Introduction
Cholangiocarcinoma (CCA) is a diverse epithelial
malignancy originating in the cholangiocytes, the epithelial cells
of the bile duct. The incidence of CCA has increased globally over
the past few decades (1). Chronic
infection and inflammation due to liver fluke infection, sclerosing
cholangitis and hepatitis C and B virus infections serve a key role
in cholangiocarcinogenesis, possibly through the accumulation of
genetic and epigenetic changes that result in abnormalities in
oncogenes and tumor suppressor genes (2,3). The
most commonly mutated genes in CCA, including KRAS proto-oncogene
GTPase (KRAS), tumor protein p53 (TP53), B-Raf
proto-oncogene, serine/threonine kinase (BRAF), BRCA1
associated protein 1(BAP1), and SMAD family member 4
(SMAD4), are associated with cell signaling pathways (for
example, MAPKs signaling and TGF-β signaling), cell cycle control
and chromatin dynamics (2,4). Previous studies have demonstrated that
KRAS point mutation in intrahepatic CCA (iCCA) may affect
patient prognosis (5). Furthermore,
mutations in KRAS and TP53 in mature cholangiocytes
and hepatocytes may cause iCCA (6).
A combination of mitogen-activated protein kinase kinase 1/2 and
BRAF inhibitors has been shown to be effective in patients
with iCCA harboring the BRAF V600E mutation (7). Activation of the transforming growth
factor-β/Smad4 signaling pathway accelerates CCA cell invasion and
migration via the epithelial-mesenchymal transition (EMT) (8). Knockdown of BAP1 increases CCA
cell proliferation, whereas overexpression of wild-type BAP1
significantly inhibits cell proliferation, suggesting that
BAP1 exhibits tumor suppressive effects (9). Despite significant efforts to elucidate
the pathogenesis of CCA, the precise molecular mechanisms involved
remain unclear. Therefore, the aim of the present study was to
investigate the molecular mechanisms involved in the pathogenesis
of CCA and to identify potential therapeutic targets.
In recent years, microarray technology has attracted
attention due to its ability to rapidly and simultaneously quantify
the expression levels of several genes, and is particularly
suitable for the screening of differentially expressed genes (DEGs)
(10). Several microarray-based
studies on CCA have identified numerous DEGs (11,12).
However, previous reports were limited to independent microarray
analysis or single cohort studies (13). Therefore, in order to identify more
accurate and practical biomarkers, the present study analyzed two
mRNA microarray datasets (GSE26566 and GSE89749), downloaded from
the Gene Expression Omnibus (GEO), and screened for DEGs between
cholangiocarcinoma and normal bile duct samples. Gene Ontology (GO)
and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway
enrichment analyses were subsequently performed and a
protein-protein interaction (PPI) network was constructed. The
results obtained in the present study may aid the early diagnosis
and treatment of CCA.
Materials and methods
Microarray data
The current study aimed to elucidate the potential
key candidate genes in CCA. Two gene expression profiles, GSE26566
(11) and GSE89749 (12), were downloaded from the GEO database
(ncbi.nlm.nih.gov/geo). The GSE26566
dataset consisted of an mRNA expression profile of 104 CCA and 6
normal bile duct samples, while the GSE89749 dataset included 118
CCA and 2 normal bile duct samples. The normal bile duct samples
were obtained from healthy controls. The samples in the GSE26566
dataset were obtained from three countries (Australia, Belgium and
the United States of America) and the samples in GSE89749 dataset
were obtained from ten countries (Singapore, Romania, Thailand,
Italy, France, Korea, Brazil, Taiwan, China and Japan).
DEGs screening
The DEGs between CCA and normal bile duct samples
were identified using GEO2R (www.ncbi.nlm.nih.gov/geo/geo2r), an interactive online
tool used to identify DEGs by comparing samples from the GEO
database. The cut-off criteria for the selection of DEGs were
P<0.01 and a |log fold-change|≥1. Default settings were used as
the screening criteria throughout the entire bioinformatics
analysis process, as has been reported in previous studies
(14,15).
GO and KEGG analyses of DEGs
GO (geneontology.org) and KEGG (www.genome.jp/kegg) enrichment analyses of the DEGs
were performed using the Database for Annotation, Visualization and
Integrated Discovery (DAVID version 6.8; david.ncifcrf.gov) with P<0.05 as the cut-off
criterion. DAVID provides a comprehensive set of gene functional
annotation information to extract biological information (16).
PPI network of DEGs
The PPI network was constructed using Cytoscape
version 3.7.0 software (www.cytoscape.org) (17) based on the Search Tool for the
Retrieval of Interacting Genes (STRING version 11.0; string-db.org). STRING is a system for searching for
interactions between proven and predicted proteins (18). In the present study, genes with a
combined score of >0.4 were considered significant. Sub-modules
of the PPI network were analyzed using the Cytoscape plug-in
Molecular Complex Detection (MCODE version 1.5.1) (19) with the criteria set as follows: MCODE
score >4, node score cut-off=0.2, degree cut-off=2, max
depth=100 and k-core=2. GO and KEGG analyses for the genes in the
most significant sub-module were subsequently performed using
Metascape version 3.5 (metascape.org)
(20).
Hub gene selection and analysis
The top 10 genes ranked by network degree were
selected as the hub genes. The corresponding proteins may be key
candidate proteins with important physiological regulatory
functions. GO and KEGG analyses for the hub genes were subsequently
performed using Metascape version 3.5. A network of the genes and
their co-expression genes was analyzed using cBioPortal (cbioportal.org), as described previously (21,22). The
University of California, Santa Cruz Cancer Genomics Browser
(genome-cancer.ucsc.edu) was used for the
hierarchical clustering of the hub genes, and for examining the
association between changes in the hub genes and the Child-pugh
classification grade or days to death (23). Survival analyses were performed to
assess the prognostic value of the hub genes identified in the
present study in CCA. Kaplan-Meier analysis in the cBioPortal
online platform was used for overall survival rate and disease-free
survival analyses of the hub genes based on a dataset from The
Cancer Genome Atlas (TCGA; cancergenome.nih.gov/) which contained 51 CCA
samples.
