Introduction
Lung cancer remains the leading cause of
cancer-associated mortalities worldwide. Small cell lung cancer and
non-small cell lung cancer (NSCLC) are the two main types of lung
cancer. NSCLC includes adenocarcinoma, squamous cell cancer and
large cell lung cancer (1).
Chemotherapy, radiotherapy, targeted therapy and immunotherapy are
the main treatment strategies for NSCLC. However, NSCLC is often
diagnosed at a late stage; consequently, the five-year survival
rate is low (2). Therefore,
investigating the etiology and prognostic factors of NSCLC is
important. Squamous cell lung cancer is strongly associated with
smoking. While lung adenocarcinoma is associated with smoking, this
type of cancer also occurs in non-smokers (3). Non-smoking lung adenocarcinoma is
strongly associated with the female sex (4). Specific molecules (CXCR2 and
PPBP) and pathways (cell adhesion molecules and CAMs)
play important roles in the pathogenesis of lung adenocarcinoma in
non-smoking female patients (5).
The prevalence of lung adenocarcinomas in
non-smoking females is higher than that in non-smoking males,
suggesting the sex hormones may be involved in tumorigenesis
(3). In vitro studies have
revealed that estrogen promotes the proliferation of NSCLC cells
through estrogen receptor-mediated signaling pathways, whereas,
anti-estrogens exhibit the opposite effect (6,7).
Downregulation of estrogen receptor β (ERβ) inhibits cell growth in
lung adenocarcinoma (8).
17β-estradiol upregulates the expression of interleukin-16 through
the ERβ signaling pathway and promotes the progression of lung
adenocarcinoma (9). Previous studies
have demonstrated that EGFR (epidermal growth factor
receptor) and HER2 (human epidermal growth factor receptor
2) mutations, and anaplastic lymphoma kinase rearrangements are
more commonly observed in lung cancer in non-smokers compared with
that in smokers (10,11). Tumor protein P53 and breast cancer
types 1 and 2 susceptibility protein variants are likely to
contribute to the development of lung adenocarcinoma in non-smoking
females (12). Osteopontin (OPN),
hypoxia inducible factor-1 and several energy metabolism-associated
proteins have been associated with estrogen receptor function
(13). However, the pathogenesis and
prognostic factors of non-smoking female patients with lung
adenocarcinoma remain unclear.
In the present study, bioinformatics analysis was
used to explore estrogen receptor-associated genes that are related
to prognosis in non-smoking female patients with lung
adenocarcinoma. The results may improve the understanding of the
pathogenic and prognostic factors associated with lung
adenocarcinoma in non-smoking females.
Materials and methods
Analysis of microarray data and
RNA-sequencing data
Microarray data and the corresponding clinical data
for non-smoking female patients with lung adenocarcinoma from the
GSE32863 (14) and GSE75037
(15) datasets, both datasets of 24
non-smoking female patients, were downloaded from the Gene
Expression Omnibus (GEO) (ncbi.nlm.nih.gov/geo/) based on the platform of
GPL6884 Illumina Human WG-6 v3.0 expression beadchip (Illumina,
Inc.). Data for 48 non-smoking female patients with lung
adenocarcinoma detected using a microarray chip in the GEO database
(GSE32863 and GSE75037) and from 160 non-smoking female patients
with lung adenocarcinoma detected using RNA-sequencing in the The
Cancer Genome Atlas (TCGA) database (portal.gdc.cancer.gov; last updated on July 2017) were
also downloaded. The SVA package (version 3.32.1; www.bioconductor.org/help/search/index.html?q=sva/) in
Bioconductor (version 3.9; www.bioconductor) was used to normalize the gene
expression profile data.
Identification of differentially
expressed genes (DEGs)
The Linear Models for Microarray Data (LIMMA)
package (version 3.1; www.bioconductor.org/help/search/index.html?q=limma)
in Bioconductor was used to identify DEGs between samples from
non-smoking female patients with lung adenocarcinoma and samples of
adjacent non-cancerous lung tissue. Adjusted P-values and
fold-change (FC) values were calculated. The DEGs screening
criteria were an adjusted P<0.05 and absolute value of
Log2FC >2. The DEGs at the intersection of the
datasets (DEGs in GEO and TCGA) were selected for subsequent
investigation. The pheatmap package (version 1.0.12; http://cran.r-project.org/web/packages/pheatmap/index.html)
was used to draw the heat map.
Enrichment analyses of DEGs
The Database for Annotation, Visualization and
Integrated Discovery (version 6.8; david.ncifcrf.gov) was used to perform functional
enrichment analysis of the DEGs in non-smoking female patients with
lung adenocarcinoma. Gene Ontology (GO; version 6.8; www.geneontology.org) and Kyoto Encyclopedia of Genes
and Genomes (KEGG; version 6.8; www.genome.ad.jp/kegg/) pathway analyses were
conducted using the WEB-based GEne SeT AnaLysis Toolkit (www.webgestalt.org). P<0.05 was considered to
indicate strong enrichment in the annotation categories.
Analysis of protein-protein
interaction (PPI) networks
The Search Tool for the Retrieval of Interacting
Genes/Proteins (STRING) (version 3.6.0; string-db.org/cgi/input.pl) provides experimental and
predicted interactions among proteins. STRING analyses were
performed to form a PPI network, with the criterion of a combined
score of >0.4.
