Introduction
Non-small cell lung cancer (NSCLC) is a leading
cause of cancer-associated mortality worldwide, accounting for ~85%
of lung cancer cases (1,2). An estimated 1.8 million new lung cancer
cases and 1.6 million lung cancer-associated mortalities were
reported worldwide in 2012 (3).
Advances in genomic research and targeted therapy have greatly
improved the development of therapeutic strategies and the clinical
outcomes of patients with NSCLC (4,5).
Activation of specific tyrosine kinases, including those derived
from epidermal growth factor receptor (EGFR) mutations or the
rearrangement of the anaplastic lymphoma kinase (ALK) gene,
promotes development in the corresponding molecular inhibitors in
EGFR mutation and ALK translocation positive subgroups (6–8). In
addition to EGFR and ALK, a number of other oncogenic driver genes,
such as ROS1, BRAF, MET, RET and Erb-B2 receptor tyrosine kinase 2
(ERBB2) have been identified as relevant targets in NSCLC (9).
Next-generation sequencing (NGS) has led to
considerable advances in the comprehensive analysis of genomic
alterations in cancer research and clinical application (10,11). At
present, the detection of genomic alterations in tumors is
primarily reliant on cancer tissue analysis. Despite the benefits
and therapeutic insights offered by tissue sample testing, spatial
and temporal tumor heterogeneity, and insufficient and unacquirable
specimens can lead to limitations in the use of tissue-based
sequencing (12,13). Circulating tumor DNA (ctDNA),
released into the plasma from apoptotic and necrotic tumor cells
derived from primary tumors and metastatic lesions, are comprised
of tumor-specific sequence alterations and may be an alternative
target to tissue sample biopsies (14–16). An
increasing body of evidence has indicated that ctDNA testing could
be utilized for patients without acquirable tissue samples to
screen for genetic variations and thus guide treatment decisions in
patients with NSCLC (17–19).
Despite the increasing utilization of plasma ctDNA
mutations in guiding clinical decisions, few studies have evaluated
whether the frequency of multiple genomic alterations observed
following ctDNA sequencing is similar to that observed following
tissue sequencing in NSCLC (17,20).
Therefore, in order to investigate the associations in the
frequency of comprehensive genomic alterations between ctDNA and
tissue profiling in Chinese patients with NSCLC, the present study
analyzed 59 plasma ctDNA samples and 40 tissue samples via targeted
NGS. The results provide important insights into the potential
benefits of ctDNA profiling in patients with NSCLC in clinical
practice.
Materials and methods
Patients and sample collection
The present study enrolled 99 patients with NSCLC
who were baseline or previously treated at the Hunan Cancer
Hospital (Changsha, China) between September 2017 and April 2018.
The specimens included 59 plasma and 40 tissue samples. The
inclusion criteria for the patients were as follows: i) Patients
were diagnosed with NSCLC by histopathology or cytology; ii) tissue
specimens were confirmed by qualified pathologists, from Hunan
Cancer Hospital who were blinded to the study and ensured >30%
tumor content; and iii) at least 150 ng and 15 ng DNA from each
tissue and ctDNA sample, respectively, were successfully extracted.
Patients who had other malignant tumors and serious mental illness
were excluded. Written informed consent was provided by all
participants. All experiments were approved by and performed in
accordance with the relevant guidelines and regulations of the
Committee of Medical Ethics of Hunan Cancer Hospital.
Targeted deep sequencing
ctDNA was isolated from ≥2 ml plasma with a QIAamp
Circulating Nucleic Acid kit (Qiagen GmbH) according to the
manufacturer's protocol. Tissue DNA was extracted using the QIAamp
Genomic DNA kit (Qiagen GmbH). The quality of the DNA and DNA
quantification were assessed using the Agilent 2100 BioAnalyzer
(Agilent Technologies, Inc.) and Qubit ds DNA HS assay kit (Thermo
Fisher Scientific, Inc.).
Sequencing libraries were constructed according to
the Illumina standard library construction instructions (Illumina,
Inc.). Genomic DNA extracted from tissue samples was first sheared
to 250 bp with an ultrasonoscope. The fragmented DNA was subjected
to end-repairing, A-tailing and ligation to the adapters with
barcode sequences. Subsequently, PCR was performed and the
resulting products were purified with AMPure XP magnetic beads
(Agencourt AMPure XP kit; Beckman Coulter, Inc.) according to the
manufacturer's protocol. The various libraries were then hybridized
with a nine-gene panel, which was enriched for the coding regions
and selected introns of genes with known relevance to NSCLC. The
target-enriched libraries were then pooled and sequenced on an
Illumina HiSeq2500 NGS platform (Illumina, Inc.). The sequencing
depth was >10,000×. Reads were aligned to the human genomic
reference sequences (hg19) using the Burrows-Wheeler alignment
(BWA) tool (21). Local realignment
and base quality score recalibration were conducted using GATK
software (version 2.3; software.broadinstitute.org/gatk) (22). MuTect2 (version 1.1.1; software.broadinstitute.org/cancer/cga/mutect)
with the recommended parameters was used to identify
single-nucleotide variants (SNVs) and small insertions or deletions
(INDELs) (23). Copy number variants
(CNV) calling was performed with CONTRA software (version 2.0.4;
contra-cnv.sourceforge.net) (24).
The cancer genome Atlas (TCGA)
datasets
Multiplatform genomics data was previously published
in The Cancer Genome Atlas (TCGA; cancergenome.nih.gov). In total, 1,144 NSCLC cases
comprising 660 patients with lung adenocarcinoma (AD) and 484
patients with lung squamous cell carcinoma (SC) were included in
TCGA cohort with gene mutation data (25).
Detection of tumor serum markers
Tumor serum markers were detected by a C12
multi-tumor marker protein chip system (Shanghai HealthDigit Co.,
Ltd.). Based on the manufacturer's instruction, 100 µl of the
different calibrators, quantitative quality control products and
serum specimen were transferred to the reaction well of the chip,
which was subsequently incubated at 37°C for 30 min with agitation.
The mixture was discarded and the well was washed four times with
the wash solution. Next, 100 µl reaction solution was added to the
well and incubated at 37°C for 30 min with agitation. The mixture
was discarded and the well was washed four times with the wash
solution. Subsequently, detection solution A and B were added to
the surface of the chip and incubated at room temperature for 1.5
min. The chip was placed into the biochip scanner (HD-2001A;
Shanghai HealthDigit Co., Ltd.) to analyze each tumor serum
marker.