Results
DEGs screening
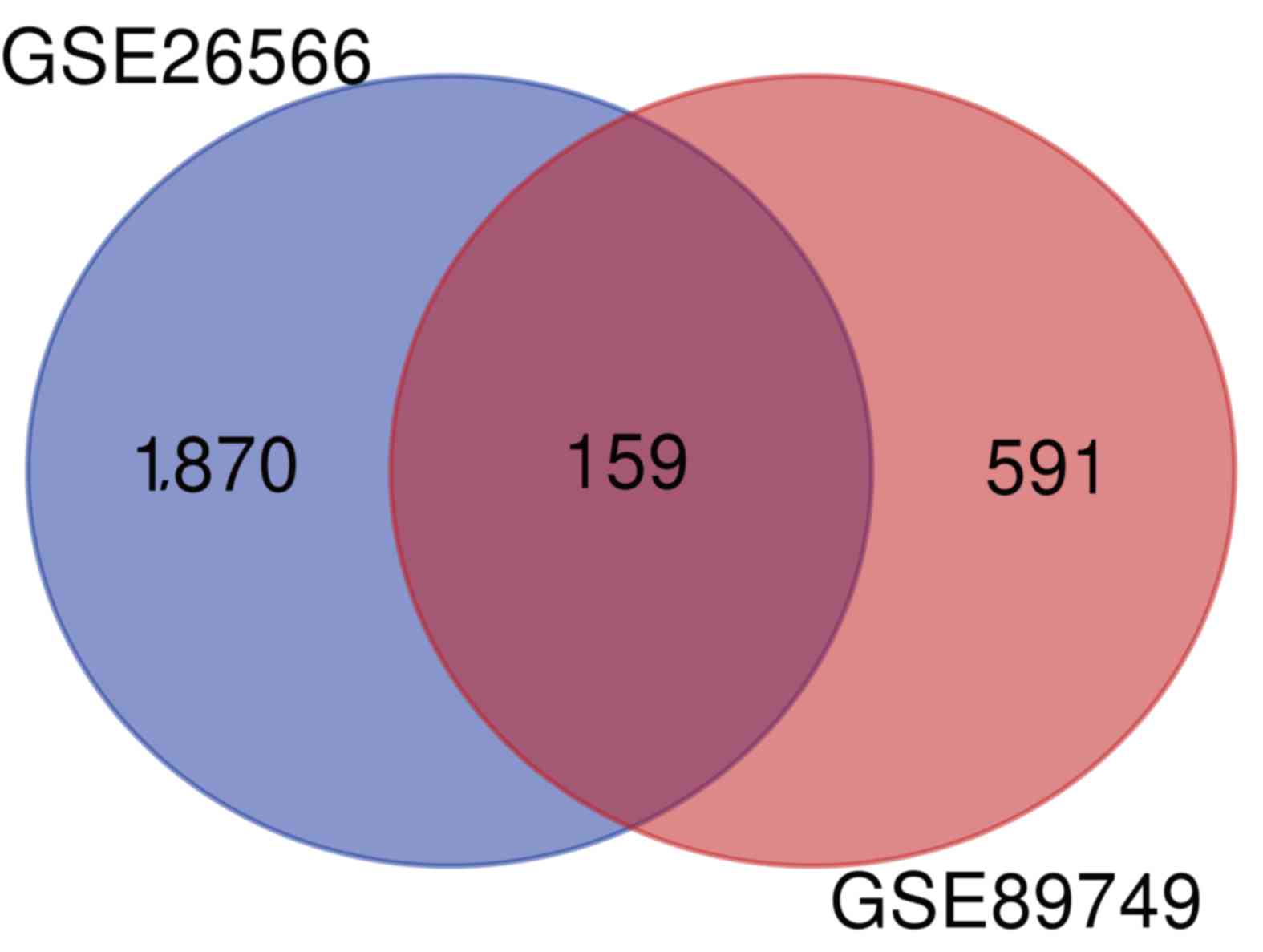
A total of 1,870 and 591 DEGs between CCA and normal
bile duct samples were identified in the GSE26566 and GSE89749
datasets, respectively. The overlap between the two datasets
included 159 DEGs (135 upregulated and 24 downregulated genes;
Fig. 1).