The DEGs, ESR1 (estrogen receptor 1),
ESR2 (estrogen receptor 2) and GPER (G
protein-coupled estrogen receptor) were used as queries in the
STRING database and the resultant PPI network was subsequently
visualized using Cytoscape software (version 3.6.0; cytoscape.org). CytoHubba (a plug-in in Cytoscape;
version 1.6; http://apps.cytoscape.org/apps/cytohubba) was used to
identify the estrogen receptor-related hub genes (the top 20 genes
with the most connections in the PPI network).
DEGs prognosis analysis
The Kaplan-Meier plotter (kmplot.com)
(16) is an online database
including clinical and expression data. The median expression level
of each gene was used to divide patients into high and low groups.
The Kaplan-Meier plotter was used to identify the hub genes with
significant effects on prognosis. P<0.05 was considered to
indicate a statistically significant difference.
Statistical analysis
Statistical analysis was performed using R (version
3.4.3; www.r-project.org). For the microarray
and RNA-sequencing data analysis, the LIMMA package in Bioconductor
was used to identify the DEGs. The thresholds for identifying DEGs
were P<0.05 and a false discovery rate <2. The SVA package
was used for batch normalization of GSE32863 and GSE75037. The log
rank test was used to compare the survival trend. P<0.05 was
considered to indicate a statistically significant difference.
Results
Identification of DEGs
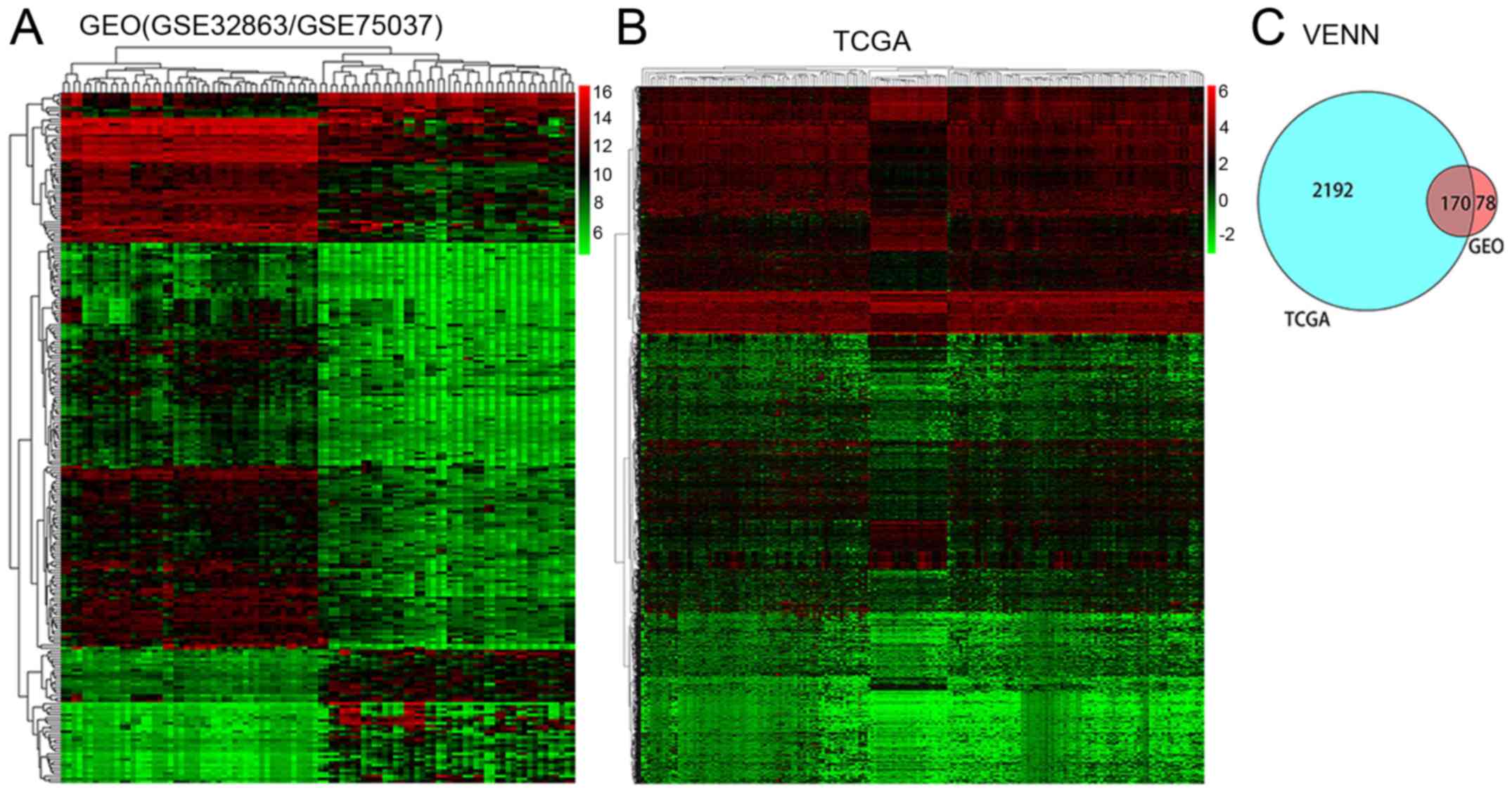
According to the screening criteria, 248 DEGs (57
upregulated and 191 downregulated) were identified in the GEO data,
and 2,362 DEGs (1,773 upregulated and 589 downregulated) were
identified in the TCGA data. The DEGs at the intersection of the
two databases were selected for further investigation, revealing
170 DEGs between lung adenocarcinoma and normal lung tissues from
non-smoking female patients. All 248 DEGs in the GEO database and
the 2,362 DEGs in the TCGA database were visualized using a heat
map, and a Venn diagram was used to present the DEGs at the
intersection of the two databases (Fig.
