Introduction
Primary hepatic carcinoma (PHC) is one of the most
common malignancies in China (1). An
estimated 466,000 new PHC cases, and 422,000 PHC-associated
mortalities occurred in China in 2015, with only 10% of patients
surviving >5 years (2).
Furthermore, due to the lack of sensitive markers for early
diagnosis, only 10–20% of patients with PHC are suitable for
surgical resection, local ablation and other potential therapies
(3). The current biomarkers that are
used for the auxiliary diagnosis of PHC include alpha fetoprotein
(AFP), AFP-L3 isoform ratio, glypican-3, des-γ-carboxy-prothrombin,
golgi protein 73 and alpha L-fucus glycosidase. However, these
markers, even when utilized together, are not sufficient for the
identification or early diagnosis of PHC. It is therefore necessary
to identify novel early diagnostic markers and therapeutic targets
for use in liver cancer diagnosis and treatment (4).
Abnormal tumor-suppressor gene inactivation or
oncogene activation are leading causes of liver cancer, and a
growing number of studies have indicated that circular RNAs
(circRNAs) are associated with a variety of cellular activities
(5). circRNAs are a large class of
endogenous, non-coding RNAs that feature covalently closed
continuous loops with highly conserved sequences; they have been
revealed to serve an important role in the regulation of gene
expression during and after transcription (6). The majority of circRNAs derive from
exons, localize in the cytoplasm, and exhibit tissue/developmental
stage-specific expression (7).
circRNAs can be detected in human blood samples and exhibit higher
expression levels than corresponding linear mRNAs (8). As previously reported, circRNAs have
been demonstrated to serve a role in the occurrence and progression
of a variety of tumor types, including esophageal squamous cell and
basal cell carcinoma, as well as colon, ovarian and breast cancers
(9–13). These studies clearly demonstrate the
potential use of circRNAs as tumor biomarkers (14–16). It
has recently been reported that hsa_circ_0001649, hsa_circ_0005075,
hsa_circ_0000130, hsa_circ_0004018 and hsa_circ_0001445 may
represent potential biomarkers for hepatocellular carcinoma (HCC),
and potentially influence PHC tumor occurrence and metastasis
(17–21). circRNAs act as a sponge for microRNAs
(miRNAs), regulating downstream mRNA genes through a competing
endogenous RNA (ceRNA) network (22). Compared with miRNAs and long
non-coding RNAs, circRNAs have not been extensively researched in
HCC, with only a few studies focusing on their differential
expression in liver cancer tissues. However, to the best of our
knowledge, their expression in plasma has not yet been determined,
and the use of ceRNA networks and their potential application in
clinical diagnosis are yet to be elucidated.
In the present study, comprehensive circRNA
expression profiles were analyzed in PHC tissues and paired
adjacent normal tissues using circRNA chip screening.
Differentially expressed (DE) circRNAs were further validated in
PHC tissues and plasma samples using reverse
transcription-quantitative (RT-q) PCR. Significant downregulation
of hsa_circ_0003056 in PHC tissues and hsa_circ_0067127 in plasma
was demonstrated. The ceRNA networks of hsa_circ_0003056 and
hsa_circ_0067127 were constructed and analyzed using gene ontology
(GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway
analyses.
Materials and methods
Patients and specimens
A total of 14 PHC tissue specimens and paired
adjacent normal tissues, along with fresh plasma samples from 35
PHC patients and 32 healthy donors, were obtained between October
2016 and August 2018 at The Second Hospital of Nanjing (Nanjing,
China). Non-tumorous tissues were removed 2.0 cm from the tumor
edge and no obvious malignant cells were identified by a
pathologist. Following dissection, all tissues were preserved in
RNA storage solution (Shanghai SangonBiotech Co., Ltd.) and stored
at −80°C until further use. All tissues were confirmed by
pathological examination and none of the patients received
neoadjuvant therapy. The baseline characteristics and
clinicopathological features of each patient were collected from
the hospital digital health care system, and are summarized in
Table I. The research procedures of
the present study were approved by the Ethics Committee of the
Second Hospital of Nanjing, and written informed consent was
obtained from all enrolled participants.
 | Table I.Baseline characteristics of PHC
patients and stratified analyses of hsa_circ_0003056 and
has_circ_0067127 expression in PHC plasma. |
Table I.