Statistical analysis
SPSS statistical software (version 21.0; IBM Corp.)
was used to analyze the data. Differences in continuous variables
were assessed using unpaired Student's t-test. Associations between
different genotypes and clinical characteristics were analyzed
using Fisher's exact test or χ2 test. Correlations
between variables were assessed using Spearman's rank correlation
coefficient. Patterns of co-alteration or mutual exclusivity
between EGFR and other driver genes were analyzed by Fisher's exact
test. The odds ratio (OR) is a statistic that quantifies the
strength of the association between two events. In the current
study, EGFR mutations and other driver gene mutations were
classified as events A and B, respectively. The odds ratio was
calculated as the ratio of the odds of A in the presence of B and
the odds of A in the absence of B. P<0.05 was considered to
indicate a statistically significant result.
Results
NGS-based tissue and ctDNA assays to
identify genomic alterations among 99 patients with NSCLC
In the present study, a total of 99 patients
diagnosed with NSCLC were enrolled. NGS was performed for
specimens, including 59 plasma ctDNA and 40 tissue samples. Among
the patients, the median age at diagnosis was 56 years (range,
39–83 years), 73.7% of the patients were male, 64.6% of the
patients were smokers, 58.6% of the patients were diagnosed with
AD, and 67.7% of the patients had stage IV cancer. The clinical and
pathological characteristics of the patients are listed in Table I.
 | Table I.Clinicopathological characteristics
among 99 patients with non-small cell lung cancer. |
Table I.
Clinicopathological characteristics
among 99 patients with non-small cell lung cancer.
| Characteristic | Total | Tissue, n=40 | cthNA, n=59 | P-value |
|---|
| Age, years, median
(range) | 56 (39–83) | 56 (46–77) | 58 (39–83) | 0.534 |
| Sex, n (%) |
|
|
| 0.112 |
|
Male | 73 (73.7) | 26 (65.0) | 47 (79.7) |
|
|
Female | 26 (26.3) | 14 (35.0) | 12 (20.3) |
|
| Smoking status, n
(%) |
|
|
| 0.068 |
|
Yes | 64 (64.6) | 24 (60.0) | 40 (67.8) |
|
| No | 27 (27.3) | 15 (37.5) | 12 (20.3) |
|
|
Unknown | 8 (8.1) | 1 (2.5) | 7 (11.9) |
|
| Histological type,
n (%) |
|
|
| 0.195 |
| AD | 58 (58.6) | 28 (70.0) | 30 (50.8) |
|
| SC | 36 (36.4) | 10 (25.0) | 26 (44.1) |
|
|
LCC | 1 (1.0) | 0 (0.0) | 1 (1.7) |
|
|
ASC | 4 (4.0) | 2 (5.0) | 2 (3.4) |
|
| Clinical stage, n
(%) |
|
|
| 0.417 |
| I | 1 (1.0) | 0 (0.0) | 1 (1.7) |
|
| II | 3 (3.0) | 2 (5.0) | 1 (1.7) |
|
|
III | 19 (19.2) | 6 (15.0) | 13 (22.0) |
|
| IV | 67 (67.7) | 30 (75.0) | 37 (62.7) |
|
|
Unknown | 9 (9.1) | 2 (5.0) | 7 (11.9) |
|
All 99 samples were profiled by targeted sequencing
with a panel of nine of the most common driver genes [EGFR, ALK,
ROS1, BRAF, MET, RET, ERBB2, KRAS and tumor protein 53 (TP53)] in
NSCLC. A total of 38 out of 40 (95.0%) tissue samples and 38 out of
59 (64.4%) ctDNA samples exhibited at least one genetic alteration.
TP53 was the most commonly mutated gene (56.6%), followed by EGFR
(36.8%). Compared with TCGA data, significantly more somatic
mutations were observed in EGFR and ERBB2 (P<0.001), and
significantly fewer mutations in TP53 (P=0.045) were identified
among the cohort of Chinese patients with NSCLC (Table II). Although the KRAS mutation rate
according to TCGA was higher than that of the cohort in the present
study, no significant difference was observed (P=0.179; Table II). Further analysis demonstrated
that the frequency of TP53 alterations was markedly higher in SC
than in AD (P=0.009; Table III),
and the frequency of EGFR alterations was associated with being
female, AD and non-smokers (P<0.001, P=0.001 and P<0.001,
respectively; Table III). KRAS
alterations were more likely to occur in male patients and smokers,
although no statistically significant differences were observed
(P=0.086 and P=0.105, respectively). ERBB2 alterations were more
frequently observed in non-smokers (P<0.001). Furthermore, in
the present study, no ERBB2 alterations were observed in patients
with localized NSCLC, and statistical analysis revealed that ERBB2
alterations were potentially associated with metastatic stage in
NSCLC (P=0.108; Table III).
 | Table II.Frequency of somatic mutations among
patients with non-small cell lung carcinoma in the present cohort
and TCGA cohort. |
Table II.
Frequency of somatic mutations among
patients with non-small cell lung carcinoma in the present cohort
and TCGA cohort.
|
| Frequency, % |
|
|---|
|
|
|
|
|---|
| Somatic
mutations | TCGA (n=1,144) | The present cohort
(n=76) | P-value |
|---|
| TP53 | 67.7 | 56.6 | 0.045 |
| EGFR | 10.2 | 36.8 | <0.001 |
| KRAS | 19.4 | 13.2 | 0.179 |
| ERBB2 | 2.3 | 9.2 | <0.001 |
| BRAF | 6.1 | 5.3 | 0.762 |
| MET | 3.0 | 5.3 | 0.267 |
 | Table III.Analysis between genetic alterations
and clinicopathological characteristics. |
Table III.