GO and KEGG enrichment analysis of
DEGs
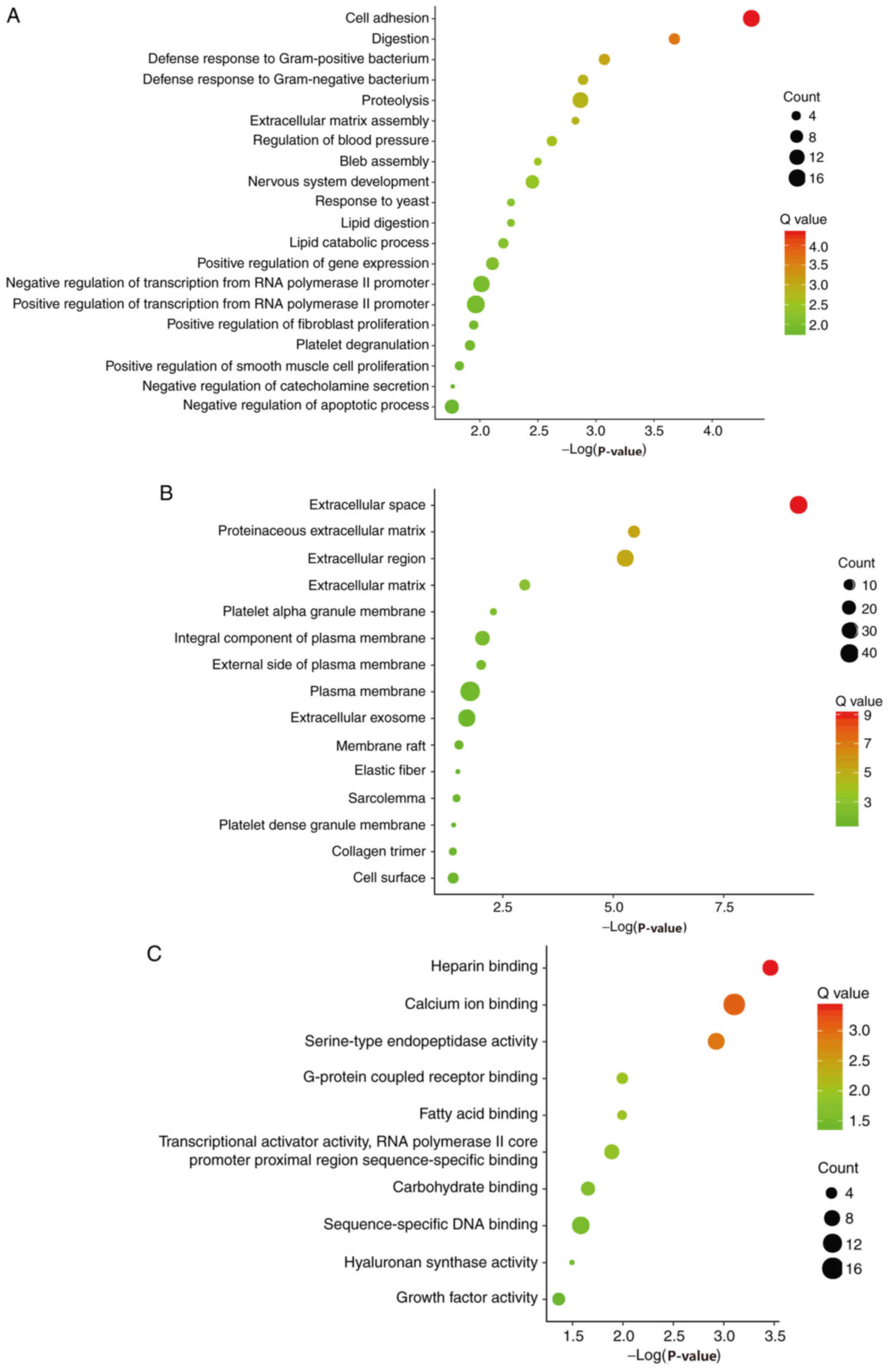
Candidate DEGs were subjected to function and
pathway enrichment analysis in DAVID. GO analysis included
biological processes (BP), cell components (CC) and molecular
functions (MF). BP results suggested that the DEGs were mainly
enriched in ‘cell adhesion’, ‘digestion’ and ‘defense response to
gram-positive bacteria’ (Fig. 2A).
CC results revealed that the DEGs were mainly enriched in
‘extracellular space’, ‘proteinaceous extracellular matrixes’ and
‘extracellular regions’ (Fig. 2B).
MF results suggested that the DEGs were mainly enriched in ‘heparin
binding’, ‘calcium ion binding’ and ‘serine-type endopeptidase
activity’ (Fig. 2C). The KEGG
pathway analysis suggested that the DEGs were mainly associated
with ‘pancreatic secretion’, ‘fat digestion and absorption’ and
‘protein digestion and absorption’ (Fig.
2D).
PPI network of the DEGs
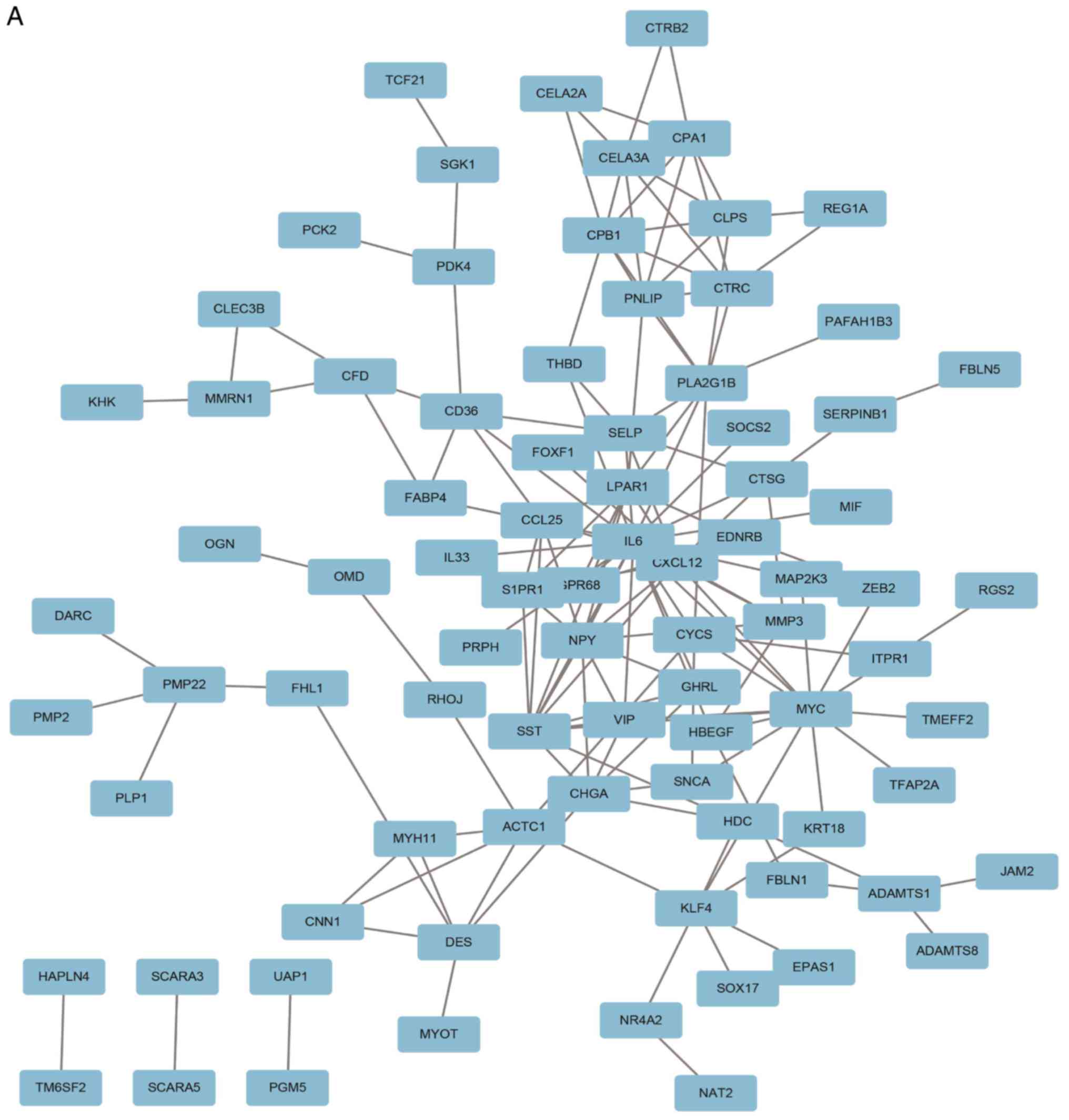
A PPI network containing 84 nodes and 155 edges was
constructed using Cytoscape software based on the STRING database
(Fig. 3A). The most significant
sub-module was extracted from the PPI network complex using MCODE