1). The top 10 (by fold change) upregulated and downregulated
DEGs in the GEO and TCGA databases are presented in Table I.
 | Table I.The top 10 (by fold change)
upregulated and downregulated DEGs in the GEO and the TCGA
databases. |
Table I.
The top 10 (by fold change)
upregulated and downregulated DEGs in the GEO and the TCGA
databases.
| A, Top 10
upregulated DEGs |
|---|
|
|---|
| GEO | TCGA |
|---|
| GCNT3, MMP11, CST1,
CEACAM5, SPINK1, SPP1, CRABP2, COMP, EEF1A2, ATP10B | MAGEA4, PSG3, PSG1,
HOXC12, REG4, PSG5, TFF2, FGB, CGB5, PSG4 |
|
| B, Top 10
downregulated DEGs |
|
| GEO | TCGA |
|
| LOC401286, ITLN2,
MCEMP1, FABP4, CA4, FAM107A, GKN2, HBB, CLEC3B, SCGB1A1 | SLC6A4, SLC6A5,
FABP4, AGER, UMOD, ITLN2, CYP1A2, FREM3, OR6K3, CHRM1 |
DEGs enrichment analysis
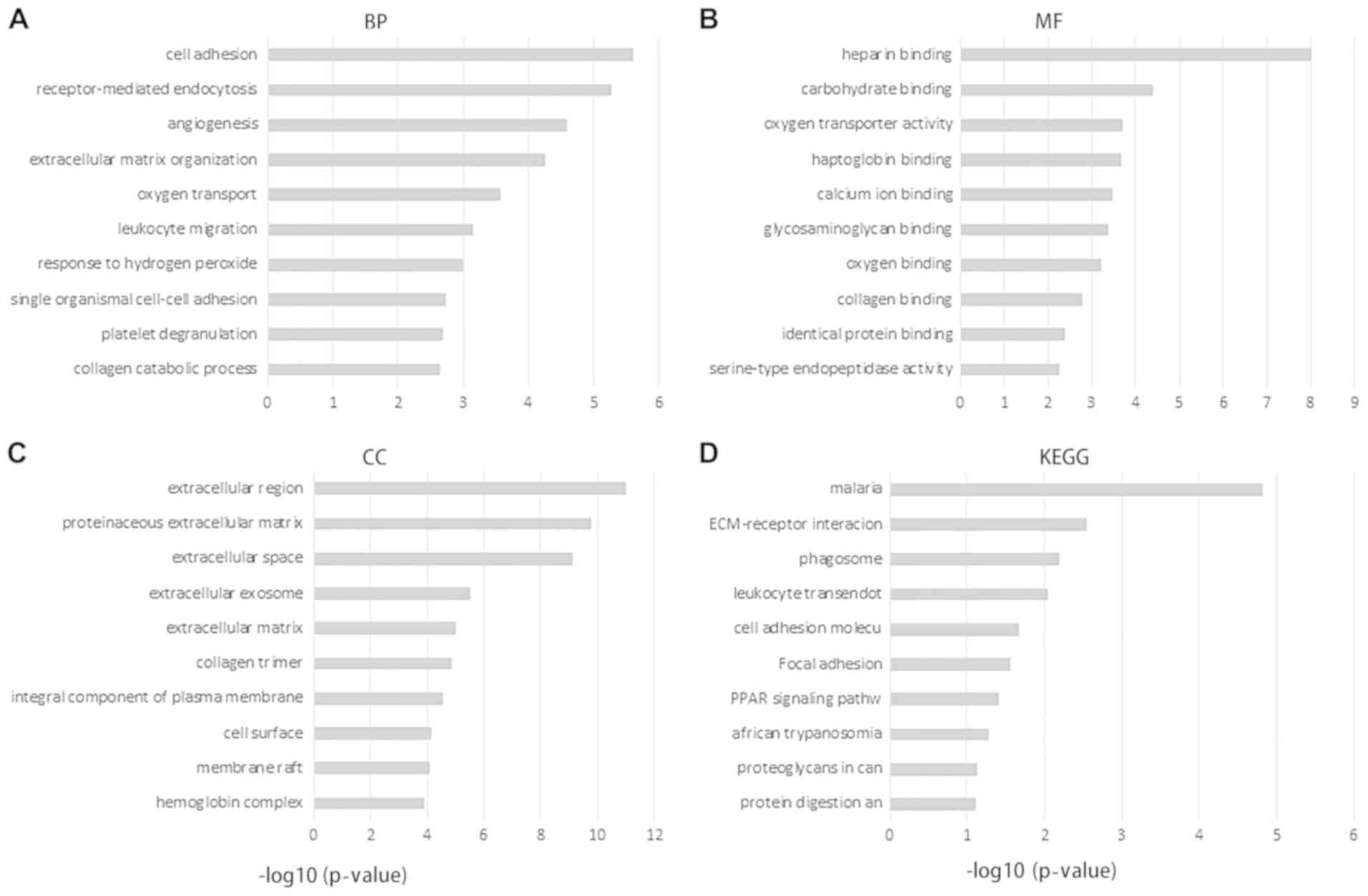
The 170 DEGs were grouped into BP (biological
process; 56 BP terms were significantly enriched), MF (molecular
function; 17 MF terms were significantly enriched) and CC (cellular
component; 23 CC terms were significantly enriched) categories. The
most enriched GO terms in the BP category were ‘cell adhesion’,
‘receptor-mediated endocytosis’ and ‘angiogenesis’. The most
enriched GO terms in the MF category were ‘heparin binding’ and
‘carbohydrate binding’. The most enriched GO terms in the CC
category were ‘extracellular region’, ‘proteinaceous extracellular
matrix’ and ‘extracellular space’ (Fig.
