
MTT assay
In total, ~1×103 KYSE450 and TE-1
cells/well were seeded onto a 96-well plate and incubated
overnight. Subsequently, 20 µl MTT solution was added per well, and
incubated for 4 h. The liquid was removed from the plate, and 150
µl DMSO was added per well. All plates were read at 490 nm.
Colony-formation assay
The proliferation of cells was analyzed using a
plate colony-formation assay. Briefly, KYSE450 and TE-1 cells
transfected with miR-544 mimic and E2F5 plasmid (300 cells/well)
were added to each well of the 6-well plates and cultured for 10
days in RPMI-1640 medium (HyClone; GE Healthcare Life Sciences)
containing 10% FBS (Gibco; Thermo Fisher Scientific, Inc.). The
cells were washed with phosphate-buffered saline (PBS) twice and
fixed using 0.1% crystal violet for 20 min at room temperature.
Images of the colonies were captured and the number of colonies was
counted under a light microscope (Olympus Corporation)
(magnification, ×100).
Flow cytometry analysis of
apoptosis
KYSE450 and TE-1 cells transfected with miR-544
mimic and E2F5 plasmids were treated with or without 15 µM
cisplatin for 24 h at 37°C, followed by determination of the
apoptotic rate. Apoptotic rates were determined using an annexin V
and propidium iodide Apoptosis Detection kit (cat. no. C1062M;
Beyotime Institute of Biotechnology), according to the
manufacturer's protocol. The apoptotic rate was determined using a
FACSAria flow cytometer and analyzed by the FASCDiva version 4.1
software (BD Biosciences).
Target prediction
The TargetScanHuman database and TargetScanHuman
Release version 7.1 software (http://www.targetScan.org) were used to predict the
potential target genes of miR-544. TargetScan target gene
prediction software identified the 617–624 site at the 3′-end of
the 3′-UTR of E2F5 mRNA as a possible site of action of
miR-544.
Dual-luciferase assay
Wild-type (wt) and mutated (mut) putative
miR-544-binding sites in E2F5 3′-untranslated region (UTR) were
cloned into the downstream region of the luciferase gene in the
pGL3-REPORT luciferase vector (Invitrogen; Thermo Fisher
Scientific, Inc.). For the reporter assay, the cells were
co-transfected with wt or mut pGL3-E2F5-3′-UTR vectors and miR-544
mimics using Lipofectamine 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.). After 24 h, Luciferase activities were measured
with a Dual Luciferase Reporter Assay kit (Promega Corporation),
according to the manufacturer's protocol. Data were normalized
against the activity of the Renilla luciferase activity.
Statistical analysis
All statistical analyses were performed using Prism
GraphPad version 6.0 (GraphPad Software, Inc.). Data of more than
two groups were analyzed using one-way analysis of variance with
Tukey's post hoc test. Correlations between miR-544 and E2F5 mRNA
levels were analyzed using Pearson's correlation analysis. The
statistical analysis of two unpaired groups was evaluated using an
unpaired Student's t-test. Statistical analysis of miR-544 and E2F5
expression between ESCC tissues and paired adjacent normal tissues
was evaluated using a paired Student's t-test. P<0.05 was
considered to indicate a statistically significant difference. The
data were presented as the mean ± standard deviation.
