Introduction
Gastric cancer was one of the four most common
malignancies and the third leading cause of cancer-associated
mortality in the world in 2014 (1,2).
Although the incidence of gastric cancer has decreased in recent
years (1,2), the prognosis of patients remains
unsatisfactory due to the lack of effective early detection methods
and efficient prognostic markers. In the early stages of gastric
cancer, patients experience no specific symptoms, and the majority
of patients with gastric cancer are diagnosed at advanced stages,
thereby missing the optimal treatment time and, subsequently,
resulting in a 5 year survival rate of <20% (3). Therefore, the development of novel
molecular markers and therapeutic targets for the treatment of
gastric cancer is essential.
The results of molecular genetics studies
demonstrated that gastric cancer is a malignant tumour, and that
gastric cancer occurrence and development is a multi-stage process,
with multiple genes and multiple factors involved in a complex
process; these genes include oncogenes, tumour suppressor genes and
DNA mismatch repair genes (4,5). STAM
binding protein-like 1 (STAMBPL1) is a key member of the COP9
signalosome subunit 5/serine protease 27/proteasome 26S subunit
non-ATPase 7 (JAMM) family, and a previous study reported that
STAMBPL1 was closely associated with tumour development (5). A number of studies reported that
STAMBPL1 (also known as AMSHLP) has a positive effect on NF-κB
activation, and that it is required for optimal Tax-induced
activation of canonical and noncanonical NF-κB pathways which are
the key component of inflammation development (6,7).
However, the effects and mechanism of STAMBPL1 remain unclear. In
the present study, STAMBPL1 expression in gastric cancer tissues at
different stages was evaluated, and the association between
STAMBPL1 protein expression and gastric cancer stages in clinical
settings was analysed. Furthermore, the present study demonstrated
how STAMBPL1 knockdown affected the biological activities of AGS
gastric cancer cells, including proliferation, apoptosis, invasion
and migration, as revealed by experiments investigating the NF-κB
pathway in vitro.
Materials and methods
Clinical data and samples
Adjacent normal (5 cm away from cancer tissue) and
gastric cancer tissues were collected from patients with gastric
cancer (n=36) who were treated at The First Affiliated Hospital of
Bengbu Medical College between December 2016 and November 2017. The
tissues were collected from female (n=16) and male (n=20) patients
aged between 30 and 52 years (mean age, 45±4.5 years). The subjects
included 24 stage I–II and 12 stage III–IV (8) patients with gastric cancer. All
patients were treated for the first time and none of the patients
received treatment prior to radical surgery. The adjacent normal
and cancer tissues were fixed by 10% formaldehyde at room
temperature for ≥24 h. The present study was approved by the Ethics
Committee of The First Affiliated Hospital of Bengbu Medical
College. All patients provided written informed consent. Following
surgery, the tissues were quickly stored in 10% formaldehyde for
fixation until further use.
Haematoxylin and eosin (H&E)
staining
The tissues were fixed in 10% formaldehyde at room
temperature and embedded in paraffin. The wax blocks were sliced
into sections of 4 µm thickness. The sections of different tissues
were stained by H&E at room temperature to 6 h, and were
subjected to pathological classification by TNM staging (8). The sections were observed under a light
microscope. No cancer cells were detected in the adjacent normal
tissues.
Immunohistochemical (IHC)
staining
The tissues were fixed in 10% formaldehyde at room
temperature and embedded in paraffin. The paraffin blocks were
sliced into 5 µm sections. The sections were incubated at 60°C in
an oven overnight, dewaxed using a gradient alcohol series (100, 95
and 80%) and endogenous peroxidase activity was blocked using 3%
H2O2 at room temperature for 2 h. The
remaining steps of IHC staining were performed according to the
manufacturer's instructions of the IHC kit (Wuhan Boster Biological
Technology, Ltd.). The primary antibody against STAMBPL1 (1:500;
cat. no. ab229144; Abcam) was added and incubated at 4°C overnight.
Following washing with PBS at room temperature, secondary rabbit
immunoglobulin G antibody (1:5,000; cat. no. ab205718; Abcam) was
added for 1 h at room temperature. Subsequently, the sections were
observed under a light microscope, and the images were analysed
using Image-Pro Plus version 6.0 image analysis software (Version
X; Media Cybernetics, Inc.) to measure the STAMBPL1 protein
expression in different tissues.
Cell lines and cell culture
Gastric cancer cell lines (AGS and MGC80-3) were
purchased from Nanjing KeyGen Biotech Co., Ltd. RPMI 1640 medium
(Abcam) containing 10% FBS (Abcam) and 100 U/ml penicillin and 100
µg/ml streptomycin (Invitrogen; Thermo Fisher Scientific, Inc.) was
used to cultivate gastric cancer cell lines at 37°C with 5%
CO2.