2).
KEGG pathway enrichment analysis (8 KEGG terms were
significantly enriched) revealed that the majority of the DEGs were
enriched in pathways including ‘malaria’ and ‘ECM-receptor
interaction’ (Fig. 2).
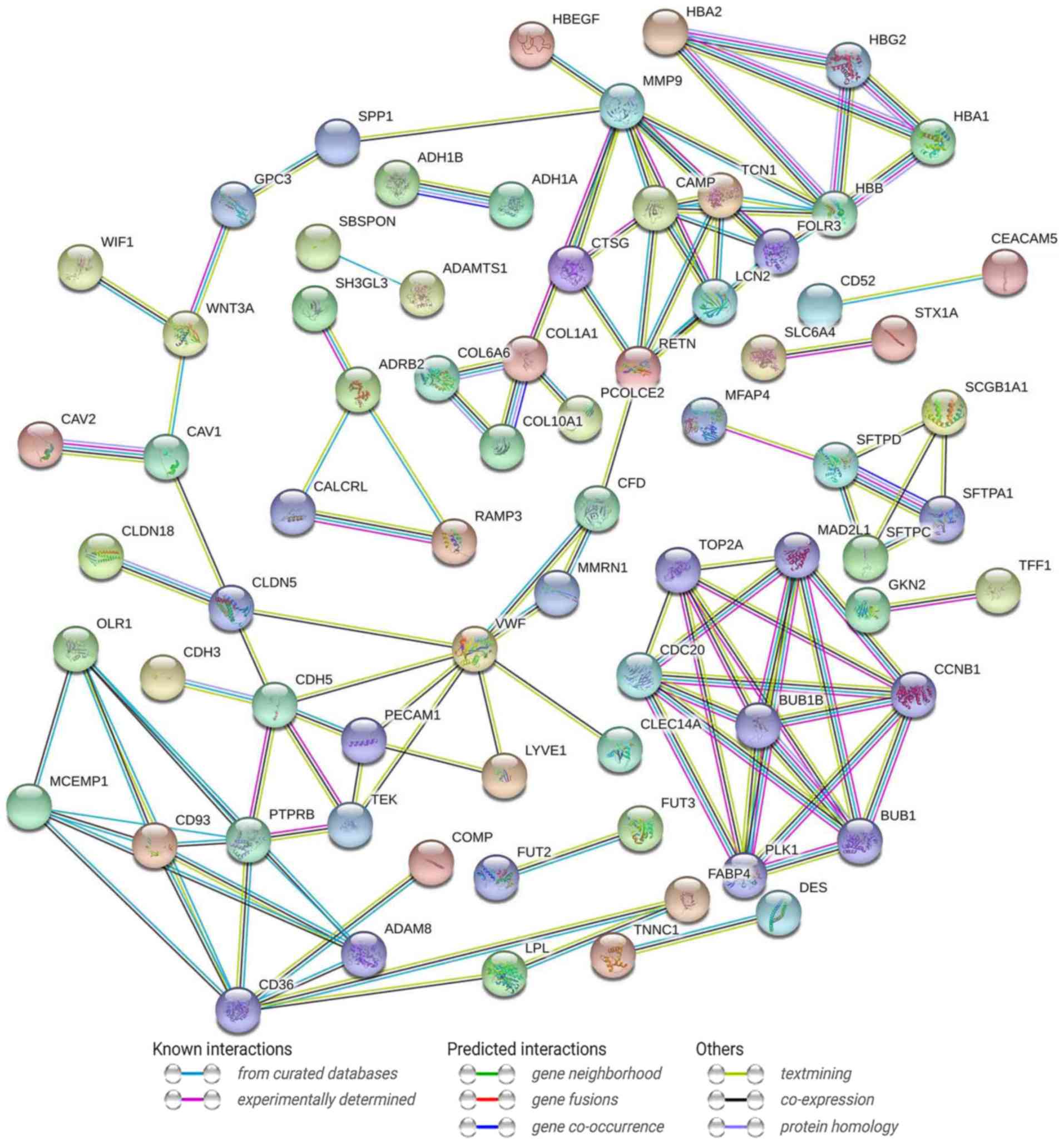
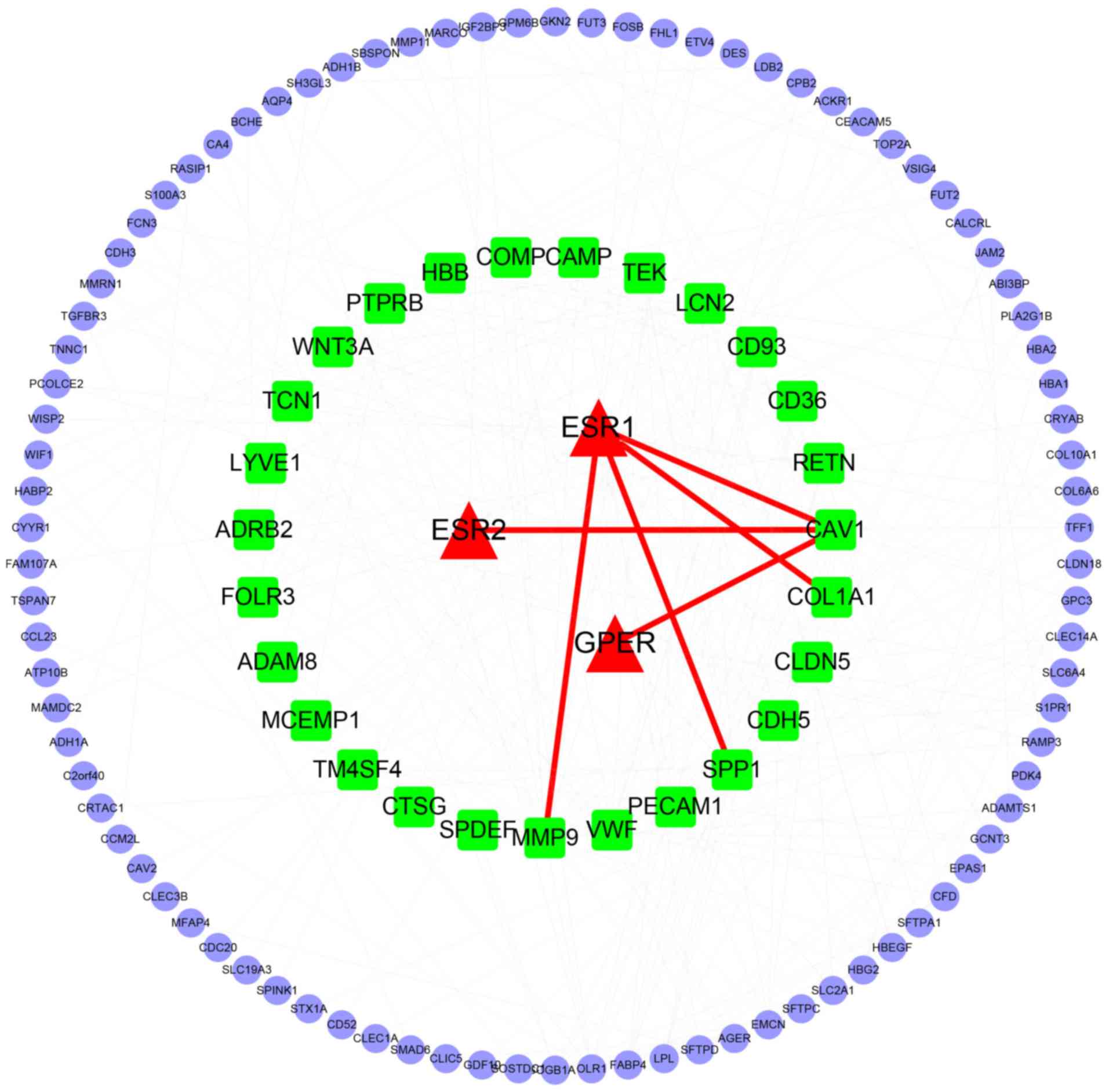
PPI network analysis
A PPI network was constructed to understand the
biological significance of the DEGs. The PPI network consisted of
124 nodes and 266 interactions (Fig.
3). The network between the estrogen receptor (ESR1, ESR2 and
GPER) and DEGs was also constructed. In addition, a network of hub
genes associated with estrogen receptor was constructed. The PPI
network analysis identified 27 DEGs that were considered as hub
genes in the network. There were four hub genes that were closely
associated with the estrogen receptor, including caveolin 1
(CAV1), matrix metalloproteinase 9 (MMP9),
SUMO-1-specific protease 1 (SPP1) and collagen type I α 1
chain (COL1A1), as presented in Fig. 4.
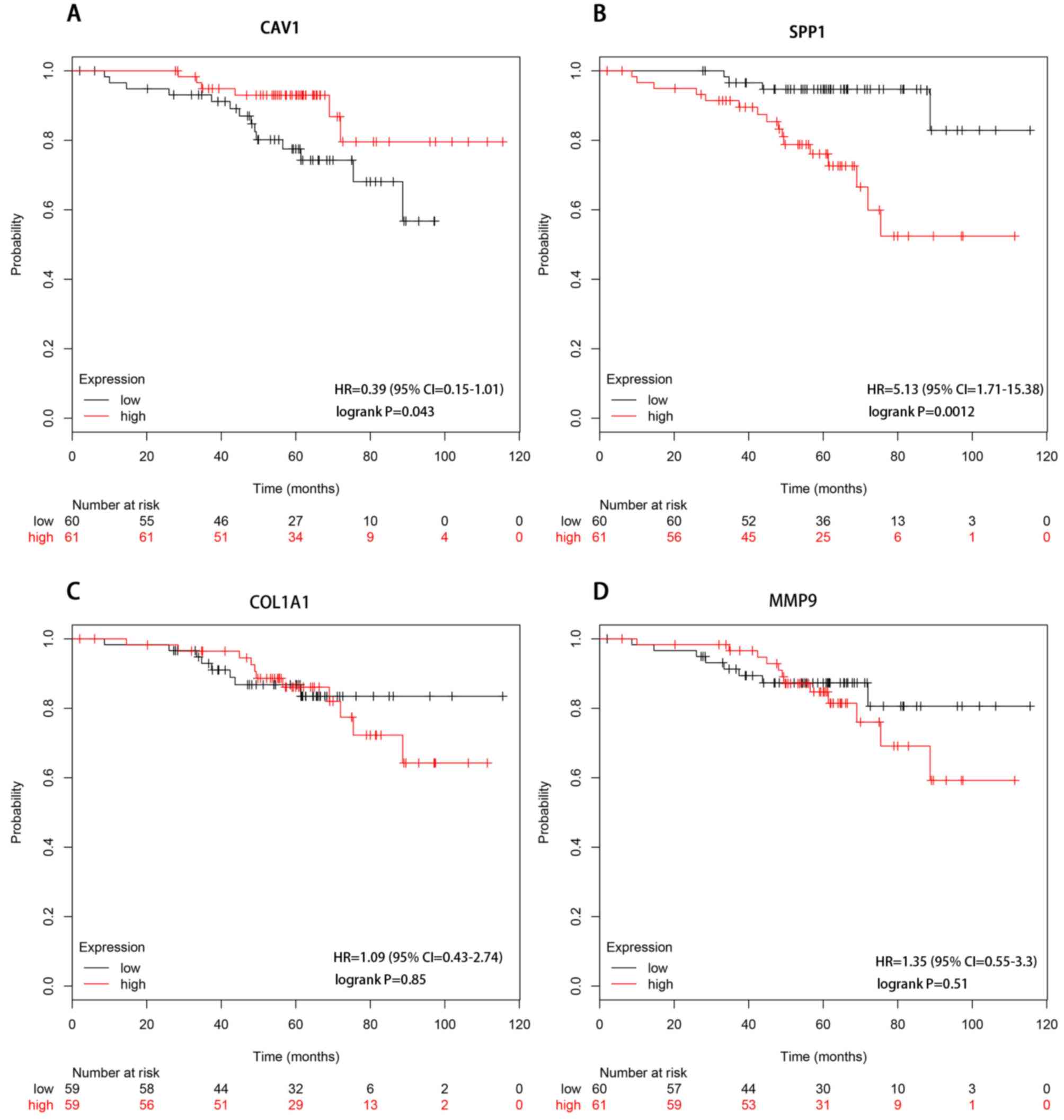
Kaplan-Meier survival analysis
Kaplan-Meier curves were used to assess the effect
of estrogen receptor-associated hub genes on the overall survival
(OS) of 121 non-smoking females with lung adenocarcinoma. Low
expression of SPP1 and high expression of CAV1 were
associated with improved OS. However, there was no significant
association between MMP9 or COL1A1 expression status
and OS (Fig. 5).
Discussion
The pathogenesis and prognostic factors for lung
adenocarcinoma in non-smoking females remain controversial
(3). Previous studies have suggested
that estrogen and its receptor (ER) may serve important roles.
In vitro studies have revealed that the ER promotes NSCLC
vasculogenic mimicry and cell invasion (17). The ER is also activated in