Results
Expression of miR-544 and E2F5 in ESCC
tumors and cell lines
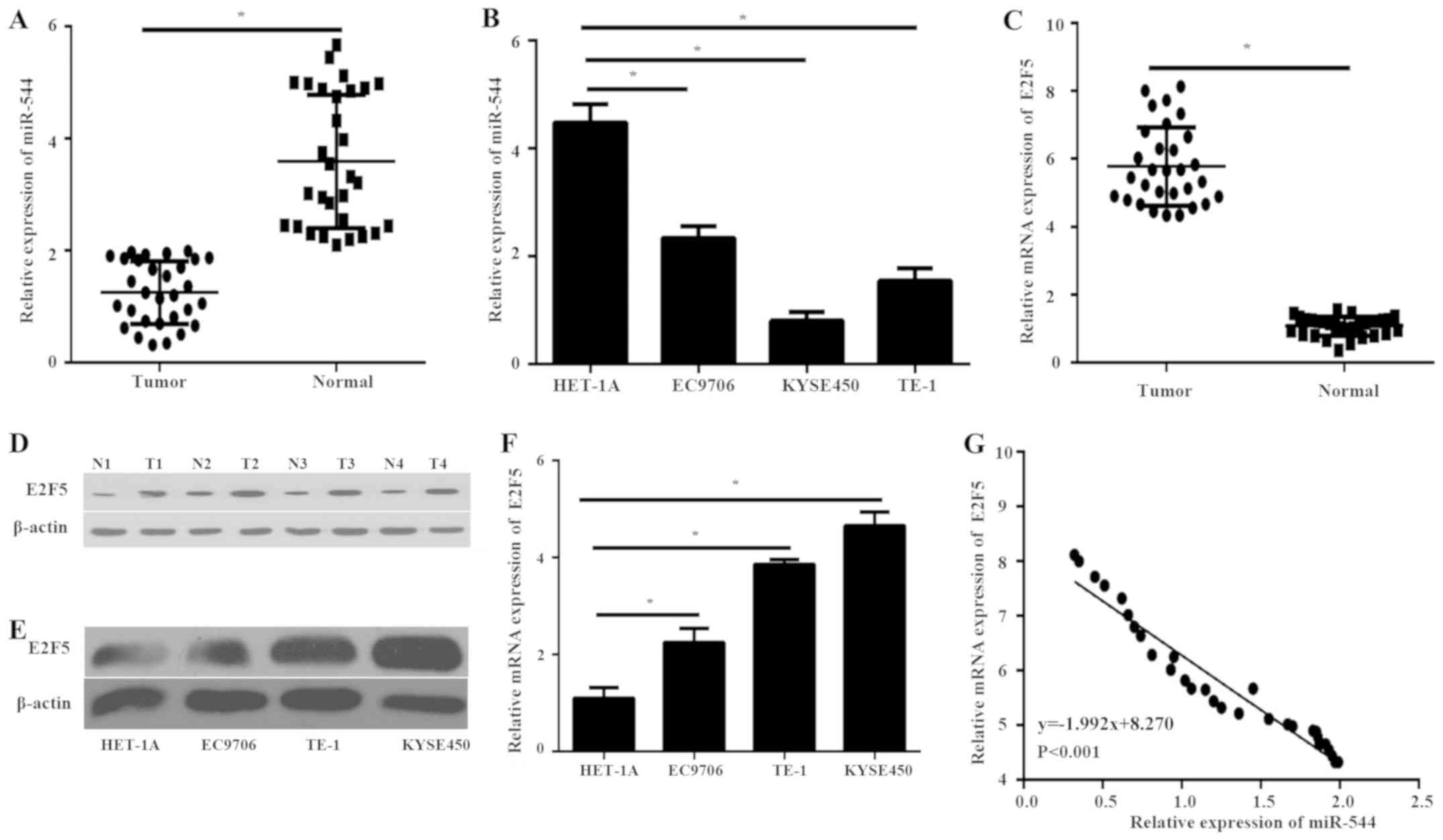
RT-qPCR was performed in order to detect the
expression levels of miR-544 and E2F5 in 30 ESCC tissues and
corresponding normal tissues. The expression level of miR-544 in
ESCC tissues was significantly decreased compared with the normal
tissues (P<0.05; Fig. 1A).
Furthermore, the expression level of miR-544 in ESCC cell lines was
also significantly decreased compared with that in normal
esophageal epithelial HET-1A cells (P<0.05; Fig. 1B). However, the mRNA level of E2F5
expression was significantly higher in ESCC tissues compared with
normal tissues (P<0.05; Fig. 1C).
The western blot analysis further demonstrated that the protein
expression of E2F5 was upregulated in ESCC tissues compared with
normal tissues (Fig. 1D). In
addition, the protein and mRNA level of E2F5 expression was
significantly higher in ESCC cell lines compared with that in
HET-1A cells (P<0.05; Fig. 1E and
F). Furthermore, Pearson's correlation analysis demonstrated a
significant negative correlation between the expression of miR-544
and E2F5 mRNA expression in ESCC tissues (P<0.001; Fig. 1G). These findings suggest that
miR-544 may play a critical role in the progression of ESCC and
have internal correlation with E2F5 in ESCC.