(Fig. 3B). The functions of the
genes in the aforementioned sub-module were analyzed using the
online tool Metascape. The sub-module consisted of 7 nodes and 17
edges, which were mainly associated with ‘cell chemotaxis’,
‘G-protein coupled receptor signaling pathway’ and ‘positive
regulation of response to external stimulus’ (Fig. 3C).
Hub gene selection and analysis
The top 10 genes ranked by network degree were
selected as hub genes and are presented in Table I. KEGG pathway analysis revealed that
hub genes were mainly associated with the activation of the
phosphoinositide 3-kinase-protein kinase B (Akt) signaling pathway
(Table SI). A network of the hub
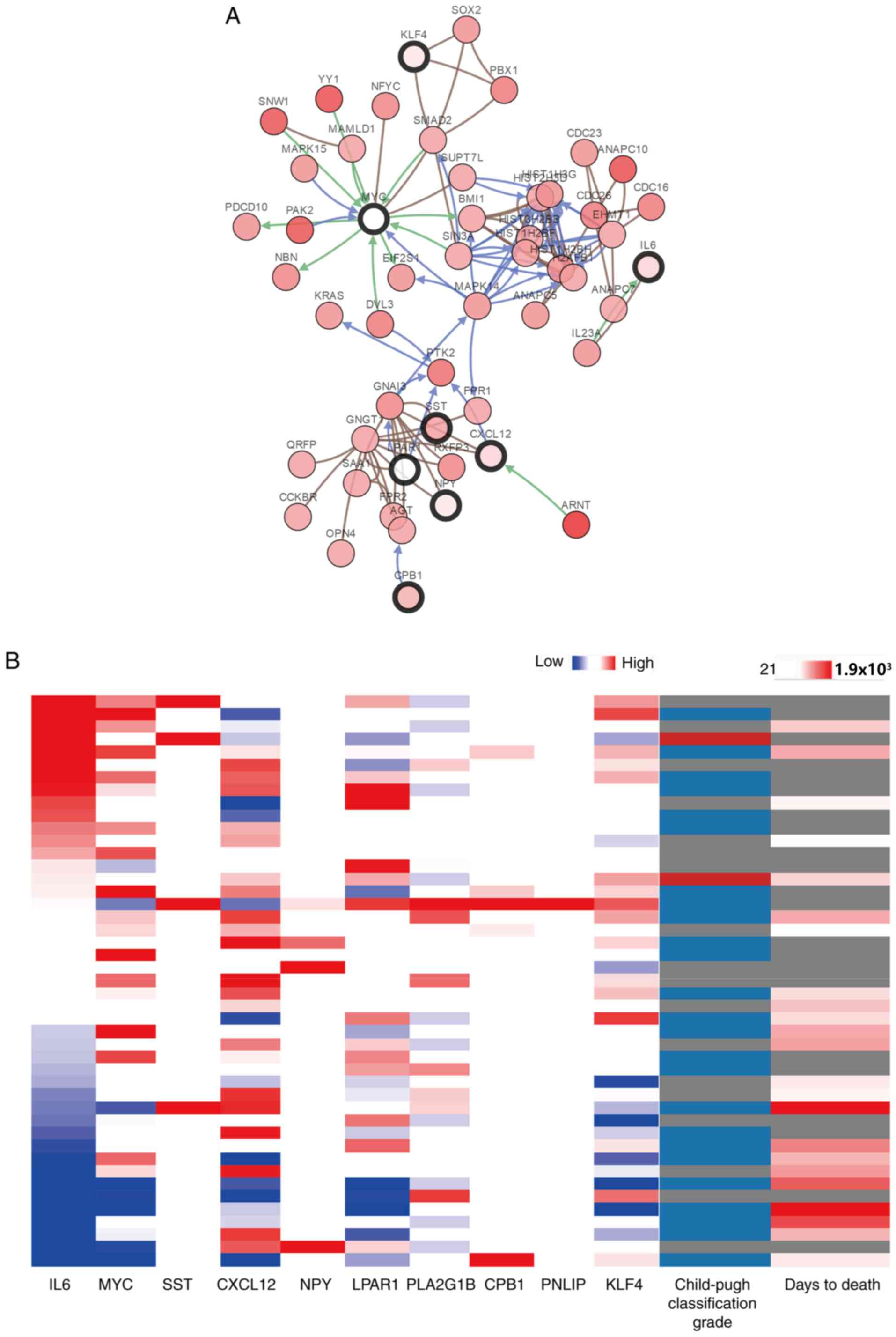
genes and their co-expression genes was analyzed using cBioPortal
(Fig. 4A). PLA2G1B and
PNLIP did not interact with other genes in the network,
thus, only eight hub genes appeared in the network. Hierarchical
clustering revealed that among the patients with CCA with
upregulated hub genes, the patients were classified as grade B
using the Child-pugh classification grade, and the days to death
value was relatively small (Fig.