Baseline characteristics of PHC
patients and stratified analyses of hsa_circ_0003056 and
has_circ_0067127 expression in PHC plasma.
|
|
|
|
|
Hsa_circ_0003056 |
Hsa_circ_0067127 |
|---|
|
|
|
|
|
|
|
|---|
| Clinicopathological
parameters | Chips (n=3) | PHC tissues
(n=11) | PHC plasma
(n=35) | Mean ± SD | P-value | Mean ± SD | P-value |
|---|
| Sex |
|
|
|
|
|
|
|
|
Male | 3 | 7 | 27 | 2.41±0.56 | 0.66 | 0.31±0.07 | 0.08 |
|
Female | 0 | 4 | 8 | 1.89±1.12 |
| 0.70±0.25 |
|
| Age, years |
|
|
|
|
|
|
|
|
<60 | 3 | 11 | 20 | 3.13±0.80 | 0.04 | 0.40±0.11 | 0.56 |
|
≥60 | 0 | 0 | 15 | 1.17±0.27 |
| 0.30±0.10 |
|
| HBsAg |
|
|
|
|
|
|
|
|
Negative | 0 | 1 | 9 | 1.32±0.44 | 0.25 | 0.39±0.16 | 0.82 |
|
Positive | 3 | 10 | 26 | 2.63±0.64 |
| 0.35±0.09 |
|
| Cirrhosis |
|
|
|
|
|
|
|
|
Negative | 1 | 8 | 25 | 1.83±0.67 | 0.56 | 0.51±0.16 | 0.3 |
|
Positive | 2 | 3 | 10 | 2.47±0.64 |
| 0.32±0.09 |
|
| AFP, ng/ml |
|
|
|
|
|
|
|
|
<200 | 3 | 6 | 24 | 2.20±0.58 | 0.78 | 0.46±0.10 | 0.05 |
|
≥200 | 0 | 5 | 11 | 2.50±0.98 |
| 0.16±0.04 |
|
| Tumor number |
|
|
|
|
|
|
|
|
Single | 3 | 8 | 13 | 3.03±1.16 | 0.25 | 0.42±0.14 | 0.64 |
|
Multiple | 0 | 3 | 22 | 1.85±0.39 |
| 0.34±0.09 |
|
| Tumor diameter |
|
|
|
|
|
|
|
| <5
cm | 2 | 6 | 21 | 2.14±0.62 | 0.72 | 0.40±0.12 | 0.61 |
| ≥5
cm | 1 | 5 | 14 | 2.51±0.84 |
| 0.32±0.09 |
|
| TNM stage |
|
|
|
|
|
|
|
|
I–II | 3 | 11 | 19 | 2.53±0.83 | 0.61 | 0.42±0.12 | 0.38 |
|
III–IV | 0 | 0 | 16 | 2.01±0.47 |
| 0.29±0.09 |
|
circRNA expression profiles
A circRNA chip (Arraystar Human circRNAs Array V2;
Zhejiang Kangchen Biotech Co., Ltd.) containing 13,617 probes
specific to human circRNA splicing sites was used. A total of three
pairs of tumor tissues and paired non-tumorous tissues from
patients with PHC were analyzed using the circRNA chips, according
to the manufacturer's protocol. The chips were then scanned using
the Agilent Scanner G2505C (Agilent Technologies, Inc.), and the
raw data were extracted using Agilent Feature Extraction software
(version 11.5.1.1; Agilent Technologies, Inc.). Quantile
normalization of the raw data and subsequent data processing were
performed using the R software limma package 3.40.6 (http://www.bioconductor.org/packages/release/bioc/html/limma.html).
Subsequently, low intensity filtering was performed, and circRNAs
that appeared in ≥3 out of 6 samples, and had flags in ‘P’ or ‘M’
(‘All Targets Value’) were selected for further analyses. circRNAs
that exhibited fold changes ≥1.5 and P≤0.05 were selected as the
significant DE circRNAs. circRNA/miRNA interactions were predicted
using Arraystar's home-made miRNA target prediction software, which
was derived by modifying an existing software package from
TargetScan (23) (http://www.targetscan.org/) and miRanda (24) (http://www.microrna.org/microrna/home.do). The miRNA
support vector regression scores were identified and used to
construct a ‘Top 5’ circRNA-miRNA-mRNA network for DE circRNAs.