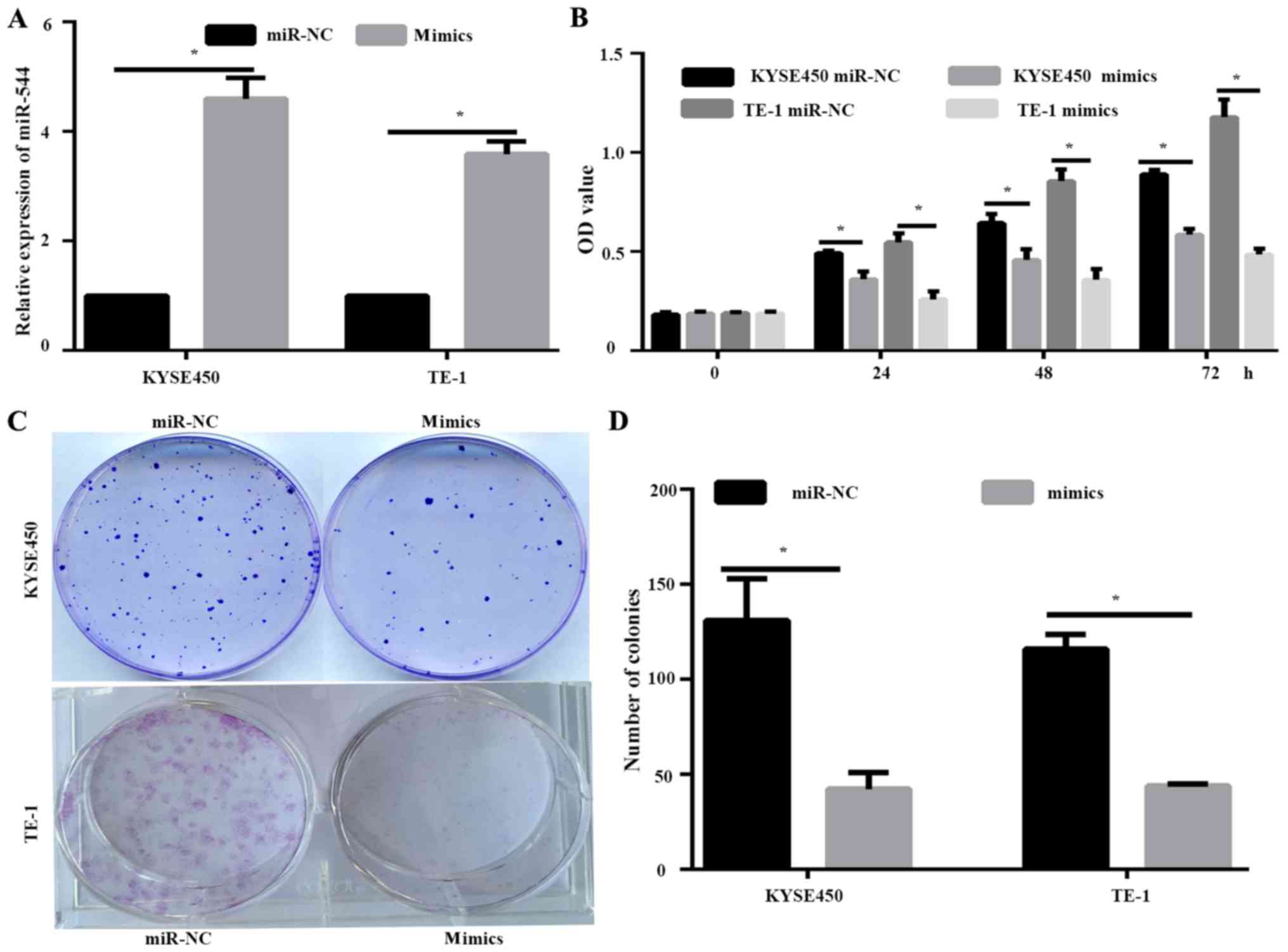
miR-544 overexpression inhibits cell
proliferation of ESCC cells
KYSE450 and TE-1 cells expressed lower levels of
miR-544 compared with EC9706 cell lines, and were therefore
selected for the further study. KYSE450 and TE-1 cells were
transfected with miR-NC or miR-544 mimic. RT-qPCR demonstrated that
the expression of miR-544 in the mimic-transfected cells was
significantly higher compared with the miR-NC group (P<0.05;
Fig. 2A). Overexpression of miR-544
resulted in decreased cell proliferation of KYSE450 and TE-1 cells,
as determined by MTT assay (P<0.05; Fig. 2B). Furthermore, the inhibition of
proliferation by miR-544 mimic was further demonstrated by
colony-formation assay (P<0.05; Fig.
2C).