4B). The expression of MYC and LPAR1 did not an
alteration in the TCGA dataset. As a result, the survival curves of
8 out of the 10 hub genes are presented in the current study. The 6
hub genes that did not have a significant effect on patient outcome
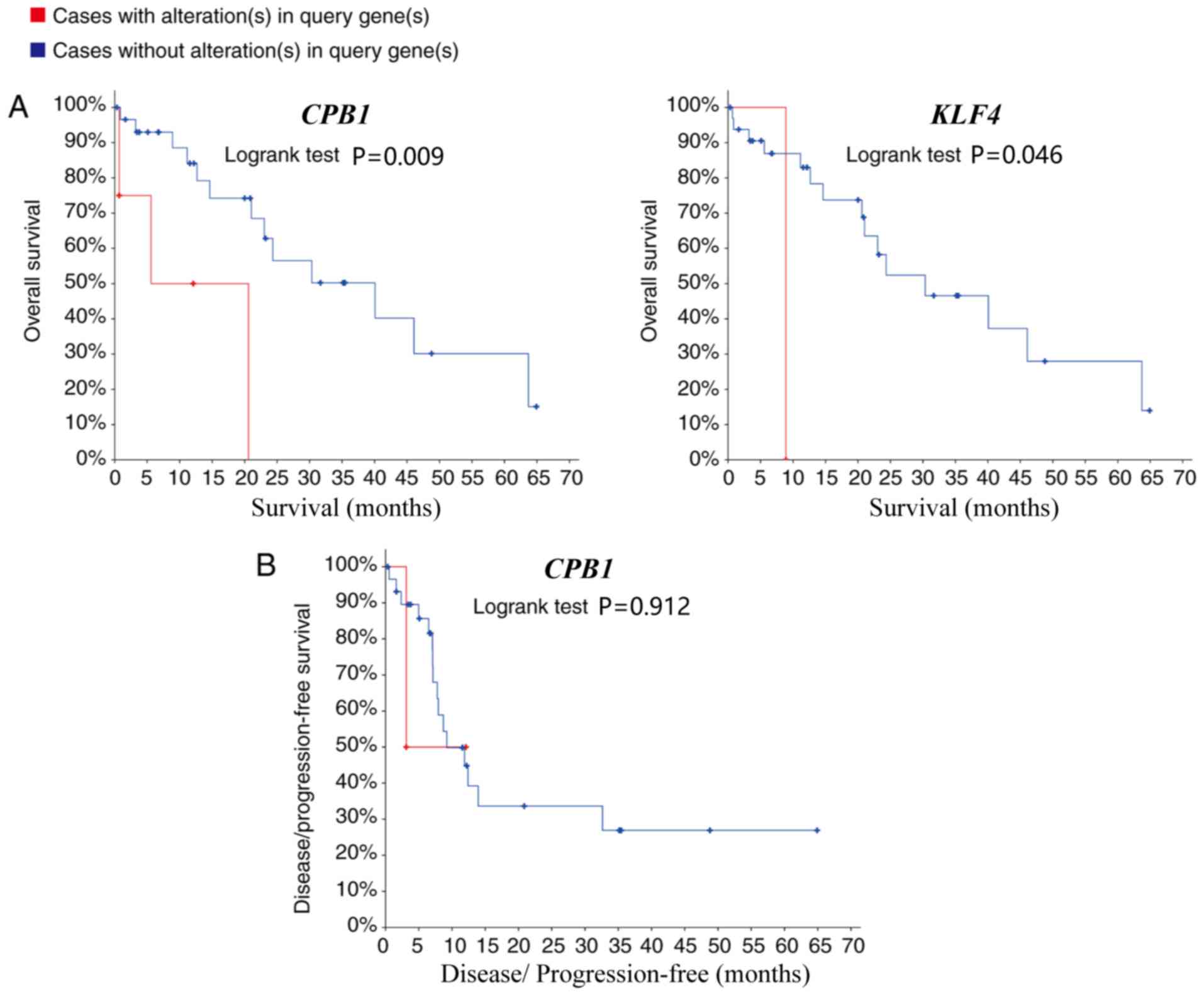
are presented in Fig. S1. Patients
with CCA with carboxypeptidase B1 (CPB1) and KLF4
(Kruppel like factor 4) upregulation, had lower overall survival
rates compared with patients without alterations (P=0.008 and 0.046
respectively; Fig. 5A). However,
whilst CPB1 upregulation was significantly associated with
lower overall survival, it was not associated with disease-free
survival (P=0.912; Fig. 5B). The
Kaplan-Meier estimate could not be used for the disease-free
survival analysis of KLF4 due to the lack of clinical data
on the ‘disease-free survival time’ of patients with the
upregulation.
 | Table I.Functions of the 10 hub genes. |
Table I.