Reverse transcription-quantitative PCR
(RT-qPCR) assay
The AGS and MGC80-3 cells were collated, and total
RNA was extracted using TRIzol kits (Takara Bio, Inc.). GoScript™
Reverse Transcription System (Promega Corporation) was used for RT
of 2 µg total RNA to cDNA. The reaction parameters were as follows:
37°C for 10 min, 42°C for 45 min and 70°C for 5 min, followed by
cooling on ice for 5 min. Subsequently, 4 µl GoScript™ 5X reaction
buffer, 1.7 µl MgCl2 (final concentration 2 mM), 1 µl
0.5 mM dNTPs, 0.3 µl ribonuclease inhibitor (20 U), 1 µl reverse
transcriptase and ddH2O to a total of 15 µl was added. Following
mixing, the samples were incubated at 42°C for 60 min and
inactivated at 70°C for 15 min. RT-qPCR was performed using
SYBR® Green Master Mix (Promega Corporation). The qPCR
was performed in a total volume of 10 µl and included the
following: 5 µl GoTaq 2X Master Mix, 0.5 µl forward primer (10 µM),
0.5 µl reverse primer (10 µM), 2 µl cDNA (diluted 10-fold) and 2 µl
ddH2O. The primers were purchased from Nanjing KeyGen
Biotech Co., Ltd. The sequences of primers used were as follows:
STAMBPL1 forward 5′-AGGCAGAAAGGAAGCGGATTG-3′ and reverse,
5′-TTGCTGACTTCGCATTTGACC-3′; and GAPDH (reference gene) forward,
5′-TGACTTCAACAGCGACACCCA-3′ and reverse
5′-CACCCTGTTGCTGTAGCCAAA-3′. The thermocycling conditions were as
follows: Pre-denaturation at 94°C for 2 min, followed by 35 cycles
of 94°C for 45 sec and 55°C for 60 sec and a final extension at
65°C for 20 sec. The relative expression of the gene was calculated
using the 2−∆∆Cq (9)
method.
Cell grouping and transfection
AGS cells were assigned to the short hairpin RNA
(sh)Ctrl group, which was transfected with empty vector, and the
shSTAMBPL1 group, which was transfected with 2 ng/ml shSTAMBPL1
(Shanghai GeneChem Co., Ltd.) by Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol to knockdown STAMBPL1. At 48 h, the cells
were collected for use in further experiments.
Celigo cell count assay
Logarithmic growth period cells of different groups
were collected and digested using 0.05% pancreatic enzyme to
suspend the AGS cells in RPMI 1640 medium, followed by cell
counting. AGS cells (100 µl; 1×106 cells/ml) were added
to each well of a 6-well plate and cultured at 37°C with 5%
CO2. Starting the next day, the Celigo assay was
performed every day for 5 consecutive days.
MTT assay
AGS cell proliferation was measured using an MTT
assay kit (Beijing Dingguo Changsheng Biotechnology Co., Ltd.). A
total of 2×103 cells were added to each well of a
96-well plate. The cells were allowed to adhere and 20 µl MTT
solution (5 mg/ml) was added at different time points (days 1, 2,
3, 4 and 5) and incubated for 4 h at room temperature.
Subsequently, 100 µl DMSO was added to the wells to stop the
reaction. Cell proliferation was finally measured for different
cell groups at different time points at 490 nm.
Cell cloning test
The logarithmic AGS cells of the shCtrl and
shSTAMBPL1 groups were collected and suspended in the medium. The
cells were inoculated into a 6-well plate at a density of 1,000
cells/well. Following inoculation, the cells of different groups
were continually cultured for 14 days at room temperature.
Subsequently, they were washed by PBS, followed by the addition of
1 ml 4% paraformaldehyde at room temperature to a final
concentration of 2% to fix the AGS cells for 45 min at room
temperature and washing by PBS. Next, 1,000 µl of 4% crystal violet
dye was added to the reaction mix for 15 min at room temperature.
ddH2O was used to wash the cells three times, followed
by the counting of the cloned cell numbers and capturing of images
under a light microscope at ×200 magnification.
Cell apoptosis Annexin
V-allophycocyanin (APC) assay
AGS cell apoptosis rates of the shCtrl and
shSTAMBPL1 groups were measured using an Flow Cytometry apoptosis
kit (cat. no. 88-8007; eBioscience; Thermo Fisher Scientific, Inc.)
according to the manufacturer's instructions. In brief, the cells
of the experimental groups were collected and centrifuged for 5 min
at 1,300 × g at 4°C, the supernatant was removed, and pre-cooled
D-Hanks (pH 7.2–7.4) was used to wash the cells at 4°C. Binding
buffer (1X) was used to wash the cells, followed by centrifugation
for 3 min at 1,300 × g at 4°C to remove the supernatant. Then, the
cells were collected, and 200 µl 1X Binding Buffer was added to
prepare the suspension. Subsequently, 10 µl Annexin V-APC was added
for staining and allowed to react for 15 min in the dark at room
temperature. Subsequently, cell apoptosis was measured by flow
cytometry analysis (FACScalibur; BD Biosciences).
Transwell assay
Matrigel was diluted with fresh medium with 10% FBS
at a 1:6 ratio (by volume) after refrigerating at 4°C overnight,
and the Transwell insert was placed in a 24-well plate.
Subsequently, 50 µl Matrigel was added and allowed to dry prior to
further use. The cells were digested by adding trypsin to the
single cell suspension, and the cell density was adjusted with 10%
FBS. To each well, 1×105 cells in 600 µl RPMI 1640
medium were added to the upper chamber, and RPMI 1640 medium with
10% FBS was added to the lower chamber, followed by incubation for
24 h at room temperature. Subsequently, the Transwell chambers were
removed and the culture medium was collected, washed with PBS three
times, fixed with 95% ethanol and stained with 0.5% crystal violet
for 10 min at room temperature., with a final washing step of
washing with PBS three times. Using a cotton swab, the cells were
gently wiped off the membrane of the upper chamber, and five fields
were randomly selected to count the numbers of cells under a light
microscope.
Wound healing assay
AGS cells of the shCtrl and shSTAMBPL1 groups were
collected, the medium of the 6-well plate was removed and the cells
were washed three times with PBS. A 200-µl pipette tip was used to
draw a straight line across the centre of the plate along the
longitudinal axis, followed by washing twice with PBS and the
addition of medium to continue culturing. After 0, 24 and 48 h,
five fields were randomly selected to count the number of cells
that had migrated into the wound area under an inverted light
microscope and images were captured. The experiment was repeated
three times.