Analysis between genetic alterations
and clinicopathological characteristics.
|
| TP53 | EGFR | KRAS | ERBB2 |
|---|
|
|
|
|
|
|
|---|
| Characteristic | Freq, % | P-value | Freq, % | P-value | Freq, % | P-value | Freq, % | P-value |
|---|
| Sex |
|
|
|
|
|
|
|
|
|
Male | 57.4 | 0.819 | 20.4 | <0.001 | 20.4 | 0.086 | 14.8 | 0.503 |
|
Female | 54.5 |
| 77.3 |
| 4.5 |
| 9.1 |
|
| Age, years |
|
|
|
|
|
|
|
|
|
≤60 | 52.5 | 0.532 | 45.0 | 0.061 | 10.0 | 0.129 | 10.0 | 0.410 |
|
>60 | 60.0 |
| 23.3 |
| 23.3 |
| 16.7 |
|
| Histological
type |
|
|
|
|
|
|
|
|
| AD | 46.8 | 0.009 | 48.9 | 0.001 | 14.9 | 0.684 | 12.8 | 0.804 |
| SC | 77.8 |
| 11.1 |
| 18.5 |
| 14.8 |
|
| Smoking |
|
|
|
|
|
|
|
|
|
Yes | 58.0 | 0.663 | 20.0 | <0.001 | 20.0 | 0.105 | 2.0 | <0.001 |
| No | 52.4 |
| 71.4 |
| 4.8 |
| 38.1 |
|
| Clinical stage |
|
|
|
|
|
|
|
|
|
I+II+III | 57.1 | 1.000 | 35.7 | 1.000 | 21.4 | 0.511 | 0.0 | 0.108 |
| IV | 57.1 |
| 35.7 |
| 14.3 |
| 16.1 |
|
Overall, 50.5% (50/99) of the patients with NSCLC
harbored at least one targetable alteration (data not shown).
Regarding pathological type, the frequency of the targetable
alterations between patients with AD (63.8%) and SC (30.6%) were
significantly different (P=0.002; Table
IV). The landscape of genomic mutations demonstrated that an
EGFR mutation was seen alongside each other gene (MET, ERBB2, KRAS
and ALK) mutation in only one patient each (1%). In addition, the
present study demonstrated that EGFR amplification coexisted with
EGFR mutations, including SNVs and INDELs, but was mutually
exclusive with the EGFR T790M mutation. Furthermore, no patients
with concurrent ERBB2 mutation and amplification were observed. All
four patients that possessed the EGFR T790M mutation also harbored
other EGFR mutations (Fig. 1). To
investigate the patterns of concurrent alterations and mutual
exclusivity, pairwise associations between somatic events were
examined. Mutual exclusivity was observed between alterations in
EGFR and TP53 [odds ratio (OR)=0.324; P=0.020; Table V] and between those in EGFR and KRAS
(OR=0.091; P=0.008) in patients with NSCLC. Furthermore, potential
mutual exclusivity between EGFR and ERBB2 alterations was also
observed, but this was not statistically significant (OR=0.161;
P=0.059; Table V).
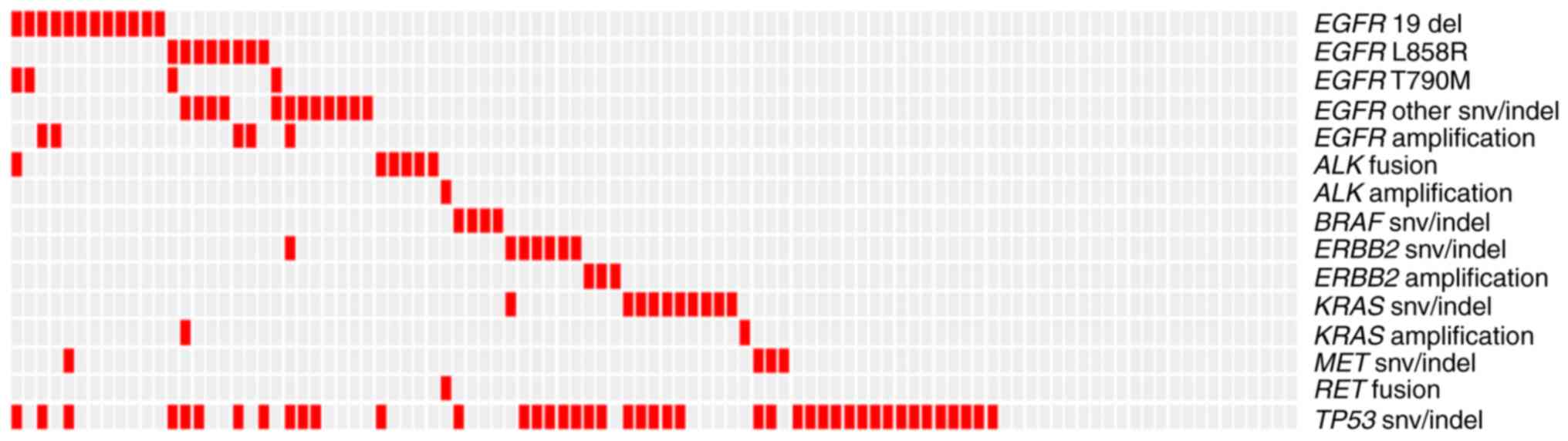 | Figure 1.Landscape of genomic alterations
among 99 patients with non-small cell lung cancer. Red boxes
indicate alterations. EGFR, epidermal growth factor receptor; del,
small deletion; snv, single nucleotide variant; ALK, anaplastic
lymphoma kinase; indel, small insertion or deletion; BRAF, v-raf
murine sarcoma viral oncogene homolog B1; KRAS, Kirsten rat sarcoma
viral oncogene homolog; MET, mesenchymal-epithelial transition
factor; RET, rearranged during transfection; TP53, tumor protein
53. |
 | Table IV.Analysis of frequency of targetable
alterations between AD and SC. |
Table IV.
Analysis of frequency of targetable
alterations between AD and SC.
| Histological
type | Frequency of
targetable alterations, % | P-value |
|---|
| AD | 63.8 | 0.002 |
| SC | 30.6 |
|
 | Table V.Patterns of association between
somatic events in non-small cell lung carcinoma. |
Table V.