Functions of the 10 hub genes.
| Gene symbol | Full name | Function |
|---|
| IL-6 | Interleukin 6 | Encodes a cytokine
that functions in inflammation and the maturation of B cells |
| MYC | MYC
proto-oncogene | Proto-oncogene that
encodes a nuclear phosphoprotein that plays a role in cell cycle
progression, apoptosis and cellular transformation |
| SST | Somatostatin | Inhibits the
release of numerous secondary hormones |
| CXCL12 | CXC motif chemokine
ligand 12 | Plays a role in a
number of cellular functions, including embryogenesis, immune
surveillance, inflammation response, tissue homeostasis and tumor
growth and metastasis |
| NPY | Neuropeptide Y | Influences several
physiological processes, including cortical excitability, stress
response, food intake, circadian rhythms and cardiovascular
function |
| LPAR1 | Lysophosphatidic
acid receptor 1 | Encodes the
lysophosphatidic acid receptor, mediate diverse biologic functions,
including proliferation, platelet aggregation, smooth muscle
contraction, chemotaxis, and tumor cell invasion. |
| PLA2G1B | Phospholipase A2
group 1B | Encodes a secreted
member of the phospholipase A2 class of enzymes. The enzyme may be
involved in several physiological processes including cell
contraction, cell proliferation and pathological response. |
| CPB1 | Carboxypeptidase
B1 | Carboxypeptidase B1
is a highly tissue-specific protein and is a useful serum marker
for acute pancreatitis and dysfunction of pancreatic
transplants. |
| PNLIP | Pancreatic
lipase | Encodes an enzyme
involved in the digestion of dietary fats |
| KLF4 | Kruppel like Factor
4 | The encoded zinc
finger protein is required for normal development of the barrier
function of skin. The encoded protein is thought to control the
G1-to-S transition of the cell cycle following DNA damage by
mediating the tumor suppressor gene p53. |
Discussion
CCA is the second most common hepatobiliary
malignancy after hepatocellular carcinoma (24). The main etiological factors of CCA
include primary sclerosing cholangitis, cirrhosis and hepatitis C
and B infections (25). While
chronic infection and inflammation in the bile ducts play a major
role in CCA, the molecular mechanisms involved in CCA remain poorly
understood. The most commonly mutated genes in CCA are SMAD4,
TP53, KRAS, BAP1, isocitrate dehydrogenase [NADP(+)] 1
cytosolic, isocitrate dehydrogenase [NADP(+)] 2 mitochondrial and
roundabout guidance receptor 2 (2,4). In
addition, CCA has been reported to be associated with inflammation,
the growth factor signaling pathway, cell signaling pathways and
epigenetic regulation (3,26). There are currently no effective
clinical biomarkers and targeted molecular therapies for the early
diagnosis and treatment of CCA; consequently, the 5-year survival
rate is ~10% (27). There is
therefore a requirement of the identification of diagnostic markers
for CCA. Microarray technology has previously been used to identify
novel biomarkers in various diseases and may also be applied to
uncover diagnostic markers in CCA (10).
In the present study, two microarray datasets
(GSE26566 and GSE89749) were obtained from the GEO and analyzed to
identify DEGs between CCA and normal bile duct samples. In total,
159 DEGs (135 upregulated and 24 downregulated) were identified. GO
and KEGG enrichment analyses were performed to investigate
interactions among the DEGs and a PPI network was constructed,
revealing 10 hub genes. The PPI network demonstrated that
interleukin-6 (IL-6) had the largest number of nodes (20 nodes) and
directly interacted with c-Myc, somatostatin, C-X-C motif chemokine
12 (CXCL12), neuropeptide Y (NPY) and LPAR1,
suggesting that IL-6 may serve an important role in CCA. Consistent
with the results obtained in the current study, a recent study
demonstrated that increased IL-6 expression plays a central role in
the pathogenesis and progression of CCA (28). In addition, the circulating level of
IL-6 in patients with CCA was reported to be increased compared
with healthy controls (29).
Moreover, overexpression of IL-6 promotes cell survival in
malignant cholangiocytes and enhances tumor growth (30). A previous study reported that a
combination of increased levels of leucine-rich α-2-glycoprotein 1,
carbohydrate antigen 19-9 and IL-6 in the serum could be sued to
discriminate between biliary tract cancer (CCA and gallbladder
carcinoma) and benign biliary disease (31). c-MYC is a proto-oncogene and
encodes a nuclear phosphoprotein, which primarily regulates
apoptosis, cell cycle progression and cellular transformation
(32). A previous study showed that
c-MYC is upregulated in human CCA (33). Furthermore, cyclin D1, a c-Myc target
gene, is a molecular biomarker of CCA (34). Knockdown of c-Myc
significantly reduced the extent of cholangiofibrosis and
cholangioma in vivo, highlighting the importance of
c-Myc in the progression of CCA (35). C-X-C motif chemokine receptor 4
(CXCR4) selectively binds CXCL12 and the CXCR4/CXCL12 axis has been
shown to be involved in tumorigenesis, cell proliferation and
angiogenesis in CCA (36). A recent
study suggested that serum CXCL12 levels may serve as a potential
biomarker for predicting the clinical outcome in CCA (37). However, in the present study,
elevated CXCL12 levels were not significantly associated with
disease-free or overall survival. This may be due to the cBioPortal
survival analyses performed, which were based on the association
between gene mutation and prognosis, whereas high expression in
serum is generally caused by a mutation or upregulation (38).
The association between CCA and the hub genes
NPY, LPAR1, phospholipase A2 group IB, CPB1,
pancreatic lipase and KLF4 has not been widely reported.
NPY expression has been shown to be upregulated in CCA
(39,40), therefore regulating NPY
expression may be beneficial for the treatment of CCA. Previous
research demonstrated that KLF4 and microRNA-21 play a key
role in mediating the EMT in CCA cells via the Akt/extracellular
signal-regulated protein kinase 1 and 2 signaling pathway (41). Hierarchical clustering revealed that
the hub genes identified in the present study may be used to
differentiate CCA from normal bile duct samples. Furthermore,
upregulation of CPB1 and KLF4 was associated with a
lower overall survival rate, suggesting that the aforementioned
genes may serve important roles in the progression of CCA.
In summary, the present study identified DEGs that
may be involved in the carcinogenesis or progression of CCA. A
total of 159 DEGs and 10 hub genes were identified, which following
further investigation may serve as diagnostic biomarkers and novel
therapeutic targets for CCA.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was financially supported by the
Natural Science Foundation of Ningbo City (grant no. 2017A610259),
the Huamei Research Foundation (grant no. 2017HMKY07) and the
Medicine and Health Sciences Research Foundation of Zhejiang
Province (grant no. 2019KY177). Additionally, the study was
sponsored by the K.C. Wong Magna Fund of Ningbo University.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from Gene Expression Omnibus; ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE26566 and
ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE89749.