Western blotting (WB) assay
The AGS cells of shCtrl and shSTAMBPL1 groups were
collected, 200 µl Cell Lysis Buffer (cat. no. 9803; Cell Signaling
Technology, Inc.) was added to extract total protein, and the
samples were placed in an ice bath for 20 min, followed by
centrifugation at 12,000 × g for 30 min at 4°C; subsequently, the
supernatant was removed. The bicinchoninic acid assay method was
used to measure the protein concentration. Proteins were separated
using 10% SDS-PAGE with 25 µg protein per lane, and the proteins
were transferred onto polyvinylidene difluoride membranes. The
membranes were blocked with 5% skimmed milk for 2 h at room
temperature. STAMBPL1 (1:500; cat. no. ab205718; Abcam), NF-κB
(1:500; cat no. ab32536; Abcam) and GAPDH (1:1,000; cat no.
ab181602; Abcam) antibodies were added and incubated overnight at
4°C. Then, the membrane was washed three times with PBS for 5 min
and incubated with a horseradish peroxidase-conjugated anti-rabbit
IgG secondary antibody (dilution, 1:1,000; cat. no. MBS435036;
MyBioSource, Inc.) at room temperature for 2 h. Following washing
with PBS, ECL (cat. no. P0018; Beyotime Institute of Biotechnology)
was added and the membrane was placed in the Gel Imaging system to
develop. The ratio of the target stripe grey value to the internal
reference GAPDH grey scale value represented the relative
expression amount of each target protein. Quantity one version 4.62
software (Bio-Rad Laboratories, Inc.) was used for analysis.
Statistical analysis
The clinical and experimental data were analysed by
SPSS version 22.0 software (IBM Corp.). The data are expressed as
the mean ± standard deviation, and the differences among three or
more groups were analysed by one-way ANOVA followed by a post hoc
least significant difference-t test. Student's t-test was used to
analyse the differences between two groups. P<0.05 was
considered to indicate a statistically significant difference. All
experiments were repeated 3 times.
Results
Clinical data and analysis
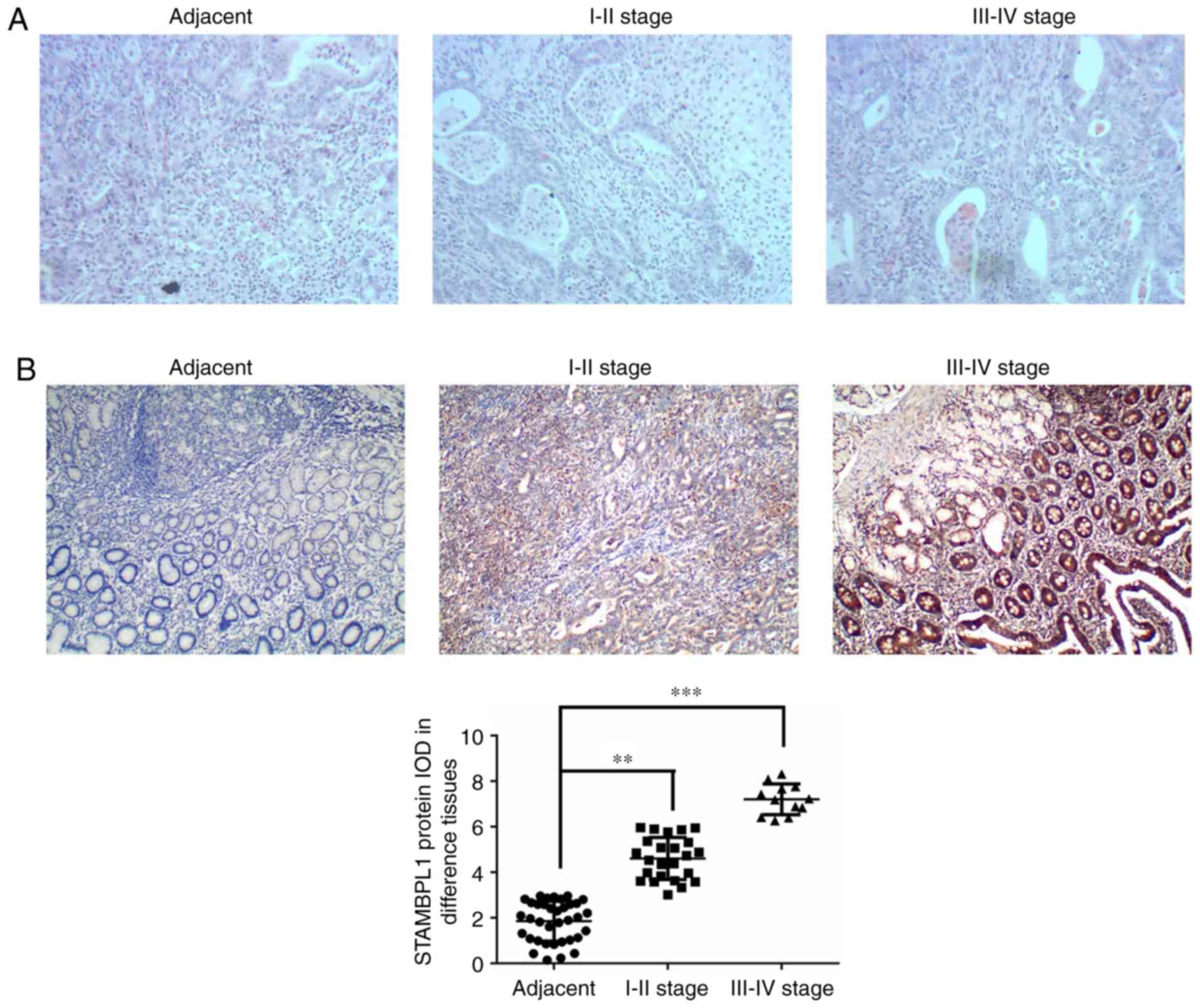
H&E staining revealed infiltration of the
gastric cancer cells compared with the adjacent normal tissues and
this was upregulated with increasing disease stage (Fig. 1A). To evaluate STAMBPL1 protein
expression in different tissues, STAMBPL1 expression was measured
by IHC staining in adjacent normal, stage I–II and stage III–IV
gastric cancer tissues. The present study demonstrated that
STAMBPL1 protein expression was significantly upregulated in stage
I–II and stage III–IV gastric cancer tissues compared with in the
adjacent normal tissues (P<0.01 and P<0.001, respectively;
Fig. 1B).