Patterns of association between
somatic events in non-small cell lung carcinoma.
|
| TP53 | KRAS | ERBB2 |
|---|
|
|
|
|
|
|---|
| EGFR | OR | P-value | OR | P-value | OR | P-value |
|---|
| Statistical
value | 0.324 | 0.020 | 0.091 | 0.008 | 0.161 | 0.059 |
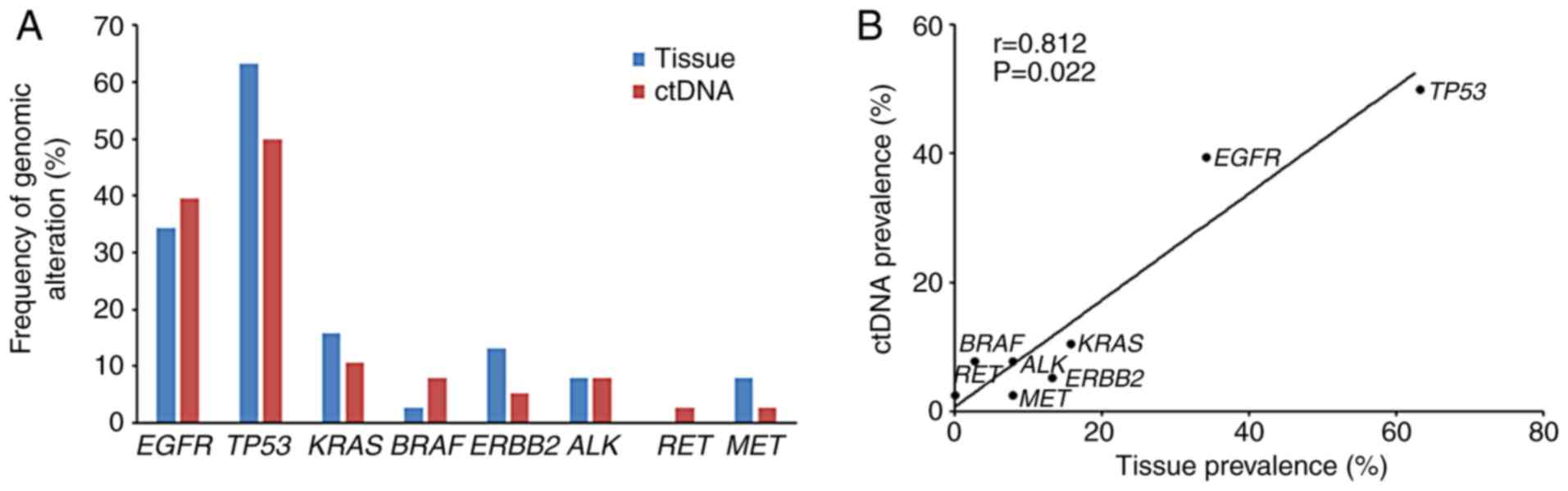
Comparison of the frequency of genomic
variants between tissue and ctDNA samples
In order to investigate the use of ctDNA sequencing,
the present study compared the prevalence of genomic alterations,
including non-synonymous SNVs, CNVs and rearrangements, between
tissue and ctDNA samples. Overall, there were marked similarities
in the frequency of variants when comparing the tissue and ctDNA
samples (Fig. 2A). Further analysis
indicated that the prevalence of alterations among the most common
NSCLC driver genes in ctDNA was positively correlated with that
observed in tumor tissues (r=0.812; P=0.022; Fig. 2B).
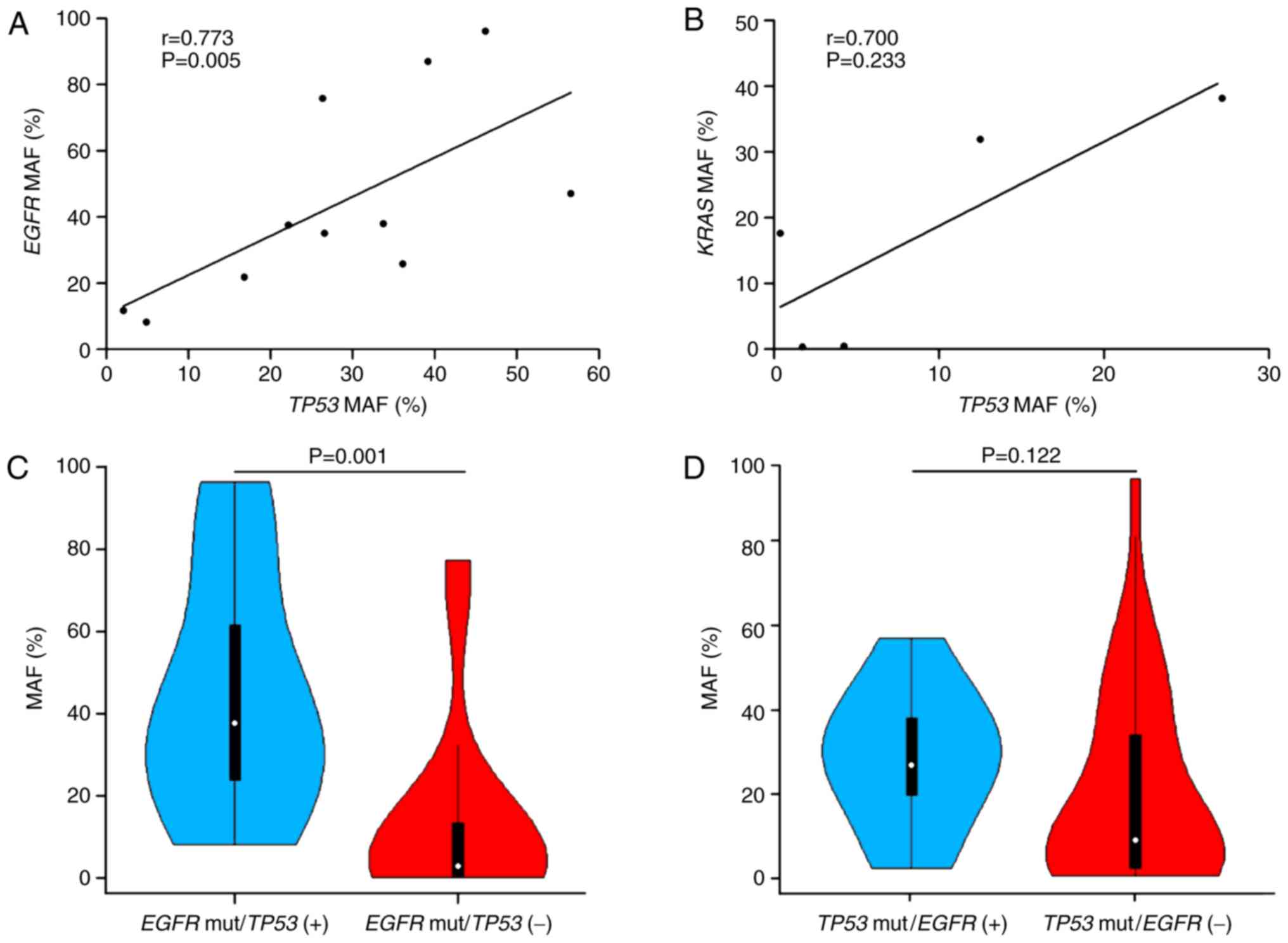
Association between EGFR and TP53
mutant allele frequencies (MAFs)
Evolutionary studies of NSCLC have demonstrated that
TP53 and EGFR mutations are the two most dominant clonal mutations
(26); thus, the present study
hypothesized that there may be a correlation between the MAFs of
TP53 and EGFR in individual patients. As expected, a significantly
linear relationship was observed (r=0.773; P=0.005; Fig. 3A). However, when the MAFs of TP53 and
KRAS were compared, no linear correlation was identified (r=0.700;
P=0.233; Fig. 3B). In addition, the
present study observed significant differences in the MAF of EGFR
between patients with and without TP53 mutations (P=0.001; Fig. 3C). By contrast, no clear difference
in the MAF of TP53 was observed between patients with and without
EGFR mutations (P=0.122; Fig.