Authors contributions
JT designed the study. RH acquired and interpreted
the datasets. MZ performed data analysis. XM and YZ performed the
survival analysis. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
CCA
|
cholangiocarcinoma
|
|
GEO
|
Gene Expression Omnibus
|
|
DEGs
|
differentially expressed genes
|
|
PPI
|
protein-protein interaction
|
|
EMT
|
epithelial-mesenchymal transition
|
|
GO
|
Gene Ontology
|
|
KEGG
|
Kyoto Encyclopedia of Genes and
Genomes
|
|
DAVID
|
Database for Annotation, Visualization
and Integrated Discovery
|
|
STRING
|
Search Tool for the Retrieval of
Interacting Genes
|
|
MCODE
|
Molecular Complex Detection
|
|
BP
|
biological process
|
|
CC
|
cell component
|
|
MF
|
molecular function
|
References
|
1
|
Saha SK, Zhu AX, Fuchs CS and Brooks GA:
Forty-Year trends in cholangiocarcinoma incidence in the U.S.:
Intrahepatic disease on the rise. Oncologist. 21:594–599. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kongpetch S, Jusakul A, Ong CK, Lim WK,
Rozen SG, Tan P and The BT: Pathogenesis of cholangiocarcinoma:
From genetics to signalling pathways. Best Pract Res Clin
Gastroenterol. 29:233–244. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Razumilava N and Gores GJ:
Cholangiocarcinoma. Lancet. 383:2168–2179. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Valle JW, Lamarca A, Goyal L, Barriuso J
and Zhu AX: New horizons for precision medicine in biliary tract
cancers. Cancer Discov. 7:943–962. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ikeno Y, Seo S, Iwaisako K, Yoh T,
Nakamoto Y, Fuji H, Taura K, Okajima H, Kaido T, Sakaguchi S and
Uemoto S: Preoperative metabolic tumor volume of intrahepatic
cholangiocarcinoma measured by 18F-FDG-PET is associated
with the KRAS mutation status and prognosis. J Transl Med.
16:952018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hill MA, Alexander WB, Guo B, Kato Y,
Patra K, ODell MR, McCall MN, Whitney-Miller CL, Bardeesy N and
Hezel AF: Kras and Tp53 Mutations cause cholangiocyte- and
hepatocyte-derived cholangiocarcinoma. Cancer Res. 78:4445–4451.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Mahipal A, Kommalapati A, Tella SH, Lim A
and Kim R: Novel targeted treatment options for advanced
cholangiocarcinoma. Expert Opin Invest Drugs. 27:709–720. 2018.
View Article : Google Scholar
|
|
8
|
Qiao P, Li G, Bi W, Yang L, Yao L and Wu
D: microRNA-34a inhibits epithelial mesenchymal transition in human
cholangiocarcinoma by targeting Smad4 through transforming growth
factor-beta/Smad pathway. BMC Cancer. 15:4692015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chan-On W, Nairismagi ML, Ong CK, Lim WK,
Dima S, Pairojkul C, Lim KH, McPherson JR, Cutcutache I, Heng HL,
et al: Exome sequencing identifies distinct mutational patterns in
liver fluke-related and non-infection-related bile duct cancers.
Nat Genet. 45:1474–1478. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Vogelstein B, Papadopoulos N, Velculescu
VE, Zhou S, Diaz LA Jr and Kinzler KW: Cancer genome landscapes.
Science. 339:1546–1558. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Andersen JB, Spee B, Blechacz BR, Avital
I, Komuta M, Barbour A, Conner EA, Gillen MC, Roskams T, Roberts
LR, et al: Genomic and genetic characterization of
cholangiocarcinoma identifies therapeutic targets for tyrosine
kinase inhibitors. Gastroenterology. 142:1021–1031.e15. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Jusakul A, Cutcutache I, Yong CH, Lim JQ,
Huang MN, Padmanabhan N, Nellore V, Kongpetch S, Ng AWT, Ng LM, et
al: Whole-Genome and epigenomic landscapes of etiologically
distinct subtypes of cholangiocarcinoma. Cancer Discov.
7:1116–1135. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zou S, Li J, Zhou H, Frech C, Jiang X, Chu
JS, Zhao X, Li Y, Li Q, Wang H, et al: Mutational landscape of
intrahepatic cholangiocarcinoma. Nat Commun. 5:56962014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Guo YC, Bao YH, Ma M and Yang WC:
Identification of key candidate genes and pathways in colorectal
cancer by integrated bioinformatical analysis. Int J Mol Sci.
18(Pii): E7222017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Fu Q, Yang F, Zhao J, Yang X, Xiang T,
Huai G, Zhang J, Wei L, Deng S and Yang H: Bioinformatical
identification of key pathways and genes in human hepatocellular
carcinoma after CSN5 depletion. Cell Signal. 49:79–86. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Huang DW, Sherman BT, Tan Q, Collins JR,
Alvord WG, Roayaei J, Stephens R, Baseler MW, Lane HC and Lempicki
RA: The DAVID gene functional classification tool: A novel
biological module-centric algorithm to functionally analyze large
gene lists. Genome Biol. 8:R1832007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Smoot ME, Ono K, Ruscheinski J, Wang PL
and Ideker T: Cytoscape 2.8: New features for data integration and
network visualization. Bioinformatics. 27:431–432. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Szklarczyk D, Morris JH, Cook H, Kuhn M,
Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, et al:
The STRING database in 2017: Quality-controlled protein-protein
association networks, made broadly accessible. Nucleic Acids Res.