Total RNA extraction and RT-qPCR
Total RNA was isolated from tumor tissues, adjacent
normal tissues and plasma samples using the AllPure Tissue kit,
HiPure Blood/Liquid RNA kit (Magen; http://www.magentec.com.cn/) and miRNeasy Serum/Plasma
kit (Qiagen, GmbH) according to the manufacturers' protocols. The
total RNA was quantified using a Colibri spectrometer (Titertek
Berthold), and RNA integrity was assessed using electrophoresis on
a denaturing agarose gel. Isolated RNA samples were stored at −80°C
prior to use. Total RNA from tissue (1.0 µg) or plasma (0.3 µg) was
reverse transcribed into first-strand cDNA using the HiScript Q RT
SuperMix for qPCR (+gDNA wiper; Vazyme) according to the
manufacturer's protocol. qPCR was performed using the
SYBR® Green Master Mix (High ROX Premixed; Vazyme), with
the Applied Biosystems™ QuantStudiot™ 3 Real-Time PCR system
(Thermo Fisher Scientific, Inc.). circRNAs were analyzed with
β-actin as the internal standard. The reactions were prepared as
follows: 10 µl qPCR SYBR® Green Master Mix (High Rox
Premixed), 0.4 µl forward primer (10 µM), 0.4 µl reverse primer (10
µM), 8.2 µl RNase free water and 1 µl cDNA. The thermocycling
conditions were as follows: 95°C for 5 min, followed by 40 cycles
of 95°C for 10 sec and 60°C for 30 sec, and a final step of 95°C
for 15 sec, 60°C for 60 sec and 95°C for 15 sec. All primers used
in the present study were designed and synthesized by Geneseed
Biotech Co., Ltd. and are listed in Table II.
 | Table II.Primer sequences for reverse
transcription-quantitative PCR. |
Table II.
Primer sequences for reverse
transcription-quantitative PCR.
| Primer name | Forward
(5′-3′) | Reverse
(5′-3′) | Product length | Regulation |
|---|
|
hsa_circ_0003056 |
atttatcacctaaagagccgt |
tcaattccttctccaccac | 119 bp | Down |
|
hsa_circ_0005090 |
gaggagctcaccatctgcta |
gagttgcagatcaccttgtcc | 131 bp | Up |
|
hsa_circ_0007646 |
ggaatgacttctctccaattttca |
aagaactgcaagaccgcaga | 152 bp | Up |
|
hsa_circ_0064557 |
gccccctttcacaggtgatct |
ttctgctccaggcgggcaat | 145 bp | Down |
|
hsa_circ_0067127 |
gtcctcccaggatctggctg |
hcgcagcttcctcactaacagc | 127 bp | Down |
Functional enrichment analyses and
ceRNA network construction
GO term enrichment analysis (http://www.geneontology.org) covers three domains:
Biological process (BP), cellular component (CC) and molecular
function (MF). KEGG pathway enrichment analyses were conducted
using the Database for Annotation, Visualization and Integrated
Discovery 6.8 (25), and the P-value
(EASE-score, Fisher-P-value or Hypergeometric-P-value) denotes the
significance of the pathway when correlated with the conditions.
miRNAs that target circRNAs were predicted by surveying for 7-mer
or 8-mer complementarity to seed region and the 3′ pairing of each
miRNA using TargetScan. For humans and mice, two databases were
used to predict the target genes of miRNAs: TargetScan 7.1
(http://www.tatgetscan.org/vert_71/)
and mirdb V5 (http://mirdb.org/miRDB). The
overlapping results of the two databases for humans and mice were
accepted. The miRNA-target interactions were then experimentally
validated using the database miraTarbase 7.0 (http://mirtarbase.mbc.nctu.edu.tw/php/index.php).
Statistical analysis
Statistical analyses were performed using SPSS
version 12.0 (SPSS, Inc.), GraphPad Prism version 5.0 (GraphPad,
Inc.) and Cytoscape 2.8.1 (https://cytoscape.org/). Data of the relative
expression of circRNAs were analyzed using the 2−ΔΔCq
method (26) and presented as the
mean ± standard deviation. Paired or unpaired Student's t-tests
were used to compare continuous variables in tissue and plasma
samples. An unpaired Student's t-test was used for stratified
analyses according to the clinicopathological features of patients
with PHC. P<0.05 was considered to indicate a statistically
significant result.