3D).
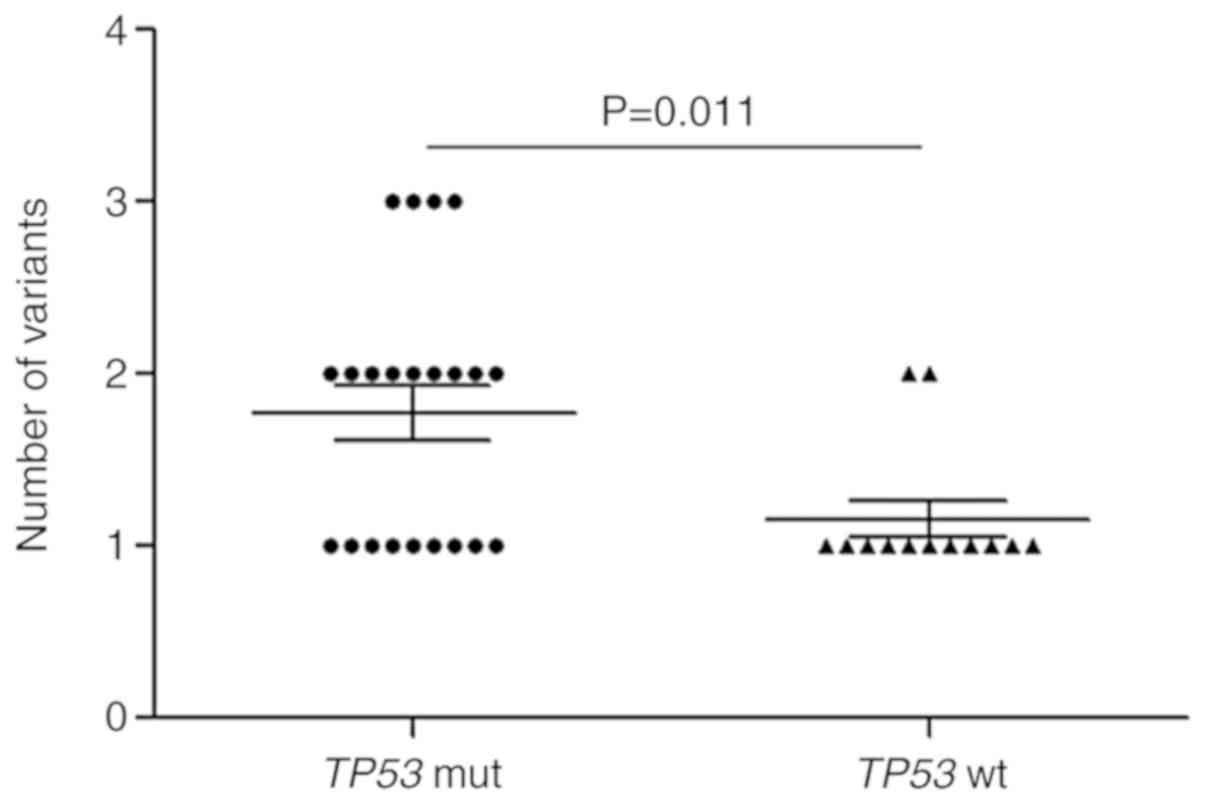
Impact of the TP53 mutation on variant
numbers in patients with NSCLC
An accumulating body of evidence has indicated that
there may be an association between the number of variants (SNVs
and INDELs) in baseline tumors and the clinical prognosis of lung
cancer (27,28). Furthermore, several studies have
demonstrated that certain mutant genes, such as TP53 and KRAS, are
associated with prognosis (29–31).
Based on these discoveries, it was speculated that mutant genes may
be associated with variant numbers in baseline tumors from patients
with NSCLC. Therefore, the number of variants in each patient was
analyzed in the context of various mutant genes. The results
revealed that patients with TP53 mutations harbored more variant
numbers when compared with patients without TP53 mutations in
baseline tumors (P=0.011; Fig. 4).
No other mutant genes were identified to be associated with variant
numbers.
Association between ctDNA-detectable
mutations and clinicopathological characteristics
Among the plasma samples, 21 out of 59 (35.6%)
samples exhibited no genomic alterations (data not shown). Based on
this finding, the present study investigated the association
between the frequency of ctDNA-detectable mutations and
clinicopathological characteristics. The results revealed that the
frequency of ctDNA-detectable mutations was significantly
associated with clinical stage (P=0.014); however, no significant
association was observed between other clinical features (such as
age, sex, histological type and smoking history) and
ctDNA-detectable mutation frequency (Table VI).
 | Table VI.Analysis between frequency of
ctDNA-detectable mutations and clinicopathological
characteristics. |
Table VI.