STAMBPL1 expression and
transfection
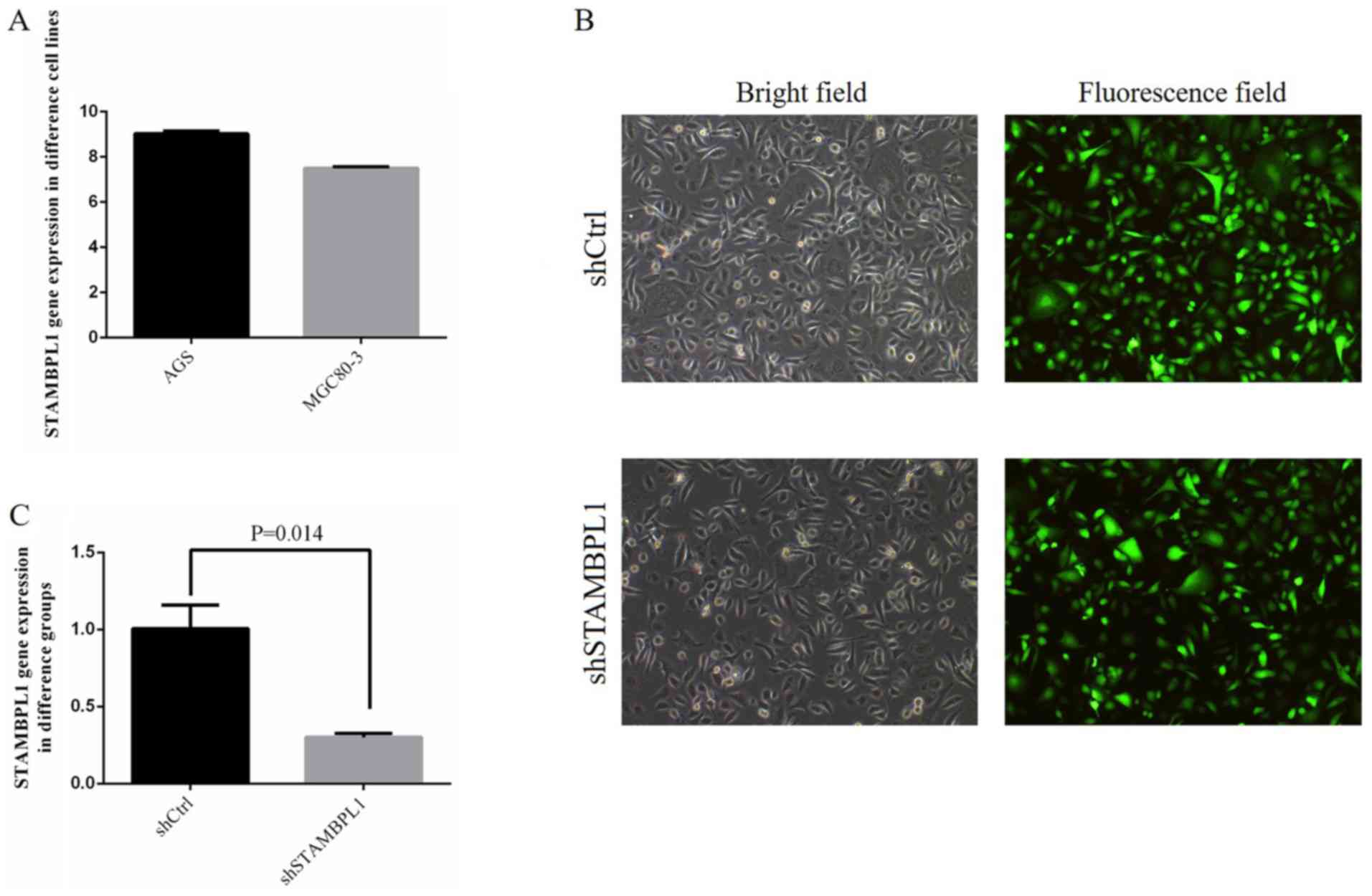
No significant differences were identified among the
AGS and MGC80-3 cells in terms of STAMBPL1 gene expression
(Fig. 2A). The STAMBPL1 gene
expression in AGS was higher compared with that in MGC80-3 cells;
therefore, the AGS cell line was selected as a representative
gastric cancer cell line.
For shCtrl and shSTAMBPL1 transfection, the
efficiency of transfection reached >80% and the cell state was
normal (Fig. 2B). Following
transfection, STAMBPL1 gene expression in the shSTAMBPL1 group was
significantly decreased compared with that in the shCtrl group
(P=0.014; Fig. 2C).