Results
Identification of DE circRNAs in PHC
tissues using circRNA chip screening
The expression patterns of circRNAs were determined
using circRNA chip screening in three pairs of PHC tissues and
adjacent non-tumorous tissues. Following microarray scanning and
normalization, significant changes were observed. A total of 155
circRNAs were indicated to be DE in PHC tissues (80 upregulated and
75 downregulated), compared with the paired non-tumorous tissues
(Table III). A differential gene
expression hierarchical cluster heat map of circRNA expression
patterns clearly distinguished PHC tissues from paired adjacent
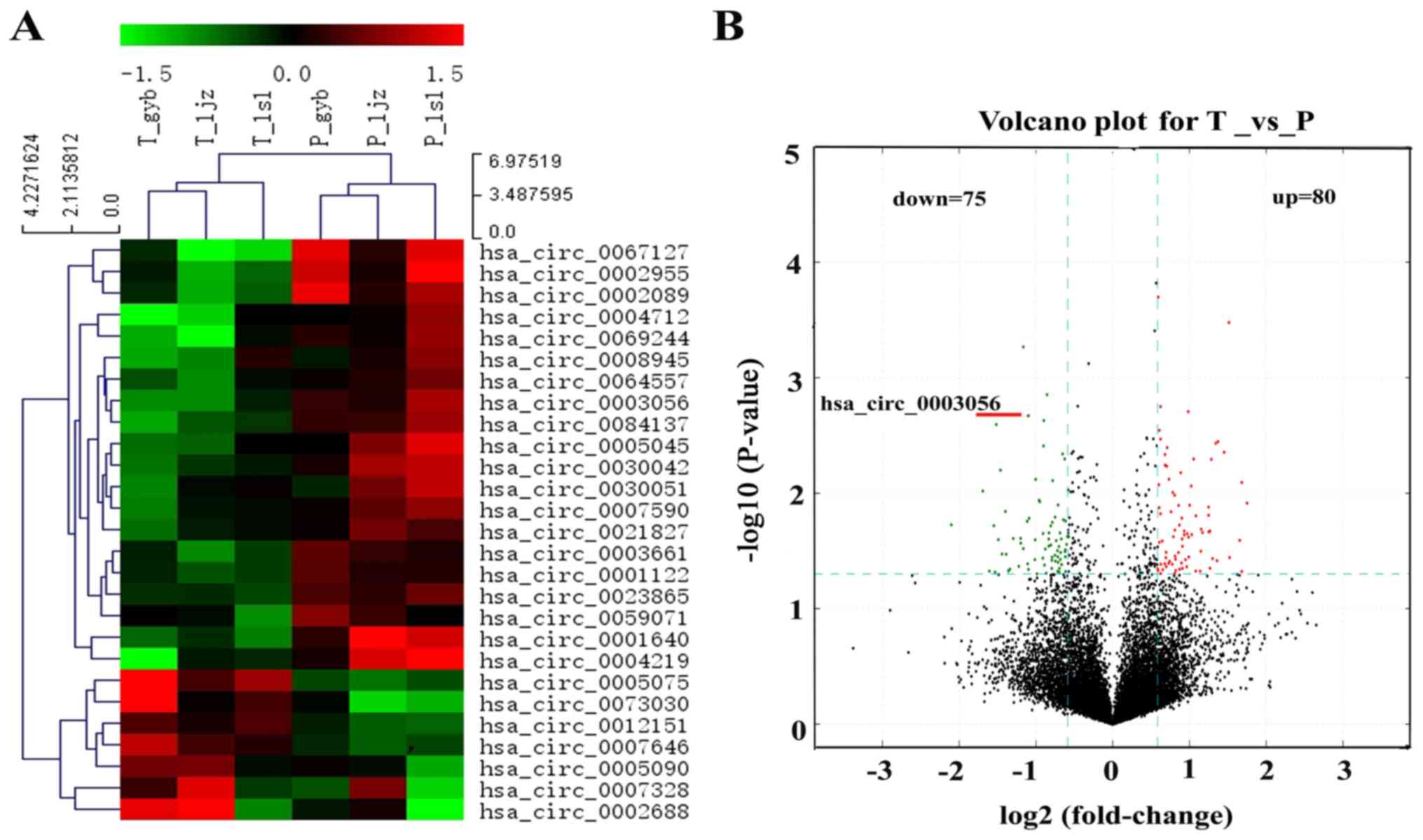
normal tissues (Fig. 1A). In the
volcano plot, green and red dots indicate the down- and
upregulation of circRNAs expression in PHC tissues, respectively
(Fig. 1B).
 | Table III.Differentially expressed
circRNAs. |
Table III.