Analysis between frequency of
ctDNA-detectable mutations and clinicopathological
characteristics.
| Characteristic | Frequency of
cthNA-detectable mutations, % | P-value |
|---|
| Sex |
|
|
|
Male | 57.4 | 0.098 |
|
Female | 83.3 |
|
| Age, years |
|
|
|
≤60 | 58.1 | 0.515 |
|
>60 | 66.7 |
|
| Histological
type |
|
|
| AD | 70.0 | 0.712 |
| SC | 65.4 |
|
| Smoking |
|
|
|
Yes | 65.0 | 0.915 |
| No | 66.7 |
|
| Stage |
|
|
|
I+II+III | 40.0 | 0.014 |
| IV | 75.7 |
|
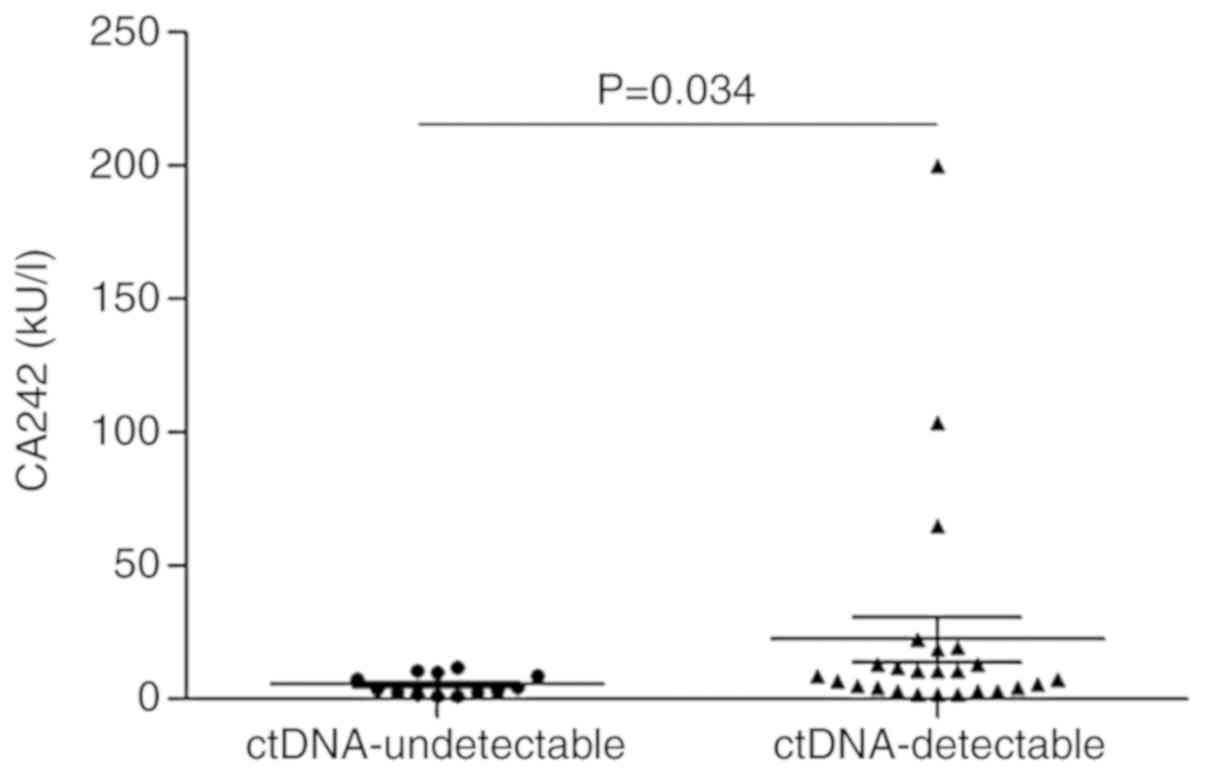
Among the patients with plasma ctDNA samples, the
levels of tumor serum markers (such as CA242, NSE, CEA, CA125 and
CA15-3) were determined in 38 patients. Further analysis revealed
that CA242 levels in patients with ctDNA-detectable mutations were
significantly higher than those observed in patients without
ctDNA-detectable mutations (P=0.034; Fig. 5). There was no significant difference
in the levels of other tumor serum markers between patients with
and without ctDNA-detectable mutations.
Discussion
Through NGS, the present study comprehensively
analyzed genomic alterations in tissue and plasma ctDNA samples
derived from Chinese patients with NSCLC. In the present study, the
associations between the frequencies of several driver gene
alterations and clinical characteristics were observed. Of note,
TP53 alterations were associated with SC, ERBB2 alterations were
associated with smoking, and KRAS alterations seemed to primarily
arise in male or smoking patients with NSCLC. In addition, ERBB2
alterations, including somatic mutations and CNVs, were only
encountered in patients exhibiting metastasis, implying that
aberrations in ERBB2 may occur later during NSCLC progression. The
differences in the prevalence of several driver gene mutations
(such as EGFR, ERBB2 and TP53) between Chinese and TCGA cohorts was
indicative of the specificity of the gene mutational spectrum in
Chinese patients with NSCLC. Furthermore, although the frequency of
targetable alterations in patients with AD was markedly higher than
in patients with SC, the data from the present study demonstrated
that a proportion of patients with SC could potentially benefit
from targeted therapy and that the detection of genomic alterations
in patients with SC is therefore required.
Multiple resistance mechanisms against
first-generation EGFR-TKI treatment, including EGFR amplification,
the EGFR T790M mutation and ERBB2 alterations, have been described
previously (32). Based on the
genomic landscape, the present study demonstrated that EGFR
amplification coexisted with EGFR somatic mutations, but was
mutually exclusive with EGFR T790M mutations; there were no cases
of simultaneous ERBB2 somatic mutations and amplification. This
implies that patients with NSCLC may not exhibit mutations and
amplification of the same genes in resistance to EGFR-TKI. Further
analysis revealed that an EGFR mutation was seen alongside each
other gene (MET, ERBB2, KRAS and ALK) mutation in only one patient
each, demonstrating the importance of multi-gene panel testing in
NSCLC. However, no co-occurrence of EGFR alterations with RET or
BRAF alterations was observed. The statistical analyses revealed
that EGFR alterations were significantly mutually exclusive with
TP53 or KRAS alterations and potentially mutually exclusive with
ERBB2 alterations. These findings are indicative of mutual
exclusivity between EGFR alterations and alterations in other
driver genes in NSCLC.
A previous study reported that EGFR and TP53 are the
two dominant clonal mutant genes in NSCLC (26). The significant linear correlation
between the MAFs of TP53 and EGFR in individual patients further
confirmed that TP53 and EGFR mutations regularly co-occur as clonal
events in the evolution of NSCLC. Notably, marked differences in
the EGFR MAFs of patients with and without TP53 mutations were
observed. A recent study suggested that the abundance of
EGFR-activating mutations was correlated with the efficacy of
EGFR-TKIs in advanced NSCLC (33).