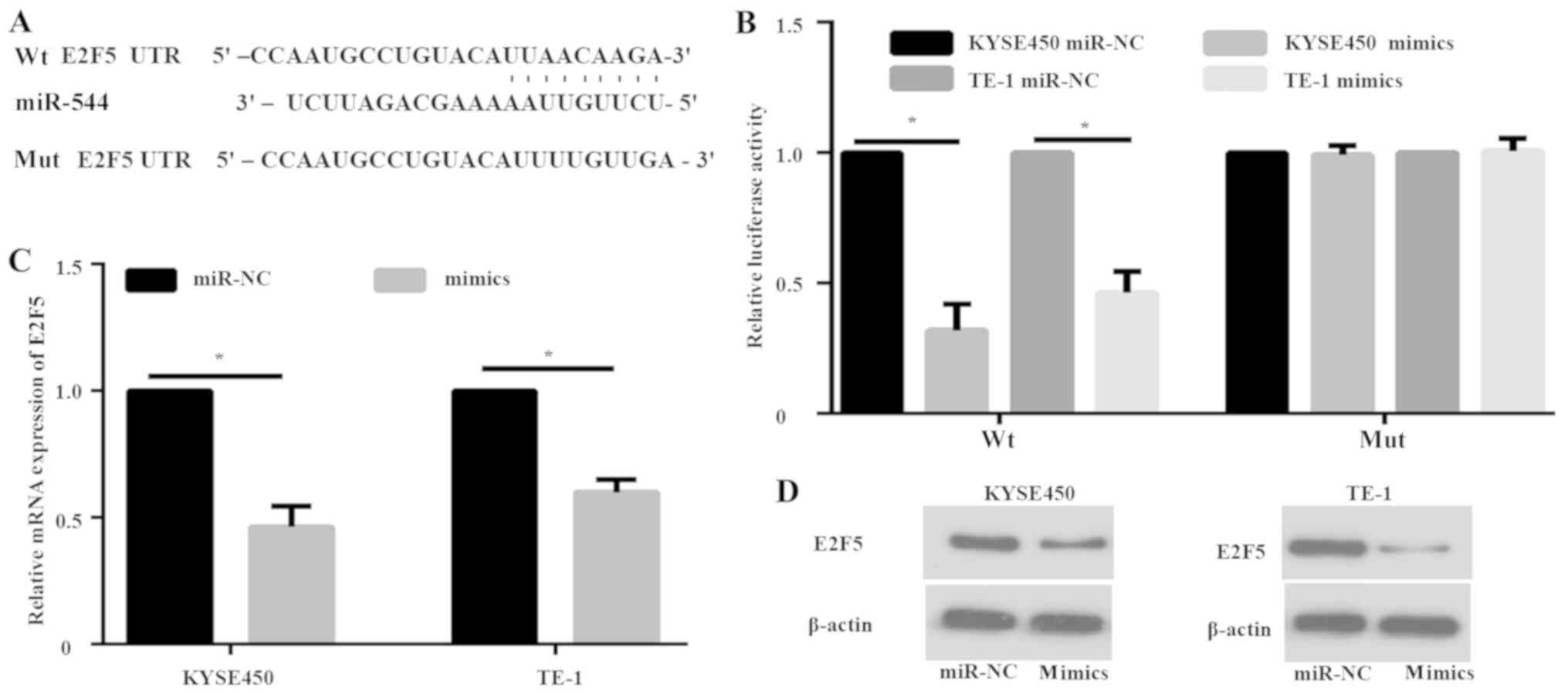
E2F5 is a direct target gene of
miR-544
To further explore the molecular mechanism of
miR-544 in ESCC progression, computational algorithms were applied
(using TargetScan) to predict potential target genes of miR-544
Dual-luciferase assays were performed to confirm this prediction.
Two luciferase reporter recombinant vectors that contained either
wt 3′UTR of E2F5 gene (wtE2F5) or E2F5 with a mut miR-544 binding
site (mutE2F5) were constructed (Fig.
3A). KYSE450 and TE-1 cells were co-transfected with wtE2F5 and
miR-544 mimics. The results of luciferase assay demonstrated that
miR-544 mimics decreased the luciferase activity of wtE2F5 compared
with the miR-NC (P<0.05; Fig.
3B). No significant difference in the luciferase activity of
mutE2F5 was identified (Fig. 3B).
Furthermore, RT-qPCR and western blot analysis showed decreased
expression of E2F5 with miR-544 overexpression at both the mRNA
(P<0.05; Fig. 3C) and protein
levels (Fig. 3D) in KYSE450 and TE-1
cells.