45(D1): D362–D368. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bader GD and Hogue CW: An automated method
for finding molecular complexes in large protein interaction
networks. BMC Bioinformatics. 4:22003. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Tripathi S, Pohl MO, Zhou Y,
Rodriguez-Frandsen A, Wang G, Stein DA, Moulton HM, DeJesus P, Che
J, Mulder LC, et al: Meta- and orthogonal integration of influenza
“OMICs” data defines a role for UBR4 in Virus budding. Cell Host
Microbe. 18:723–735. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Cerami E, Gao J, Dogrusoz U, Gross BE,
Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et
al: The cBio cancer genomics portal: An open platform for exploring
multidimensional cancer genomics data. Cancer Discov. 2:401–404.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Gao J, Aksoy BA, Dogrusoz U, Dresdner G,
Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al:
Integrative analysis of complex cancer genomics and clinical
profiles using the cBioPortal. Sci Signal. 6:pl12013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang K, Yu J, Yu XL, Han ZY, Cheng ZG,
Liu FY and Liang P: Clinical and survival outcomes of percutaneous
microwave ablation for intrahepatic cholangiocarcinoma. Int J
Hyperthermia. 34:292–297. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Rizvi S, Khan SA, Hallemeier CL, Kelley RK
and Gores GJ: Cholangiocarcinoma - evolving concepts and
therapeutic strategies. Nat Rev Clin Oncol. 15:95–111. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Bagante F, Gamblin TC and Pawlik TM:
Cholangiocarcinoma risk factors and the potential role of aspirin.
Hepatology. 64:708–710. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Rizvi S and Gores GJ: Pathogenesis,
diagnosis, and management of cholangiocarcinoma. Gastroenterology.
145:1215–1229. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Nakamura H, Arai Y, Totoki Y, Shirota T,
Elzawahry A, Kato M, Hama N, Hosoda F, Urushidate T, Ohashi S, et
al: Genomic spectra of biliary tract cancer. Nat Genet.
47:1003–1010. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Tanjak P, Thiantanawat A, Watcharasit P
and Satayavivad J: Genistein reduces the activation of AKT and
EGFR, and the production of IL6 in cholangiocarcinoma cells
involving estrogen and estrogen receptors. Int J Oncol. 53:177–188.
2018.PubMed/NCBI
|
|
29
|
Cheon YK, Cho YD, Moon JH, Jang JY, Kim
YS, Kim YS, Lee MS, Lee JS and Shim CS: Diagnostic utility of
interleukin-6 (IL-6) for primary bile duct cancer and changes in
serum IL-6 levels following photodynamic therapy. Am J
Gastroenterol. 102:2164–2170. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Meng F, Yamagiwa Y, Ueno Y and Patel T:
Over-expression of interleukin-6 enhances cell survival and
transformed cell growth in human malignant cholangiocytes. J
Hepatol. 44:1055–1065. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Sandanayake NS, Sinclair J, Andreola F,
Chapman MH, Xue A, Webster GJ, Clarkson A, Gill A, Norton ID, Smith
RC, et al: A combination of serum leucine-rich alpha-2-glycoprotein
1, CA19-9 and interleukin-6 differentiate biliary tract cancer from
benign biliary strictures. Br J Cancer. 105:1370–1378. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Stine ZE, Walton ZE, Altman BJ, Hsieh AL
and Dang CV: MYC, metabolism, and cancer. Cancer Discov.
5:1024–1039. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Boulter L, Guest RV, Kendall TJ, Wilson
DH, Wojtacha D, Robson AJ, Ridgway RA, Samuel K, Van Rooijen N,
Barry ST, et al: WNT signaling drives cholangiocarcinoma growth and
can be pharmacologically inhibited. J Clin Invest. 125:1269–1285.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yang H, Li TW, Peng J, Tang X, Ko KS, Xia
M and Aller MA: A mouse model of cholestasis-associated
cholangiocarcinoma and transcription factors involved in
progression. Gastroenterology. 141:378–388, 388.e1-e4. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yang H, Liu T, Wang J, Li TW, Fan W, Peng
H, Krishnan A, Gores GJ, Mato JM and Lu SC: Deregulated methionine
adenosyltransferase α1, c-Myc, and Maf proteins together promote
cholangiocarcinoma growth in mice and humans (double dagger).
Hepatology. 64:439–455. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhao S, Wang J and Qin C: Blockade of
CXCL12/CXCR4 signaling inhibits intrahepatic cholangiocarcinoma
progression and metastasis via inactivation of canonical Wnt
pathway. J Exp Clin Cancer Res. 33:1032014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Lee SJ, Kim JE, Kim ST, Lee J, Park SH,
Park JO, Kang WK, Park YS and Lim HY: The correlation between serum
Chemokines and clinical outcome in patients with advanced biliary
tract cancer. Transl Oncol. 11:353–357. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Li L, Lei QS, Zhang SJ, Kong LN and Qin B:
Screening and identification of key biomarkers in hepatocellular
carcinoma: Evidence from bioinformatic analysis. Oncol Rep.
38:2607–2618. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
DeMorrow S, Onori P, Venter J, Invernizzi
P, Frampton G, White M, Franchitto A, Kopriva S, Bernuzzi F,
Francis H, et al: Neuropeptide Y inhibits cholangiocarcinoma cell
growth and invasion. Am J Physiol Cell Physiol. 300:C1078–C1089.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
DeMorrow S, Onori P, Venter J,
Leyva-Illades D, Francis H, Frampton G, Pae HY, Quinn M, Onori P,
Glaser S, et al: Neuropeptide Y inhibits biliary hyperplasia of
cholestatic rats by paracrine and autocrine mechanisms. Am J
Physiol Gastrointest Liver Physiol. 305:G250–G257. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Liu CH, Huang Q, Jin ZY, Zhu CL, Liu Z and
Wang C: miR-21 and KLF4 jointly augment epithelial-mesenchymal
transition via the Akt/ERK1/2 pathway. Int J Oncol. 50:1109–1115.
2017. View Article : Google Scholar : PubMed/NCBI
|