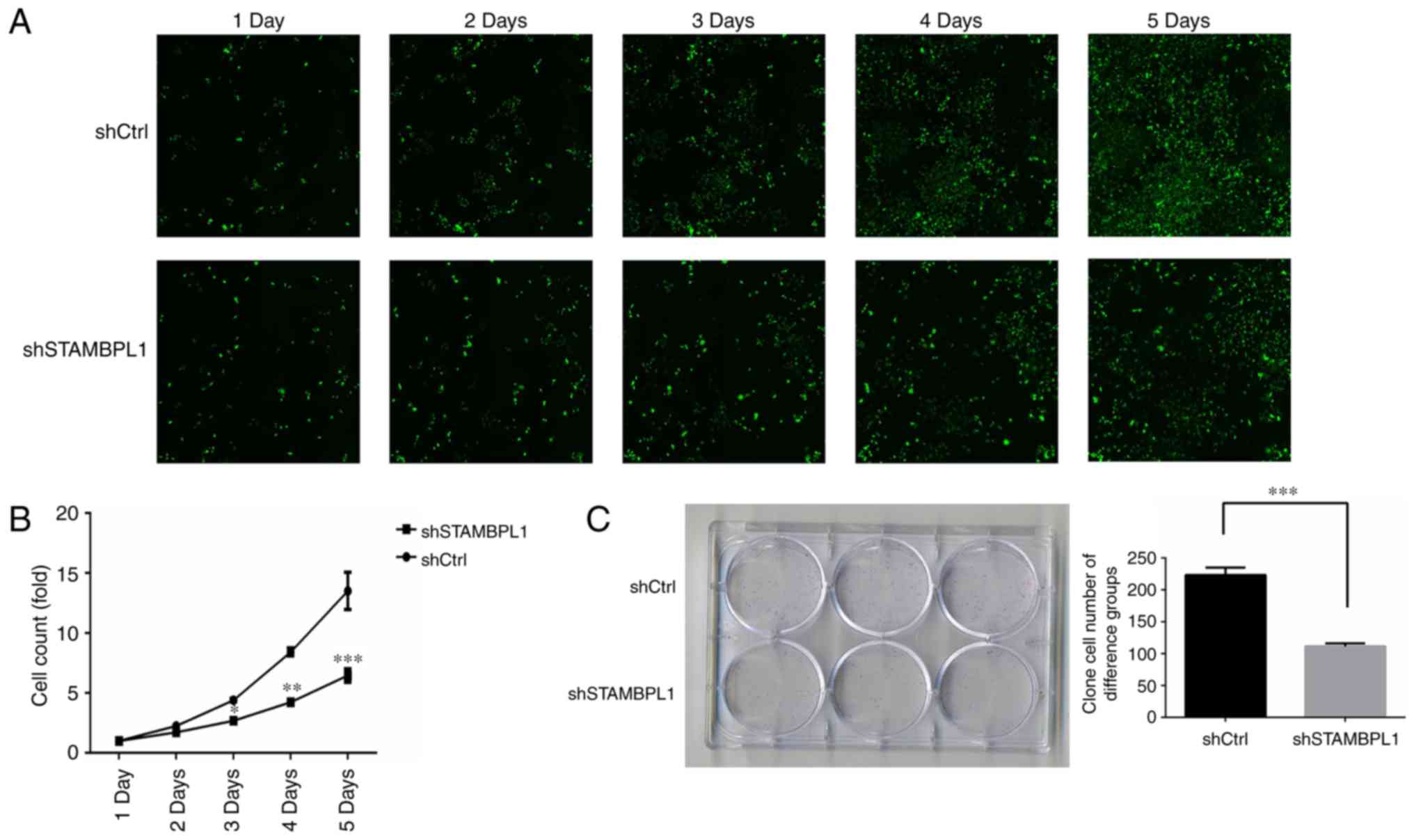
Cell proliferation
Following transfection, cell proliferation was
evaluated by a Celigo assay for 5 days. Cell proliferation was
suppressed from day 3 in the shSTAMBPL1 group (Fig. 3A). An MTT assay revealed that the
cell proliferation rates of the shSTAMBPL1 group were significantly
decreased compared with those of the shCtrl group on days 3, 4 and
5 (P<0.05, P<0.01 and P<0.001, respectively; Fig. 3B). Following transfection for 5 days,
AGS cell proliferation was evaluated using a clone test. The
results of the present study revealed that the AGS clone cell
numbers of the shSTAMBPL1 group were significantly decreased
compared with those of the shCtrl group (P<0.001; Fig. 3C). These results suggested that AGS
cell proliferation was suppressed by STAMBPL1 knockdown.