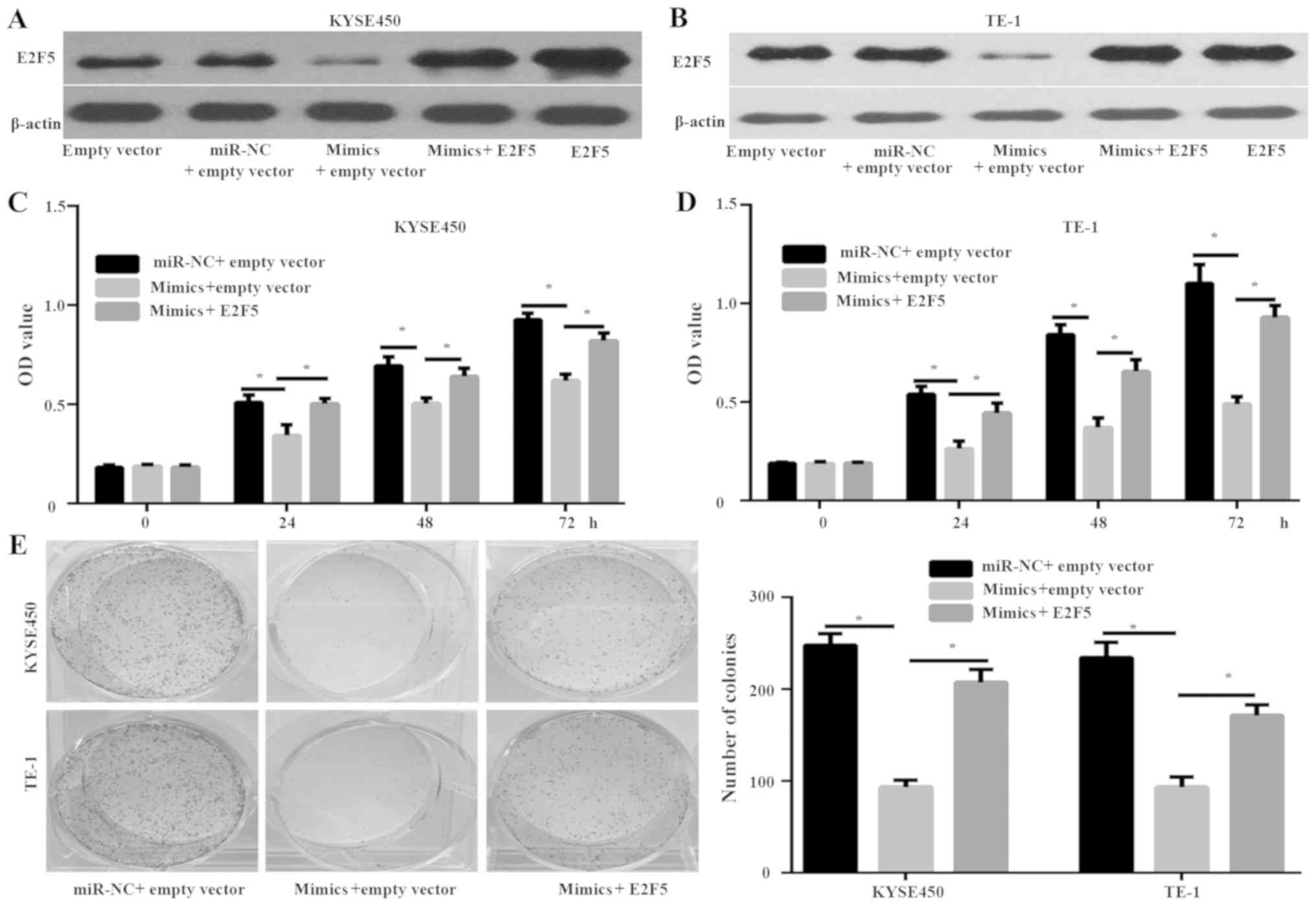
E2F5 is a functional target of miR-544
in ESCC cells
The potential of E2F5 as a functional target of
miR-544 was investigated. The KYSE450 and TE-1 cells were
co-transfected with miR-544 mimics and E2F5 plasmid (Fig. 4). Western blot analysis demonstrated
that the expression of E2F5 was significantly increased in KYSE450
and TE-1 cells that were transfected with E2F5 plasmid groups
compared with the empty plasmid groups (Fig. 4A and B). In addition, E2F5 expression
was increased in KYSE450 and TE-1 cells transfected with miR-544
mimics and E2F5 plasmid compared with that in cells transfected
with miR-544 mimics and empty plasmid (Fig. 4A and B). Furthermore, MMT (P<0.05;
Fig. 4C and D) and colony-formation
assays (P<0.05; Fig. 4E)
indicated that overexpression of E2F5 rescued the inhibitory effect
of miR-544 mimics on the proliferation of KYSE450 and TE-1 cells.
These data suggest E2F5 acts as a functional target of miR-544 in
ESCC cells.