Differentially expressed
circRNAs.
| CircRNA ID
(circBase) | Chromosome | Strand | circRNA type | Gene symbol | P-value | Fold change
(FC) | Regulation |
|---|
|
hsa_circ_0000745 | chr17 | + | Exonic | SPECC1 |
0.0212245 |
1.9054513 | Up |
| #N/A | chr1 | − | Exonic | PIK3C2B |
0.025524 |
3.1458593 | Up |
|
hsa_circ_0092125 | chrX | − | Exonic | G6PD |
0.0155316 |
2.3746559 | Up |
|
hsa_circ_0068293 | chr3 | + | Exonic | AP2M1 |
0.0373273 |
1.7639796 | Up |
|
hsa_circ_0007693 | chr1 | − | Exonic | ERI3 |
0.0404268 |
1.6569751 | Up |
|
hsa_circ_0025768 | chr12 | − | Exonic | TMTC1 |
0.0265629 |
1.5242278 | Up |
|
hsa_circ_0005075 | chr1 | − | Exonic | EIF4G3 |
0.0360363 |
2.8689281 | Up |
|
hsa_circ_0073030 | chr5 | − | Exonic | FAM169A |
0.0044122 |
2.731725 | Up |
|
hsa_circ_0008106 | chr3 | + | Exonic | LRCH3 |
0.0472836 |
2.2037281 | Up |
|
hsa_circ_0001231 | chr22 | − | Exonic | DMC1 |
0.0239959 |
1.9820607 | Up |
|
hsa_circ_0007646 | chr4 | + | Exonic | DCUN1D4 |
0.0433039 |
1.9509878 | Up |
|
hsa_circ_0008439 | chr3 | + | Exonic | LRCH3 |
0.0402462 |
1.8599736 | Up |
|
hsa_circ_0028711 | chr12 | − | Exonic | RAB35 |
0.0040302 |
1.6338613 | Up |
|
hsa_circ_0000760 | chr17 | + | Overlapping | MLLT6 |
0.0058239 |
1.6282967 | Up |
|
hsa_circ_0005873 | chr3 | + | Exonic | LRCH3 |
0.0368806 |
2.4621779 | Up |
|
hsa_circ_0002563 | chr1 | + | Exonic | KIF2C |
0.0035669 |
2.5852184 | Up |
|
hsa_circ_0076522 | chr6 | + | Exonic | ABCC10 |
0.0204543 |
2.2221132 | Up |
|
hsa_circ_0008719 | chr19 | − | Exonic | AKT2 |
0.0326238 |
1.8104931 | Up |
|
hsa_circ_0011480 | chr1 | − | Exonic | PHC2 |
0.0495597 |
1.5408582 | Up |
|
hsa_circ_0011477 | chr1 | − | Exonic | PHC2 |
0.0465285 |
1.5066423 | Up |
|
hsa_circ_0081626 | chr7 | + | Exonic | CUX1 |
0.0458484 |
1.5696195 | Up |
|
hsa_circ_0002994 | chr17 | − | Exonic | ACLY |
0.0228593 |
2.0403136 | Up |
|
hsa_circ_0043101 | chr17 | − | Exonic | NLE1 |
0.015046 |
1.539322 | Up |
|
hsa_circ_0000937 | chr19 | + | Exonic | BCKDHA |
0.024951 |
1.8707604 | Up |
|
hsa_circ_0008958 | chr1 | + | Overlapping | CIART |
0.0002 |
1.5087793 | Up |
|
hsa_circ_0003209 | chr20 | + | Exonic | TPX2 |
0.0411834 |
1.5429038 | Up |
|
hsa_circ_0043497 | chr17 | − | Exonic | MED24 |
0.0446081 |
2.4071688 | Up |
| #N/A | chr7 | + | Exonic | AP1S1 |
0.005098 |
2.4339844 | Up |
|
hsa_circ_0007328 | chr19 | + | Exonic | DOT1L |
0.0098403 |
1.7198141 | Up |
|
hsa_circ_0087080 | chr9 | − | Exonic | FBXO10 |
0.0361757 |
1.6176128 | Up |
|
hsa_circ_0028990 | chr12 | − | Exonic | KDM2B |
0.0075388 |
1.6618626 | Up |
|
hsa_circ_0001591 | chr6 | + | Overlapping | HIST1H4I |
0.0229166 |
1.9052747 | Up |
|
hsa_circ_0060456 | chr20 | + | Exonic | MYBL2 |
0.0131702 |
2.3768838 | Up |
|
hsa_circ_0017289 | chr1 | − | Exonic | SMYD3 |
0.0356072 |