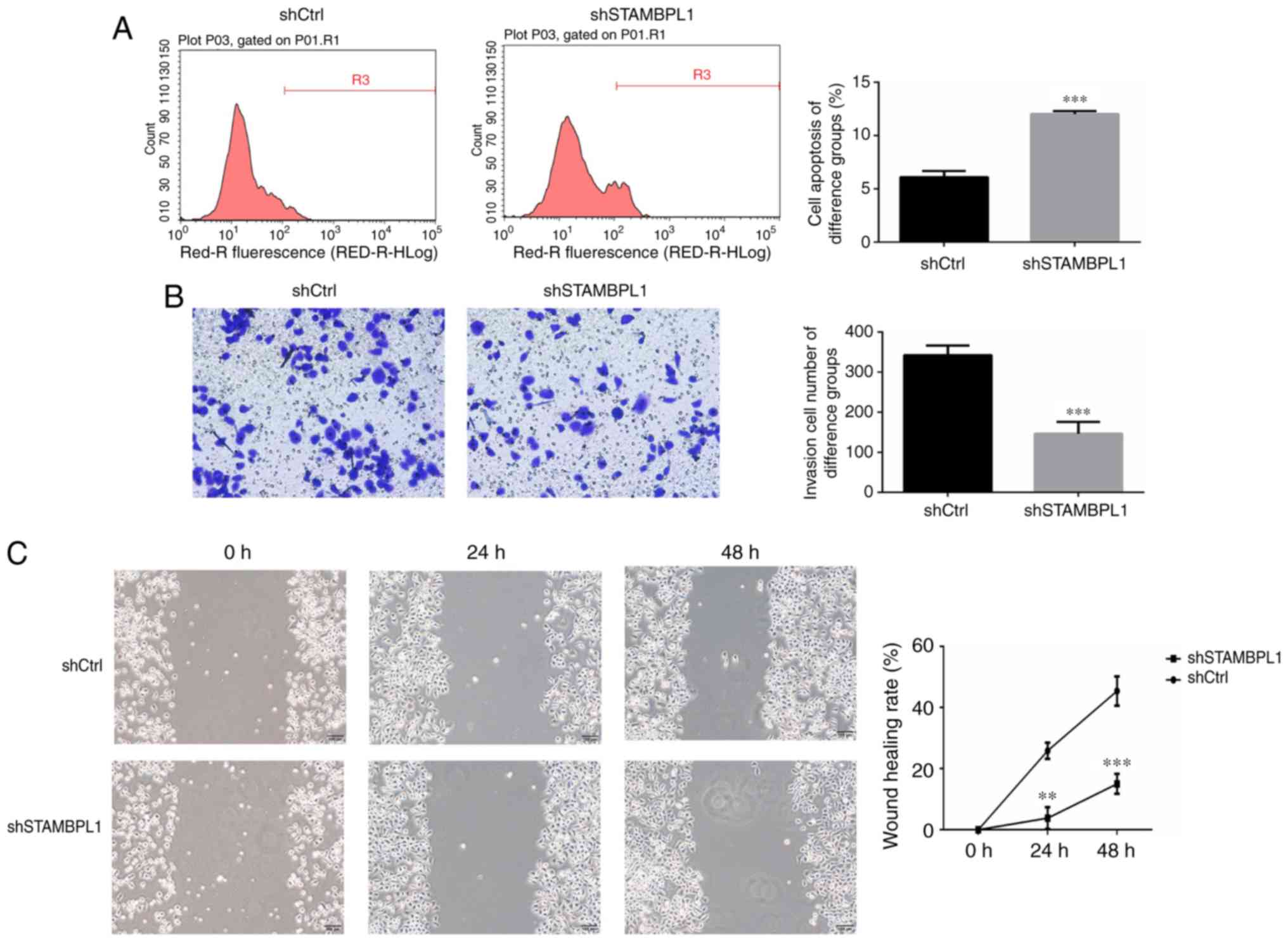
AGS cell apoptosis, invasion and
migration
The results of the Annexin V-APC assay revealed that
the AGS cell apoptosis rate was significantly increased in the
STAMBPL1 knockdown group compared with the shCtrl group
(P<0.001; Fig. 4A). AGS cell
invasion abilities of the shCtrl and shSTAMBPL1 groups were
evaluated using a Transwell assay. The results revealed that
numbers of invasive AGS cells in the shSTAMBPL1 group were
significantly decreased compared with those in the shCtrl group
(P<0.001; Fig. 4B). To evaluate
the AGS cell migration ability, the wound-healing distance in
different groups of AGS cells were measured at 0, 24 and 48 h
post-transfection. The results revealed that the wound healing rate
of the shSTAMBPL1 group was significantly decreased compared with
that of the shCtrl group after 24 and 48 h (P<0.01 and
P<0.001, respectively; Fig.
4C).
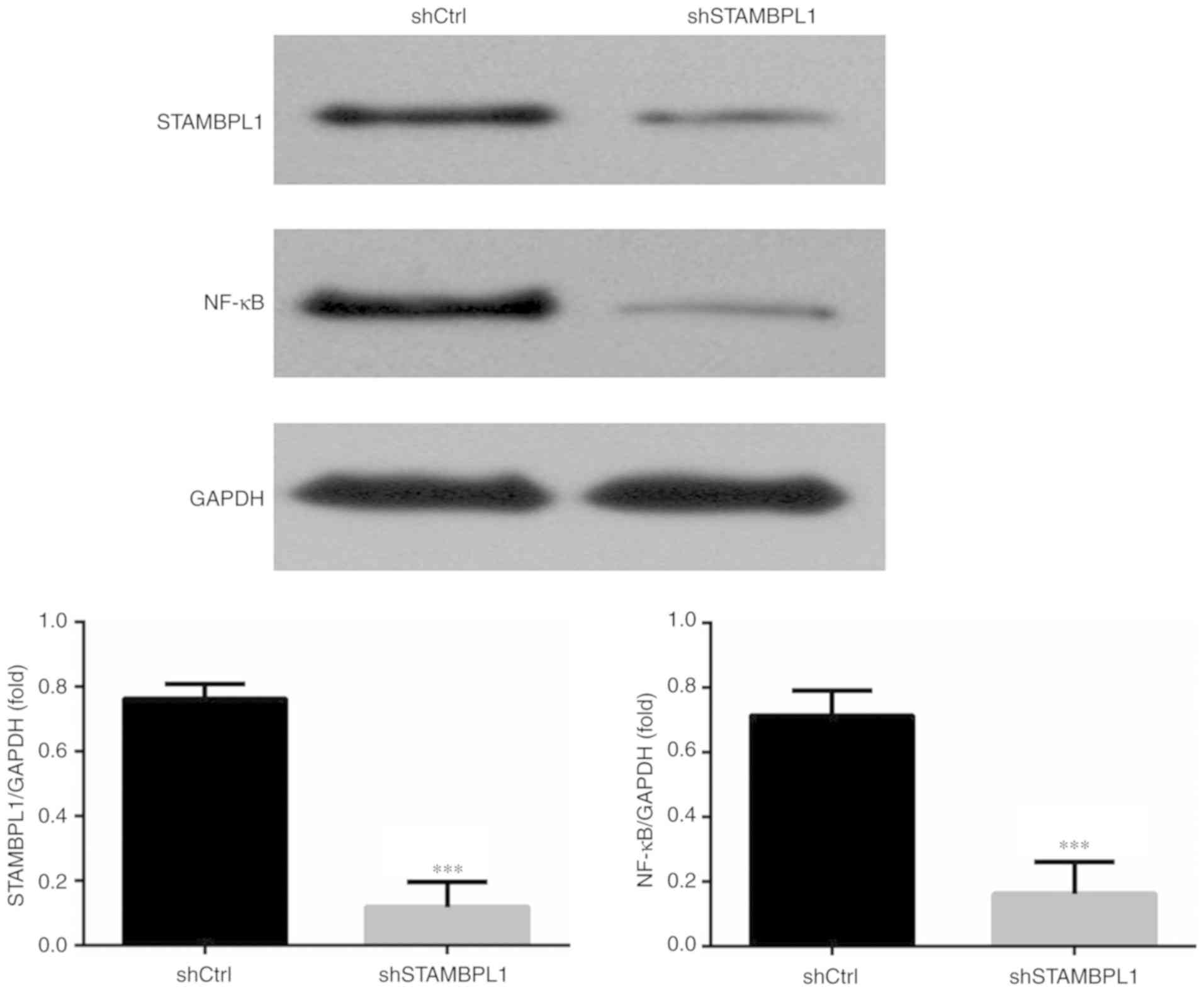
Relative protein expression levels in
the WB assay
Compared with in the shCtrl group, the STAMBPL1 and
NF-κB protein expression levels of the shSTAMBPL1 group were
significantly decreased in AGS cells (P<0.001; Fig. 5).
Discussion
In China, the incidence of gastric cancer is high,
and it was the second-most common type of cancer in 2017 (10). The early symptoms of gastric cancer
are insignificant. On discovery, it usually presents at its
advanced or metastasised stage, when surgical treatment is not
ideal. Presently, a combination of surgery and chemotherapy is used
for treatment. However, chemotherapy may result in drug resistance,
and the drug toxicity is relatively strong (11). Invasion and metastasis occur
frequently in patients with advanced gastric cancer, which affects
the therapeutic efficiency of gastric cancer. Therefore, the
molecular mechanism of the invasion and metastasis of gastric
cancer cells is of great significance for the diagnosis and
treatment of gastric cancer.
STAMBPL1 is the main factor of the JAMM family
members. Additionally, previous studies have reported high
expression levels of STAMBPL1 in cancer tissues and that knockdown
of STAMBPL1 could suppress the biological activities of leukaemia
via regulation of NF-κB activity (12–14). In
the present study, it was also demonstrated that STAMBPL1 protein
expression was higher in gastric cancer tissues compared with that
in adjacent normal tissues and that the expression levels increased
with advanced stages in the clinical samples. Furthermore,
knockdown of STAMBPL1 by shSTAMBPL1 resulted in STAMBPL1
downregulation, which in turn decreased the cell proliferation of
AGS gastric cancer cells, increasing cell apoptosis, cell invasion
and migration in vitro. The present study also revealed that