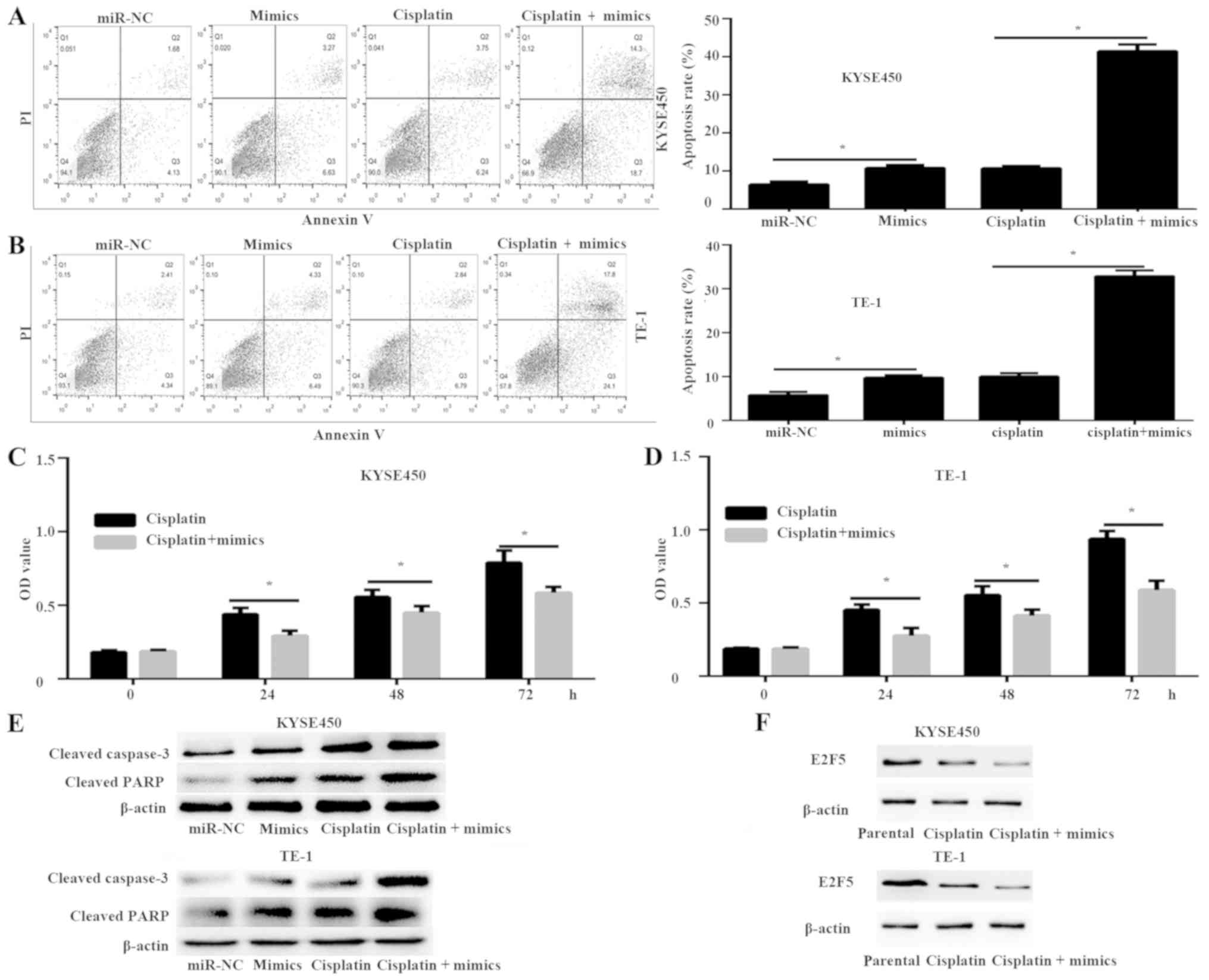
miR-544 increases the chemosensitivity
of ESCC cells to cisplatin
The present study investigated the effect of miR-544
on the sensitivity of KYSE450 and TE-1 cells to cisplatin. Flow
cytometry was used to analyze the apoptotic rate of KYSE450 and
TE-1 cells transfected with miR-544 mimics and E2F5 plasmid that
were treated with or without 15 µM cisplatin. The results of flow
cytometry demonstrated that the miR-544 mimics significantly
increased the apoptotic rate of KYSE450 and TE-1 cells compared
with the miR-NC-transfected cells, which was further enhanced by
cisplatin (P<0.05; Fig. 5A and
B). Furthermore, the MTT assay demonstrated that miR-544 mimics
and cisplatin groups significantly decreased cell proliferation
compared with the cisplatin groups in both KYSE450 and TE-1 cells
(P<0.05; Fig. 5C and D). In
addition, the expression of apoptosis-associated proteins cleaved
caspase-3 and cleaved PARP were detected. The expression of cleaved
caspase-3 and cleaved PARP was significantly enhanced in the
miR-544 mimics and cisplatin groups compared with the miR-544
mimics or cisplatin groups, respectively (Fig. 5E). These data demonstrated that
miR-544 can enhance the chemosensitivity to cisplatin. Finally, the
role of E2F5 in the enhanced chemosensitivity to cisplatin of
miR-544 mimics was investigated. The protein expression of E2F5 in
the cisplatin group was downregulated compared with the parental
group, and the miR-544 mimics and cisplatin group had decreased
expression of E2F5 compared with the cisplatin group (Fig. 5F). Overall, these data demonstrate
increased chemosensitivity of ESCC cells to cisplatin by miR-544
via E2F5.