EGFR-tyrosine kinase inhibitor mediated secondary resistance
(18). In addition, a high
expression level of ER is a significant prognostic factor for
survival in advanced NSCLC (19).
There are three types of estrogen receptors: ERα, ERβ and GPER. ERα
and ERβ are important nuclear transcription factors located in the
cell nucleus. GPER is a G-protein coupled receptor containing seven
transmembrane domains located in the cell membrane (19). Few studies have investigated the
association between ERs and lung adenocarcinoma in non-smoking
female patients (20,21).
In the present study, gene expression profiles from
the GSE32863 and GSE75037 datasets and data from the TCGA database
were analyzed using bioinformatic methods. A total of 170 DEGs
between lung adenocarcinoma and normal lung tissue samples from
non-smoking women were common to both databases. Additionally, the
GO terms and KEGG pathways associated with these DEGs, which might
significantly affect lung adenocarcinoma in non-smoking females,
were identified.
The PPI network analysis identified that 27 DEGs
were considered as hub genes in the network. A network consisting
of the ERs, DEGs and hub genes was constructed and it was revealed
that the hub genes CAV1, SPP1, MMP9 and COL1A1 are
significantly associated with ER function.
CAV1 is the main structural component of the
caveolae, which form flask-shaped invaginations that are involved
in cell signaling and transport (22). Low expression levels of CAV1
induce a hyper-proliferative state, promoting cell proliferation,
angiogenesis and tumor progression in certain tumors, suggesting
that loss of CAV1 regulation is an important step in the
acquisition of a transformed phenotype (23). Ramírez et al (24) demonstrated that ERα is present in