NF-κB expression was decreased following STAMBPL1 knockdown.
Previous studies have indicated that NF-κB serves an
important role in cell proliferation, differentiation and apoptosis
pathophysiological processes (15,16).
Cell migration and invasion serve an important role in the process
of cancer metastasis (17,18). NF-κB is a transcription factor with
various regulatory functions in the body, and is involved in the
differentiation, apoptosis and migration of tumour cells and can
promote the development of tumours (19,20).
NF-κB is usually present in the cytoplasm in an inactive manner,
and it is activated by phosphorylation following stimulation. The
abnormal activation of its pathway can cause the abnormal
expression of several tumour suppressor genes, resulting in the
inhibition of apoptosis of tumour cells, normal cell
differentiation and, notably, NF-κB overexpression leads to tumour
metastasis (21–25). The results of the present study
suggested that NF-κB expression was significantly decreased
following STAMBPL1 downregulation, which may be associated with the
antitumour effects of STAMBPL1 knockdown.
There were some limitations of the present study.
Only one gastric cancer cell line (AGS) was extensively analysed in
the present study. In future studies, the effects and mechanism of
STAMBPL1 on biological activities in two or more gastric cancer
cell lines will be assessed.
In conclusion, STAMBPL1 may be an oncogene in
gastric cancer, and knockdown of STAMBPL1 affected the regulation
of gastric cancer cell biological activities, including the
proliferation, apoptosis, invasion and migration via suppression of
NF-κB signalling in vitro.
Acknowledgements
Not applicable.
Funding
The Natural Science Research Project of Education
Office of Anhui Province (No. KJ2019A0387)
Availability of data and materials
The datasets used and/or analysed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
DJY conceived and designed the study and guaranteed
its integrity. JQ participated in the design of the experiment and
performed literature research. XJ performed clinical and
experimental studies. JL acquired and analysed the data. CXG
preformed statistical analysis and prepared the manuscript. XCY
designed the study and analysed the data.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of The First Affiliated Hospital of Bengbu Medical
College. All patients provided written informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Kanda M and Kodera Y: Recent advances in
the molecular diagnostics of gastric cancer. World J Gastroenterol.
14:9838–9852. 2015. View Article : Google Scholar
|
|
2
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Perrotti D and Neviani P: Protein
phosphatase 2A: A target for anticancer therapy. Lancet Oncol.