Discussion
The rapid proliferative ability of ESCC is a
critical factor contributing to its dismal prognosis (3); however, the molecular mechanisms
involved remain incompletely understood. Thus, the identification
of key genes that are dysregulated in ESCC tissues and the
elucidation of the mechanisms leading to aberrant expression of
genes promoting ESCC progression are essential to achieve
successful management of ESCC. Previous studies have shown that
miRNA expression is aberrant in ESCC, which suggests that miRNAs
play an important role in ESCC progression (6,7). miR-544
was recently demonstrated to function either as a tumor suppressor
or oncogene in different types of cancer (8–13,16).
However, the expression, function and molecular mechanism of
miR-544 in ESCC have not been well illustrated. Therefore,
examining the effects of miR-544 in ESCC is important.
Previous studies have shown that miR-544 targets
different genes in various cancer types. In osteosarcoma, miR-544
promotes cell proliferation by negatively regulating axin-2
(8). In hepatocellular carcinoma, it
enhances immune escape through downregulation of natural
cytotoxicity triggering receptor 1/NKp46 by targeting runt-related
transcription factor 3 (9). miR-544
also promotes the cell proliferation and metastasis of colorectal
cancer by directly targeting forkhead box O1 (10). In glioblastoma, miR-544 inhibits cell
proliferation, invasion and migration and increases cell apoptosis
by targeting Parkinsonism associated deglycase (13). Previous data have reported that E2F5
is overexpressed in various types of cancer, including breast
cancer, epithelial ovarian cancer, ESCC, prostate cancer and
hepatocellular carcinoma; it was also associated with cancer
progression and prognosis (17–21).
Certain miRNAs, such as miR-34a and miR-1179, could regulate the
function of E2F5 (22,23). However, the mechanism of E2F5 in
ESCC, as well as the correlation of miRNAs and E2F5 in ESCC, was
not completely demonstrated. The present study demonstrates, for
the first time, that miR-544 could negatively regulate E2F5 in ESCC
by using a luciferase assay, western blot analysis and RT-qPCR.
E2F5 overexpression could rescue the inhibitory effect of miR-544
mimics in ESCC cells. These data demonstrate that E2F5 is a
functional target gene of miR-544. Moreover, the expression levels
of miR-544 and E2F5 were negatively correlated in ESCC tissues. The
function of E2F5 in drug resistance is beyond the scope of the
present study; however, it will be the focus of future studies.
In summary, miR-544 is downregulated in human ESCC
and functions as a tumor suppressor by inducing cell proliferation
and drug resistance. These functions are mediated by inhibiting the
expression of its direct target gene, E2F5, an oncogene associated
with tumorigenesis in certain types of cancer. Therefore, the
miR-544/E2F5 axis may be considered as a potential therapeutic
target in ESCC.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data analyzed during the present study are
included in this published article.
Authors' contributions
FS and KW conceived and designed the experiments.
FS, CZ, DLM and KW performed all the experiments. CZ and KW wrote
and revised the manuscript. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The present study was approved by the Research
Ethics Committee of Linyi Central Hospital (Lin Yi, China). Written
informed consent was obtained from all patients prior to
enrolment.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
Statistics, 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Enzinger PC and Mayer RJ: Esophageal
cancer. N Engl J Med. 349:2241–2252. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Abnet CC, Arnold M and Wei WQ:
Epidemiology of esophageal squamous cell carcinoma.