caveolae and is stabilized by CAV1. Interactions between ERα
with CAVI were demonstrated using epitope proximity ligation
assays (25). In vitro, the
association between ERα and caveolin-1 increased in tumors that
regressed in response to estradiol (26). In addition, CAV1 is associated
with prognosis in cancer, such as in breast, colon and ovarian
carcinoma (22). In the present
study, non-smoking female patients with lung adenocarcinoma with
high CAV1 expression had an improved prognosis compared with
patients with a low CAV1 expression. The interaction between
CAV1 and ERα may therefore serve an important role in the
pathogenesis and prognosis of lung adenocarcinoma in non-smoking
female patients.
SPP1 encodes secreted phosphoprotein 1, also
known as OPN. OPN is a highly phosphorylated glycophosphoprotein
rich in aspartic acid, which facilitates cell-matrix interactions
(27). Previous studies
investigating lung cancer have reported that tumor development,
progression and metastasis are promoted by increasing the release
of OPN (28–30). In addition, OPN expression levels
were significantly associated with lung cancer differentiation and
the efficacy of platinum-based treatment (31). OPN levels may have a significant
predictive potential in estimating survival of NSCLC and high OPN
expression levels were significantly associated with poor prognosis
in NSCLC compared with low expression (32). The survival analysis of non-smoking
female patients with lung adenocarcinoma in the current study
supported the aforementioned study in NSCLC, as a low expression
level of OPN was associated with improved prognosis compared with
high expression. SPP1 may be a candidate molecular marker
associated with the pathogenesis and prognosis of lung
adenocarcinoma in non-smoking female patients.