According to the results of the present study, there may be
differences in the therapeutic effect of EGFR-TKIs in patients with
EGFR-activating mutations with and without TP53 mutations. Another
finding was the difference in the number of mutations among
baseline tumors with and without TP53 mutations. Several studies
have reported an association between the number of mutations and
prognosis, or between mutations in specific genes and prognosis
(27–31). As the present study did not obtain
complete information on the therapeutic effect and prognosis in a
large proportion of patients, determination of the efficacy of
EGFR-TKIs and prognosis in tumors with and without TP53 mutations
could not be assessed.
At present, the detection of genomic alterations
primarily relies on the analysis of cancer tissues obtained via
surgical excision or biopsy. However, tumor tissues are not always
available for all patients, and tissue biopsy of a single site may
not fully reflect the tumor genomic landscape due to tumor genetic
heterogeneity (34). Therefore, in
the present study, the use of plasma ctDNA was assessed for
patients without sufficient tissue samples to screen for genomic
alterations to guide personalized therapy. In the analysis of the
ctDNA and tissue profiles, the landscape and frequencies of genomic
alterations in ctDNA samples demonstrated a strong similarity with
those observed in tissue samples, supporting the feasibility and
use of plasma ctDNA testing among patients with NSCLC. The
association between the frequency of plasma ctDNA-detectable
mutations and clinical characteristics was also assessed in the
present study, which revealed that the frequency of
ctDNA-detectable mutations was significantly associated with
clinical stage, consistent with previous results (15). In addition, it was observed that
serum CA242 levels were significantly higher in patients with
ctDNA-detectable mutations when compared with patients without
ctDNA-detectable mutations. A prior study reported that serum CA242
levels were associated with clinical stage in NSCLC (35). Therefore, the present study provides
a novel line of evidence confirming the prior result that
ctDNA-detectable mutations are associated with clinical stage in
NSCLC.
In conclusion, the present study identified
targetable genetic alterations in 50.5% of patients with NSCLC
using targeted NGS and discovered significant correlations between
the frequency of genomic variants in tissue and plasma ctDNA
samples. In addition, the results of the present study demonstrated
that the MAF of EGFR was associated with that of TP53 in individual
tumors and that the EGFR MAF in patients with the TP53 mutation was
different to that observed in patients without the TP53 mutation.
Although further studies are required to confirm these findings,
these findings provide an improved understanding of the spectra of
genomic alterations detected by tissue and plasma ctDNA assays in
Chinese patients with NSCLC.
Acknowledgements
The abstract was presented at the 55th Meeting of
ASCO, 31 May to 4 June, 2019 in Chicago, USA and published as
abstract no. e20534 in Journal of Clinical Oncology, Vol. 37:
15_suppl.
Funding
The present study was funded by the Scientific
Research Fund Project in the Department of Science and Technology
of Hunan Province, China (grant. no. 2014SK3006).
Availability of data and materials
All data analyzed during the present study are
included in this published article.
Authors' contributions
HY, HC, HW, SC and JC conceived and designed the
project. HY, JZ, LZ, XW, YL, DY and TC collected samples and
performed the experiments; HC, HW, FL, XL and JG analyzed the data.
HY, HC, HW, SC and JC wrote the paper. All authors reviewed and
edited the manuscript, and approved the final version for
publication.
Ethics approval and consent to
participate
The present study was approved by the Committee of
Medical Ethics of Hunan Cancer Hospital (Changsha, China). All
patients provided written informed consent prior to the study
start.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Liu K, Chen HL, Wang S, Gu MM, Chen XM,
Zhang SL, Yu KJ and You QS: High expression of RIOK2 and NOB1
predict human non-small cell lung cancer outcomes. Sci Rep.
6:286662016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Yang J, Lin J, Liu T, Chen T, Pan S, Huang
W and Li S: Analysis of lncRNA expression profiles in non-small
cell lung cancers (NSCLC) and their clinical subtypes. Lung Cancer.
85:110–115. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Torr LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Tursz T, Andre F, Lazar V, Lacroix L and
Soria JC: Implications of personalized medicine-perspective from a
cancer center. Nat Rev Clin Oncol. 8:177–183. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Swanton C and Govindan R: Clinical
implications of genomic discoveries in lung cancer. N Engl J Med.
374:1864–1873. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Berge EM and Doebele RC: Targeted
therapies in non-small cell lung cancer: Emerging oncogene targets
following the success of epidermal growth factor receptor. Semin
Oncol. 41:110–125. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Travis WD, Brambilla E, Nicholson AG,
Yatabe Y, Austin JHM, Beasley MB, Chirieac LR, Dacic S, Duhig E,
Flieder DB, et al: The 2015 world health organization
classification of lung tumors: Impact of genetic, clinical and
radiologic advances since the 2004 classification. J Thorac Oncol.
10:1243–1260. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Vargas AJ and Harris CC: Biomarker
development in the precision medicine era: Lung cancer as a case
study. Nat Rev Cancer. 16:525–537. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tsao AS, Scagliotti GV, Bunn PA Jr,
Carbone DP, Warren GW, Bai C, de Koning HJ, Yousaf-Khan AU,
McWilliams A, Tsao MS, et al: Scientific advances in lung cancer
2015. J Thorac Oncol. 11:613–638. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Shyr D and Liu Q: Next generation
sequencing in cancer research and clinical application. Biol Proced
Online. 15:42013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Gagan J and Van Allen EM: Next-generation
sequencing to guide cancer therapy. Genome Med. 7:802015.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gerlinger M, Rowan AJ, Horswell S, Math M,
Larkin J, Endesfelder D, Gronroos E, Martinez P, Matthews N,
Stewart A, et al: Intratumor heterogeneity and branched evolution
revealed by multiregion sequencing. N Engl J Med. 366:883–892.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kris MG, Johnson BE, Berry LD, Kwiatkowski
DJ, Iafrate AJ, Wistuba II, Varella-Garcia M, Franklin WA, Aronson
SL, Su PF, et al: Using multiplexed assays of oncogenic drivers in
lung cancers to select targeted drugs. JAMA. 311:1998–2006. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Crowley E, Di Nicolantonio F, Loupakis F
and Bardelli A: Liquid biopsy: Monitoring cancer-genetics in the
blood. Nat Rev Clin Oncol. 10:472–484. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bettegowda C, Sausen M, Leary RJ, Kinde I,
Wang Y, Agrawal N, Bartlett BR, Wang H, Luber B, Alani RM, et al:
Detection of circulating tumor DNA in early- and late-stage human
malignancies. Sci Transl Med. 6:224ra242014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hicks JK, Saller J, Wang E, Boyle T and
Gray JE: Cell-free circulating tumor DNA supplementing tissue
biopsies for identification of targetable mutations: Implications
for precision medicine and considerations for reconciling results.