1.9878667 | Up |
|
hsa_circ_0037409 | chr16 | + | Exonic | TSC2 |
0.0212241 |
2.4079542 | Up |
|
hsa_circ_0082614 | chr7 | − | Exonic | KIAA1549 |
0.0017741 |
1.5392865 | Up |
|
hsa_circ_0004656 | chr16 | + | Exonic | SLC7A6 |
0.0019672 |
1.977973 | Up |
|
hsa_circ_0071653 | chr5 | + | Exonic | CEP72 |
0.0224188 |
1.9589469 | Up |
|
hsa_circ_0003048 | chr16 | − | Exonic | VAC14 |
0.0495136 |
1.7512677 | Up |
|
hsa_circ_0006916 | chr5 | − | Exonic | HOMER1 |
0.0127235 |
2.0032953 | Up |
|
hsa_circ_0050898 | chr19 | + | Exonic | ACTN4 |
0.0383419 |
2.5628111 | Up |
|
hsa_circ_0069370 | chr4 | − | Exonic | SEL1L3 |
0.0033967 |
1.5398487 | Up |
| #N/A | chr19 | + | Intronic | DHX34 |
0.0467045 |
1.766391 | Up |
|
hsa_circ_0089090 | chr9 | + | Exonic | NCS1 |
0.0028527 |
1.522742 | Up |
|
hsa_circ_0092297 | chr2 | − | Intronic | SMPD4 |
0.0208144 |
1.7537756 | Up |
|
hsa_circ_0051718 | chr19 | − | Exonic | LIG1 |
0.006625 |
1.8456723 | Up |
|
hsa_circ_0011279 | chr1 | + | Exonic | SERINC2 |
0.0451375 |
1.5002793 | Up |
|
hsa_circ_0088072 | chr9 | − | Exonic | PTBP3 |
0.0046249 |
1.603221 | Up |
|
hsa_circ_0017287 | chr1 | − | Exonic | SMYD3 |
0.0350386 |
1.7546953 | Up |
|
hsa_circ_0005090 | chr1 | − | Exonic | SMYD3 |
0.0144482 |
1.6936278 | Up |
|
hsa_circ_0002688 | chr4 | + | Exonic | WHSC1 |
0.0121876 |
3.363273 | Up |
| #N/A | chr5 | − | Exonic | LPCAT1 |
0.0447121 |
1.8767479 | Up |
|
hsa_circ_0006177 | chr2 | + | Intronic | AGAP1 |
0.0084361 |
1.5176364 | Up |
|
hsa_circ_0002245 | chr6 | + | Exonic | CAP2 |
0.0433245 |
1.6219694 | Up |
|
hsa_circ_0014754 | chr1 | − | Exonic | IQGAP3 |
0.0036918 |
2.5346514 | Up |
|
hsa_circ_0092324 | chr17 | − | Intronic | DNAJC7 |
0.0216354 |
2.380195 | Up |
| #N/A | chr5 | − | Exonic | CDC25C |
0.0454826 |
1.8293722 | Up |
|
hsa_circ_0041008 | chr16 | − | Exonic | FANCA |
0.0127351 |
1.8125889 | Up |
|
hsa_circ_0055855 | chr2 | − | Exonic | AFF3 |
0.0003327 |
2.8560336 | Up |
|
hsa_circ_0059760 | chr20 | + | Exonic | TM9SF4 |
0.0219282 |
2.247014 | Up |
| #N/A | chr21 | − | Exonic | HSF2BP |
0.0387871 |
1.6720688 | Up |
|
hsa_circ_0043947 | chr17 | − | Exonic | BRCA1 |
0.0408289 |
1.5983177 | Up |
| #N/A | chr4 | + | Exonic | TACC3 |
0.0080579 |
3.2048942 | Up |
|
hsa_circ_0001728 | chr7 | − | Intronic | MCM7 |
0.0318177 |
2.2162243 | Up |
|
hsa_circ_0092370 | chr1 | − | Intronic | DNAJC11 |
0.0188763 |
1.8250598 | Up |
| #N/A | chr19 | − | Exonic | CADM4 |
0.0287784 |
1.8768288 | Up |
| #N/A | chr4 | − | Exonic | SEL1L3 |
0.0412118 |
1.711422 | Up |
|
hsa_circ_0082688 | chr7 | − | Exonic | PARP12 |
0.0259324 |
1.5632151 | Up |
| #N/A | chr12 | + | Exonic | P2RX4 |
0.0057199 |
1.6028692 | Up |
|
hsa_circ_0006893 | chr3 | − | Exonic | PHC3 |
0.0133705 |
1.5220367 | Up |
|
hsa_circ_0082304 | chr7 | + | Exonic | NRF1 |