14:e229–e238. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Qu X, Yu J, Bhagat G, Furuya N, Hibshoosh
H, Troxel A, Rosen J, Eskelinen EL, Mizushima N, Ohsumi Y, et al:
Promotion of tumorigenesis by heterozygous disruption of the Beclin
I antorpharge gene. J Clin Invest. 112:1809–1820. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lavorgna A and Harhaj EW: An RNA
interference screen identifies the Deubiquitinase STAMBPL1 as a
critical regulator of human T-cell leukemia virus type 1 tax
nuclear export and NF-κB activation. J Virol. 86:3357–3369. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Shahriyar SA, Woo SM, Seo SU, Min KJ and
Kwon TK: Cepharanthine enhances TRAIL-mediated apoptosis through
STAMBPL1-mediated downregulation of survivin expression in renal
carcinoma cells. Int J Mol Sci. 22:192018.
|
|
7
|
Berman TA and Schiller JT: Human
papillomavirus in cervical cancer and oropharyngeal cancer: One
cause, two diseases. Cancer. 15:2219–2229. 2017. View Article : Google Scholar
|
|
8
|
Pang L, Wang J, Fan Y, Xu R, Bai Y and Bai
L: Correlations of TNM staging and lymph node metastasis of gastric
cancer with MRI features and VEGF expression. Cancer Biomark.
23:53–59. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ma X, Ren D, Kan J, Zheng F, Zhang S,
Zhang Y, Li Y, Liu Z, Ye L, Shen G, et al: Clinicopathological
characteristics and prognoses of elderly gastric cancer patients
after R0 resection: A multicenter study in china. J Environ Pathol
Toxicol Oncol. 37:81–91. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Mazzei MA, Bagnacci G, Gentili F, Nigri A,
Pelini V, Vindigni C, Mazzei FG, Baiocchi GL, Pittiani F, Morgagni
P, et al: Gastric cancer maximum tumour diameter reduction rate at
CT examination as a radiological index for predicting
histopathological regression after neoadjuvant treatment: A
multicentre GIRCG study. Gastroenterol Res Pract.
15:17945242018.
|
|
12
|
Lee NH, Kim M, Oh SY, Kim SG, Kwon HC and
Hwang TH: Gene expression profiling of hematologic malignant cell
lines resistant to oncolytic virus treatment. Oncotarget.
3:1213–1225. 2017.
|
|
13
|
Li CW and Chen BS: Investigating core
genetic-and-epigenetic cell cycle networks for stemness and
carcinogenic mechanisms, and cancer drug design using big database
mining and genome-wide next-generation sequencing data. Cell Cycle.
15:2593–2607. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sacco JJ, Coulson JM, Clague MJ and Urbé
S: Emerging roles of deubiquitinases in cancer-associated pathways.
IUBMB Life. 62:140–157. 2010.PubMed/NCBI
|
|
15
|
Pahl HL: Activator and target genes of
Rel/NF-κB transcription factors. Oncogene. 18:68531999. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Karin M: The beginning of the end: IkappaB
kinase (IKK) and NF-kappa B activation. J Biol Chem.
24:27339–27342. 1999. View Article : Google Scholar
|
|
17
|
Appert-Collin A, Hubert P, Crémel G and
Bennasroune A: Role of ErbB receptors in cancer cell migration and
invasion. Front Pharmacol. 24:2832015.
|
|
18
|
Lauffenburger DA and Horwitz AF: Cell
migration: A physically integrated molecular process. Cell.
9:359–369. 1996. View Article : Google Scholar
|
|
19
|
Prabhu L, Mundade R, Korc M, Loehrer PJ
and Lu T: Critical role of NF-κB in pancreatic cancer. Oncotarget.
30:10969–10975. 2014.
|
|
20
|
Shinoda K, Kuboki S, Shimizu H, Ohtsuka M,
Kato A, Yoshitomi H, Furukawa K and Miyazaki M: Pinl facilitates
NF-κB activation and promotes tumour progression in human
hepatocellular carcinoma. Br J Cancer. 3:1323–1331. 2015.
View Article : Google Scholar
|
|
21
|
Zhao M, Gao Y, Wang L, Liu S, Han B, Ma L,
Ling Y, Mao S and Wang X: Overexpression of integrin-linked kinase
promotes lung cancer cell migration and invasion via NF-κB-mediated
upregulation of matrix megalloproteinase-9. Int J Med Sci.
14:995–1002. 2013. View Article : Google Scholar
|
|
22
|
Woo JH, Park JW, Lee SH, Kim YH, Lee IK,
Gabrielson E, Lee SH, Lee HJ, Kho YH and Kwon TK: Dykellic acid
inhibits phorbol myristate acetate-induced matrix
metalloproteinase-9 expression by inhibiting nuclear factor kappa B
transcriptional activity. Cancer Res. 15:3430–3434. 2003.
|
|
23
|
Yamamoto Y and Gaynor RB: Ikappa B kinase:
Key regulators of the NF-kappa B pathway. Trends Biochem Sci.
29:72–79. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhi Y, Duan Y, Zhou X, Yin X, Guan G,
Zhang H, Dong Q and Yang K: NF-κB signaling pathway confers
neuroblastoma cells migration and invasion ability via the
regulation of CXCR4. Med Sci Monit. 21:2746–2752. 2014.
|
|
25
|
Chiu CT, Chen JH, Chou FP and Lin HH:
Hibiscus sabdariffa leaf extract inhibits human prostate cancer
cell invasion via down-regulation of Akt/NF-κB/MMP-9 pathway.
Nutrients. 24:5065–5087. 2015. View Article : Google Scholar
|