MMP9 regulates various cellular behaviors
associated with cancer cell differentiation, migration, invasion
and immune system surveillance (33). Suppression of ESR2 in breast
cancer cells may affect the expression of MMP9 though
microRNA-145 (34). In lung
adenocarcinoma, downregulation of ESR2 inhibits cell growth
though decreased expression of MMP9 (8). MMP9 may therefore be implicated
in lung adenocarcinoma in non-smoking women.
COL1A1 is dysregulated in a variety of
tumors, including breast and gastric cancer (35). COL1A1 gene expression is
inhibited by halofuginone, resulting in inhibition of the
proliferation of bladder carcinoma cells (36). Thus, COL1A1 may serve as a
potential therapeutic target for lung adenocarcinoma in non-smoking
female patients.
The present study had a number of limitations. The
expression levels of the DEGs, their functions, the hub genes and
the association between the ERs and DEGs require experimental
validation. The lack of tissues collected from newly diagnosed
patients with adenocarcinoma in non-smoking female patients and
clinical data were also a limitation and should be conducted in
future studies.
In conclusion, the present study used bioinformatics
analysis to explore the pathogenesis of lung adenocarcinoma in
non-smoking female patients and to identify prognostic biomarkers
for this disease. Additionally, the effect of genetic and molecular
effect of estrogen in non-smoking female patients with lung
adenocarcinoma was investigated. The results obtained in the
present study provide novel insights into the molecular mechanisms
of lung adenocarcinoma in non-smoking female patients.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets analyzed in the present study are
available from the GEO repository at (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=gse32863)
and (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=gse75037),
and TCGA repository at (https://portal.gdc.cancer.gov/repository).
Authors' contributions
LW and HW conceived the study and reviewed the
literature. ZZ, KX, SW and LL collected and analyzed the data. HW
wrote the paper. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Torre LA, Siegel RL and Jemal A: Lung
cancer statistics. Adv Exp Med Biol. 893:1–19. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Pirker R and Filipits M: Adjuvant therapy
in patients with completely resected non-small-cell lung cancer:
Current status and perspectives. Clin Lung Cancer. 20:1–6. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Saito S, Espinoza-Mercado F, Liu H, Sata
N, Cui X and Soukiasian HJ: Current status of research and
treatment for non-small cell lung cancer in never-smoking females.
Cancer Biol Ther. 18:359–368. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Pikor LA, Ramnarine VR, Lam S and Lam WL:
Genetic alterations defining NSCLC subtypes and their therapeutic
implications. Lung Cancer. 82:179–189. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Shi K, Li N, Yang M and Li W:
Identification of key genes and pathways in female lung cancer
patients who never smoked by a bioinformatics analysis. J Cancer.
10:51–60. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Mollerup S, Jorgensen K, Berge G and
Haugen A: Expression of estrogen receptors alpha and beta in human
lung tissue and cell lines. Lung Cancer. 37:153–159. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Tang H, Bai Y, Xiong L, Zhang L, Wei Y,
Zhu M, Wu X, Long D, Yang J, Yu L, et al: Interaction of estrogen
receptor β5 and interleukin 6 receptor in the progression of
non-small cell lung cancer. J Cell Biochem. 2018.(Epub ahead of
print).
|
|
8
|
Chen W, Xin B, Pang H, Han L, Shen W, Zhao
Z, Duan L, Cao P, Liu L and Zhang H: Downregulation of estrogen
receptor β inhibits lung adenocarcinoma cell growth. Oncol Rep.
41:2967–2974. 2019.PubMed/NCBI
|
|
9
|
Huang Q, Zhang Z, Liao Y, Liu C, Fan S,
Wei X, Ai B and Xiong J: 17β-estradiol upregulates IL6 expression
through the ERβ pathway to promote lung adenocarcinoma progression.
J Exp Clin Cancer Res. 37:1332018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Donner I, Katainen R, Sipila LJ, Aavikko
M, Pukkala E and Aaltonen LA: Germline mutations in young
non-smoking women with lung adenocarcinoma. Lung Cancer. 122:76–82.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lindeman NI, Cagle PT, Beasley MB, Chitale
DA, Dacic S, Giaccone G, Jenkins RB, Kwiatkowski DJ, Saldivar JS,
Squire J, et al: Molecular testing guideline for selection of lung
cancer patients for EGFR and ALK tyrosine kinase inhibitors:
Guideline from the college of American pathologists, international
association for the study of lung cancer, and association for
molecular pathology. J Thorac Oncol. 8:823–859. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lee K, Jung HA, Sun JM, Lee SH, Ahn JS,
Park K and Ahn MJ: Clinical characteristics and outcomes of
non-small cell lung cancer patients with HER2 alterations in korea.
Cancer Res Treat. 2019.(Epub ahead of print). View Article : Google Scholar
|
|
13
|
Ranhotra HS: Estrogen-related receptor
alpha and cancer: Axis of evil. J Recept Signal Transduct Res.
35:505–508. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Selamat SA, Chung BS, Girard L, Zhang W,
Zhang Y, Campan M, Siegmund KD, Koss MN, Hagen JA, Lam WL, et al:
Genome-scale analysis of DNA methylation in lung adenocarcinoma and
integration with mRNA expression. Genome Res. 22:1197–1211. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Girard L, Rodriguez-Canales J, Behrens C,
Thompson DM, Botros IW, Tang H, Xie Y, Rekhtman N, Travis WD,
Wistuba II, et al: An expression signature as an aid to the
histologic classification of non-small cell lung cancer. Clin
Cancer Res. 22:4880–4889. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Győrffy B, Surowiak P, Budczies J and
Lanczky A: Online survival analysis software to assess the
prognostic value of biomarkers using transcriptomic data in
non-small-cell lung cancer. PLoS One. 8:e822412013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yu W, Ding J, He M, Chen Y, Wang R, Han Z,
Xing EZ, Zhang C and Yeh S: Estrogen receptor β promotes the
vasculogenic mimicry (VM) and cell invasion via altering the
lncRNA-MALAT1/miR-145-5p/NEDD9 signals in lung cancer. Oncogene.