Gastroenterology. 154:360–373. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Baba Y, Saeki H, Nakashima Y, Oki E,
Shigaki H, Yoshida N, Watanabe M, Maehara Y and Baba H: Review of
chemotherapeutic approaches for operable and inoperable esophageal
squamous cell carcinoma. Dis Esophagus. 30:1–7. 2017.
|
|
5
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Mei LL, Qiu YT, Zhang B and Shi ZZ:
MicroRNAs in esophageal squamous cell carcinoma: Potential
biomarkers and therapeutic targets. Cancer Biomark. 19:1–9. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Harada K, Baba Y, Ishimoto T, Shigaki H,
Kosumi K, Yoshida N, Watanabe M and Baba H: The role of microRNA in
esophageal squamous cell carcinoma. J Gastroenterol. 51:520–530.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chen M, Liu YY, Zheng MQ, Wang XL, Gao XH,
Chen L and Zhang GM: microRNA-544 promoted human osteosarcoma cell
proliferation by downregulating AXIN2 expression. Oncol Lett.
15:7076–7082. 2018.PubMed/NCBI
|
|
9
|
Pan C, Xiang L, Pan Z, Wang X, Li J, Zhuge
L, Fang P, Xie Q and Hu X: MiR-544 promotes immune escape through
downregulation of NCR1/NKp46 via targeting RUNX3 in liver cancer.
Cancer Cell Int. 18:522018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yao GD, Zhang YF, Chen P and Ren XB:
MicroRNA-544 promotes colorectal cancer progression by targeting
forkhead box O1. Oncol Lett. 15:991–997. 2018.PubMed/NCBI
|
|
11
|
Zhi Q, Guo X, Guo L, Zhang R, Jiang J, Ji
J, Zhang J, Zhang J, Chen X, Cai Q, et al: Oncogenic miR-544 is an
important molecular target in gastric cancer. Anticancer Agents Med
Chem. 13:270–275. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhu Z, Wang S, Zhu J, Yang Q, Dong H and
Huang J: MicroRNA-544 down-regulates both Bcl6 and Stat3 to inhibit
tumor growth of human triple negative breast cancer. Biol Chem.
397:1087–1095. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Jin S, Dai Y, Li C, Fang X, Han H and Wang
D: MicroRNA-544 inhibits glioma proliferation, invasion and
migration but induces cell apoptosis by targeting PARK7. Am J
Transl Res. 8:1826–1837. 2016.PubMed/NCBI
|
|
14
|
Sobin LH, Gospodarowicz MK and Wittekind
C: International union against cancer (UICC) TNM classification of
malignant tumoursWiley-Liss; New York: pp. 73–76. 2010
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Fang R, Zhao NN, Zeng KX, Wen Q, Xiao P,
Luo X, Liu XW and Wang YL: MicroRNA-544 inhibits inflammatory
response and cell apoptosis after cerebral ischemia reperfusion by
targeting IRAK4. Eur Rev Med Pharmacol Sci. 22:5605–5613.
2018.PubMed/NCBI
|
|
17
|
Ren B, Cam H, Takahashi Y, Volkert T,
Terragni J, Young RA and Dynlacht BD: E2F integrates cell cycle
progression with DNA repair, replication and G(2)/M checkpoints.
Genes Dev. 16:245–256. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kothandaraman N, Bajic VB, Brendan PN,
Huak CY, Keow PB, Razvi K, Salto-Tellez M and Choolani M: E2F5
status significantly improves malignancy diagnosis of epithelial
ovarian cancer. BMC Cancer. 10:642010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ishimoto T, Shiozaki A, Ichikawa D,
Fujiwara H, Konishi H, Komatsu S, Kubota T, Okamoto K, Nakashima S,
Shimizu H, et al: E2F5 as an independent prognostic factor in
esophageal squamous cell carcinoma. Anticancer Res. 33:5415–5420.
2013.PubMed/NCBI
|
|
20
|
Zhao J, Wu XY, Ling XH, Lin ZY, Fu X, Deng
YH, He HC and Zhong W: Analysis of genetic aberrations on
chromosomal region 8q21-24 identifies E2F5 as an oncogene with copy
number gain in prostate cancer. Med Oncol. 30:4652013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Jiang Y, Yim SH, Xu HD, Jung SH, Yang SY,
Hu HJ, Jung CK and Chung YJ: A potential oncogenic role of the
commonly observed E2F5 overexpression in hepatocellular carcinoma.
World J Gastroenterol. 17:470–477. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lin C, Hu Z, Yuan G, Su H, Zeng Y, Guo Z,
Zhong F, Jiang K and He S: MicroRNA-1179 inhibits the
proliferation, migration and invasion of human pancreatic cancer
cells by targeting E2F5. Chem Biol Interact. 291:65–71. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Li L, Wu C and Zhao Y: miRNA-34a enhances
the sensitivity of gastric cancer cells to treatment with
paclitaxel by targeting E2F5. Oncol Lett. 13:4837–4842. 2017.
View Article : Google Scholar : PubMed/NCBI
|