0.0164259 |
1.8654058 | Up |
|
hsa_circ_0001196 | chr21 | − | Exonic | WDR4 |
0.0471616 |
2.1165344 | Up |
|
hsa_circ_0068894 | chr4 | + | Exonic | WHSC1 |
0.0476107 |
3.2029375 | Up |
|
hsa_circ_0087047 | chr9 | + | Exonic | ZCCHC7 |
0.0103933 |
1.7475255 | Up |
|
hsa_circ_0002071 | chr12 | − | Exonic | GOLGA3 |
0.0050376 |
2.0771037 | Up |
|
hsa_circ_0012166 | chr1 | + | Exonic | KIF2C |
0.0324916 |
1.6186238 | Up |
|
hsa_circ_0008777 | chr20 | − | Exonic | AURKA |
0.0376056 |
1.9286644 | Up |
|
hsa_circ_0005219 | chr20 | + | Exonic | STK35 |
0.0086497 |
2.0307961 | Up |
|
hsa_circ_0012151 | chr1 | − | Exonic | ERI3 |
0.0237599 |
1.7283688 | Up |
|
hsa_circ_0037526 | chr16 | + | Exonic | CCNF |
0.0156714 |
2.1195943 | Up |
|
hsa_circ_0002198 | chr6 | + | Exonic | PDE7B |
0.0301888 |
2.2417711 | Down |
|
hsa_circ_0004712 | chr6 | + | Exonic | PDE7B |
0.0455082 |
2.5301703 | Down |
|
hsa_circ_0039783 | chr16 | + | Exonic | CBFB |
0.0005419 |
2.2404601 | Down |
| #N/A | chr3 | − | Intronic | MAGI1 |
0.0456285 |
2.1403102 | Down |
|
hsa_circ_0008945 | chr14 | − | Exonic | NIN |
0.017864 |
1.7247068 | Down |
| #N/A | chr10 | − | Exonic | ANK3 |
0.0335636 |
2.7176467 | Down |
|
hsa_circ_0003661 | chr11 | + | Exonic | ANKRD42 |
0.0446637 |
1.7336621 | Down |
|
hsa_circ_0030051 | chr13 | − | Exonic | ELF1 |
0.03574 |
1.731437 | Down |
|
hsa_circ_0000720 | chr16 | + | Exonic | PLCG2 |
0.0304322 |
1.8390616 | Down |
|
hsa_circ_0089372 | chr9 | + | Exonic | ADAMTS13 |
0.0252263 |
2.8096697 | Down |
|
hsa_circ_0002089 | chr11 | + | Exonic | ARHGEF12 |
0.0063054 |
2.7586096 | Down |
|
hsa_circ_0078223 | chr6 | − | Exonic | LATS1 |
0.0447052 |
1.6186692 | Down |
| #N/A | chr5 | − | Exonic | MYO10 |
0.0190623 |
2.9255358 | Down |
|
hsa_circ_0059071 | chr2 | + | Exonic | FARP2 |
0.0414464 |
1.5860201 | Down |
|
hsa_circ_0005801 | chr10 | − | Exonic | TM9SF3 |
0.0396484 |
1.6371578 | Down |
|
hsa_circ_0003357 | chr10 | + | Exonic | ADD3 |
0.0499515 |
1.9810488 | Down |
|
hsa_circ_0069323 | chr4 | − | Exonic | GPR125 |
0.0114958 |
1.9450245 | Down |
|
hsa_circ_0004219 | chr8 | + | Exonic | DOCK5 |
0.009565 |
3.2333123 | Down |
|
hsa_circ_0002955 | chr11 | + | Exonic | ARHGEF12 |
0.0472601 |
3.0446992 | Down |
|
hsa_circ_0017248 | chr1 | − | Exonic | AKT3 |
0.0481372 |
1.6202917 | Down |
|
hsa_circ_0042339 | chr17 | − | Exonic | SHMT1 |
0.0289513 |
1.6262584 | Down |
| #N/A | chr2 | − | Exonic | SMC6 |
0.0354823 |
1.6429599 | Down |
|
hsa_circ_0003056 | chr22 | − | Exonic | PITPNB |
0.0021306 |
2.1416189 | Down |
|
hsa_circ_0001041 | chr2 | − | Exonic | EIF2AK3 |
0.0281227 |
1.7488955 | Down |
|
hsa_circ_0000550 | chr14 | + | Antisense | SLC10A1 |
0.0143731 |
2.6344379 | Down |
|
hsa_circ_0003640 | chr4 | + | Exonic | METAP1 |
0.002348 |
1.8630256 | Down |