Lung Cancer. 111:135–138. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Xu S, Lou F, Wu Y, Sun DQ, Zhang JB, Chen
W, Ye H, Liu JH, Wei S, Zhao MY, et al: Circulating tumor DNA
identified by targeted sequencing in advanced-stage non-small cell
lung cancer patients. Cancer Lett. 370:324–331. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Pisapia P, Pepe F, Smeraglio R, Russo M,
Rocco D, Sgariglia R, Nacchio M, De Luca C, Vigliar E, Bellevicine
C, et al: Cell free DNA analysis by SiRe® next
generation sequencing panel in non small cell lung cancer patients:
Focus on basal setting. J Thorac Dis. 9 (Suppl 13):S1383–S1390.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Mayo-de-Las-Casas C, Jordana-Ariza N,
Garzón-Ibañez M, Balada-Bel A, Bertrán-Alamillo J, Viteri-Ramírez
S, Reguart N, Muñoz-Quintana MA, Lianes-Barragan P, Camps C, et al:
Large scale, prospective screening of EGFR mutations in the blood
of advanced NSCLC patients to guide treatment decisions. Ann Oncol.
28:2248–2255. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chen K, Zhang J, Guan T, Yang F, Lou F,
Chen W, Zhao M, Zhang J, Chen S and Wang J: Comparison of plasma to
tissue DNA mutations in surgical patients with non-small cell lung
cancer. J Thorac Cardiovasc Surg. 154:1123–1131.e2. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kimura K and Koike A: Ultrafast SNP
analysis using the Burrows-Wheeler transform of short-read data.
Bioinformatics. 31:1577–1583. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
McKenna A, Hanna M, Banks E, Sivachenko A,
Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly
M and DePristo MA: The genome analysis toolkit: A MapReduce
framework for analyzing next-generation DNA sequencing data. Genome
Res. 20:1297–1303. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Cibulskis K, Lawrence MS, Carter SL,
Sivachenko A, Jaffe D, Sougnez C, Gabriel S, Meyerson M, Lander ES
and Getz G: Sensitive detection of somatic point mutations in
impure and heterogeneous cancer samples. Nat Biotechnol.
31:213–219. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
24
|
Li J, Lupat R, Amarasinghe KC, Thompson
ER, Doyle MA, Ryland GL, Tothill RW, Halgamuge SK, Campbell IG and
Gorringe KL: CONTRA: Copy number analysis for targeted
resequencing. Bioinformatics. 28:1307–1313. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Campbell JD, Alexandrov A, Kim J, Wala J,
Berger AH, Pedamallu CS, Shukla SA, Guo G, Brooks AN, Murray BA, et
al: Distinct patterns of somatic genome alterations in lung
adenocarcinomas and squamous cell carcinomas. Nat Genet.
48:607–616. 2016. View
Article : Google Scholar : PubMed/NCBI
|
|
26
|
Jamal-Hanjani M, Wilson GA, McGranahan N,
Birkbak NJ, Watkins TBK, Veeriah S, Shafi S, Johnson DH, Mitter R,
Rosenthal R, et al: Tracking the evolution of non-small cell lung
cancer. N Engl J Med. 376:2109–2121. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Thompson JC, Yee SS, Troxel AB, Savitch
SL, Fan R, Balli D, Lieberman DB, Morrissette JD, Evans TL, Bauml
J, et al: Detection of therapeutically targetable driver and
resistance mutations in lung cancer patients by next-generation
sequencing of cell-free circulating tumor DNA. Clin Cancer Res.
22:5772–5782. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Nahar R, Zhai W, Zhang T, Takano A, Khng
AJ, Lee YY, Liu X, Lim CH, Koh TPT, Aung ZW, et al: Elucidating the
genomic architecture of Asian EGFR-mutant lung adenocarcinoma
through multi-region exome sequencing. Nat Commun. 9:2162018.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
VanderLaan PA, Rangachari D, Mockus SM,
Spotlow V, Reddi HV, Malcolm J, Huberman MS, Joseph LJ, Kobayashi
SS and Costa DB: Mutations in TP53, PIK3CA, PTEN and other genes in
EGFR mutated lung cancers: Correlation with clinical outcomes. Lung
Cancer. 106:17–21. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Molina-Vila MA, Bertran-Alamillo J, Gascó
A, Mayo-de-las-Casas C, Sánchez-Ronco M, Pujantell-Pastor L,
Bonanno L, Favaretto AG, Cardona AF, Vergnenègre A, et al:
Nondisruptive p53 mutations are associated with shorter survival in
patients with advanced non-small cell lung cancer. Clin Cancer Res.
20:4647–4659. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Mascaux C, Iannino N, Martin B, Paesmans
M, Berghmans T, Dusart M, Haller A, Lothaire P, Meert AP, Noel S,
et al: The role of RAS oncogene in survival of patients with lung
cancer: A systematic review of the literature with meta-analysis.
Br J Cancer. 92:131–139. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Rotow J and Bivona TG: Understanding and
targeting resistance mechanisms in NSCLC. Nat Rev Cancer.
17:637–658. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Li X, Cai W, Yang G, Su C, Ren S, Zhao C,
Hu R, Chen X, Gao G, Guo Z, et al: Comprehensive analysis of
EGFR-mutant abundance and its effect on efficacy of EGFR TKIs in
advanced NSCLC with EGFR mutations. J Thorac Oncol. 12:1388–1397.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Sholl LM, Aisner DL, Varella-Garcia M,
Berry LD, Dias-Santagata D, Wistuba II, Chen H, Fujimoto J, Kugler
K, Franklin WA, et al: Multi-institutional oncogenic driver
mutation analysis in lung adenocarcinoma: The lung cancer mutation
consortium experience. J Thorac Oncol. 10:768–777. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Pujol JL, Cooper EH, Lehmann M, Purves DA,
Dan-Aouta M, Midander J, Godard P and Michel FB: Clinical
evaluation of serum tumor marker CA 242 in non-small cell lung
cancer. Br J Cancer. 67:1423–1429. 1993. View Article : Google Scholar : PubMed/NCBI
|