38:1225–1238. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Fu S, Liu C, Huang Q, Fan S, Tang H, Fu X,
Ai B, Liao Y and Chu Q: Estrogen receptor β1 activation accelerates
resistance to epidermal growth factor receptor-tyrosine kinase
inhibitors in non-small cell lung cancer. Oncol Rep. 39:1313–1321.
2018.PubMed/NCBI
|
|
19
|
Lund-Iversen M, Scott H, Strøm EH, Theiss
N, Brustugun OT and Grønberg BH: Expression of estrogen
receptor-alpha and survival in advanced-stage non-small cell lung
cancer. Anticancer Res. 38:2261–2269. 2018.PubMed/NCBI
|
|
20
|
Kuo LC, Cheng LC, Lee CH, Lin CJ, Chen PY
and Li LA: Estrogen and cigarette sidestream smoke particulate
matter exhibit ERα-dependent tumor-promoting effects in lung
adenocarcinoma cells. Am J Physiol Lung Cell Mol Physiol.
313:L477–L490. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chen KY, Hsiao CF, Chang GC, Tsai YH, Su
WC, Chen YM, Huang MS, Tsai FY, Jiang SS, Chang IS, et al: Estrogen
receptor gene polymorphisms and lung adenocarcinoma risk in
never-smoking women. J Thorac Oncol. 10:1413–1420. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ketteler J and Klein D: Caveolin-1, cancer
and therapy resistance. Int J Cancer. 143:2092–2104. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Senetta R, Stella G, Pozzi E, Sturli N,
Massi D and Cassoni P: Caveolin-1 as a promoter of tumour
spreading: When, how, where and why. J Cell Mol Med. 17:325–336.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ramírez CM, González M, Díaz M, Alonso R,
Ferrer I, Santpere G, Puig B, Meyer G and Marin R: VDAC and ERalpha
interaction in caveolae from human cortex is altered in Alzheimer's
disease. Mol Cell Neurosci. 42:172–183. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Watson CS, Jeng YJ, Hu G, Wozniak A,
Bulayeva N and Guptarak J: Estrogen- and xenoestrogen-induced ERK
signaling in pituitary tumor cells involves estrogen receptor-α
interactions with G protein-αi and caveolin I. Steroids.
77:424–432. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Perez White B, Molloy ME, Zhao H, Zhang Y
and Tonetti DA: Extranuclear ERα is associated with regression of
T47D PKCα-overexpressing, tamoxifen-resistant breast cancer. Mol
Cancer. 12:342013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhang X, Zhang L, Tan X, Lin Y, Han X,
Wang H, Ming H, Li Q, Liu K and Feng G: Systematic analysis of
genes involved in oral cancer metastasis to lymph nodes. Cell Mol
Biol Lett. 23:532018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Icer MA and Gezmen-Karadag M: The multiple
functions and mechanisms of osteopontin. Clin Biochem. 59:17–24.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Mountzios G, Ramfidis V, Terpos E and
Syrigos KN: Prognostic significance of bone markers in patients
with lung cancer metastatic to the skeleton: A review of published
data. Clin Lung Cancer. 12:341–349. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wei R, Wong JPC and Kwok HF: Osteopontin-a
promising biomarker for cancer therapy. J Cancer. 8:2173–2183.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ouyang X, Huang Y, Jin X, Zhao W, Hu T, Wu
F and Huang J: Osteopontin promotes cancer cell drug resistance,
invasion, and lactate production and is associated with poor
outcome of patients with advanced non-small-cell lung cancer.
OncoTargets Ther. 11:5933–5941. 2018. View Article : Google Scholar
|
|
32
|
Wang Y, Yang J, Liu H, Bi JR, Liu Y, Chen
YY, Cao JY and Lu YJ: The association between osteopontin and
survival in non-small-cell lung cancer patients: A meta-analysis of
13 cohorts. OncoTargets Ther. 8:3513–3521. 2015.
|
|
33
|
Moirangthem A, Bondhopadhyay B, Mukherjee
M, Bandyopadhyay A, Mukherjee N, Konar K, Bhattacharya S and Basu
A: Simultaneous knockdown of uPA and MMP9 can reduce breast cancer
progression by increasing cell-cell adhesion and modulating EMT
genes. Sci Rep. 6:219032016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Piperigkou Z, Franchi M, Götte M and
Karamanos NK: Estrogen receptor beta as epigenetic mediator of
miR-10b and miR-145 in mammary cancer. Matrix Biol. 64:94–111.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhang Z, Fang C, Wang Y, Zhang J, Yu J,
Zhang Y, Wang X and Zhong J: COL1A1: A potential therapeutic target
for colorectal cancer expressing wild-type or mutant KRAS. Int J
Oncol. 53:1869–1880. 2018.PubMed/NCBI
|
|
36
|
Elkin M, Miao HQ, Nagler A, Aingorn E,
Reich R, Hemo I, Dou HL, Pines M and Vlodavsky I: Halofuginone: A
potent inhibitor of critical steps in angiogenesis progression.
FASEB J. 14:2477–2485. 2000. View Article : Google Scholar : PubMed/NCBI
|