|
hsa_circ_0078328 | chr6 | − | Exonic | SYNE1 |
0.0220923 |
1.7778472 | Down |
|
hsa_circ_0036610 | chr15 | + | Exonic | PDE8A |
0.0491465 |
1.8214826 | Down |
|
hsa_circ_0005045 | chr2 | − | Exonic | RTN4 |
0.0407689 |
2.1566167 | Down |
|
hsa_circ_0006663 | chr16 | − | Exonic | LUC7L |
0.0277407 |
1.5172731 | Down |
|
hsa_circ_0073649 | chr5 | − | Exonic | DTWD2 |
0.0126395 |
1.6366199 | Down |
| #N/A | chr13 | + | Exonic | EEF1DP3 |
0.0365432 |
1.8802713 | Down |
|
hsa_circ_0067127 | chr3 | − | Exonic | ALDH1L1 |
0.0187756 |
4.2915721 | Down |
|
hsa_circ_0038409 | chr16 | − | Exonic | LOC100271836 |
0.0381125 |
1.731821 | Down |
|
hsa_circ_0071311 | chr4 | + | Exonic | KIAA0922 |
0.007598 |
2.0057656 | Down |
|
hsa_circ_0002771 | chr16 | − | Exonic | PARN |
0.0286315 |
1.7820343 | Down |
|
hsa_circ_0005471 | chr17 | − | Exonic | NCOR1 |
0.0039017 |
1.8690309 | Down |
|
hsa_circ_0069244 | chr4 | − | Exonic | LDB2 |
0.0246507 |
2.4543633 | Down |
|
hsa_circ_0015164 | chr1 | − | Exonic | SLC19A2 |
0.0118506 |
1.9309544 | Down |
|
hsa_circ_0006539 | chr8 | + | Exonic | RBPMS |
0.0471379 |
2.564619 | Down |
| #N/A | chr4 | − | Intronic | GPR125 |
0.0314876 |
1.6460992 | Down |
|
hsa_circ_0053070 | chr2 | + | Exonic | HADHB |
0.0223274 |
1.8756974 | Down |
|
hsa_circ_0023730 | chr11 | − | Exonic | INTS4 |
0.0474224 |
1.644243 | Down |
|
hsa_circ_0038644 | chr16 | + | Exonic | PRKCB |
0.0050378 |
1.5424169 | Down |
|
hsa_circ_0064557 | chr3 | − | Exonic | SATB1 |
0.0344432 |
1.6859964 | Down |
| #N/A | chr1 | − | Exonic | PDE4DIP |
0.0220458 |
1.6090404 | Down |
|
hsa_circ_0069681 | chr4 | − | Exonic | FRYL |
0.0374764 |
1.6037813 | Down |
|
hsa_circ_0006891 | chr11 | + | Exonic | PPFIBP2 |
0.0257878 |
1.6666432 | Down |
|
hsa_circ_0021827 | chr11 | − | Exonic | PHF21A |
0.0249203 |
1.589884 | Down |
|
hsa_circ_0001122 | chr2 | + | Exonic | FARP2 |
0.0045619 |
1.5731657 | Down |
|
hsa_circ_0013607 | chr1 | − | Exonic | RSBN1 |
0.0376341 |
2.0202019 | Down |
|
hsa_circ_0005967 | chr10 | − | Exonic | KCNMA1 |
0.0160756 |
1.6827265 | Down |
|
hsa_circ_0000446 | chr12 | − | Exonic | TAOK3 |
0.0325774 |
1.6013056 | Down |
|
hsa_circ_0070857 | chr4 | + | Exonic | KIAA1109 |
0.0418502 |
1.8511599 | Down |
|
hsa_circ_0007590 | chr13 | − | Exonic | LATS2 |
0.007774 |
1.7207037 | Down |
|
hsa_circ_0006629 | chr11 | − | Exonic | PICALM |
0.0191864 |
1.750488 | Down |
|
hsa_circ_0001640 | chr6 | − | Exonic | EPB41L2 |
0.0482272 |
2.8931722 | Down |
|
hsa_circ_0066378 | chr3 | + | Exonic | ABHD6 |
0.0429032 |
2.3329893 | Down |
|
hsa_circ_0025711 | chr12 | − | Exonic | TM7SF3 |
0.0253257 |
2.0133054 | Down |
|
hsa_circ_0023865 | chr11 | − | Exonic | CCDC90B |
0.0386574 |
1.6774031 | Down |
|
hsa_circ_0000077 | chr1 | − | Overlapping | TM2D1 |
0.0236483 |
1.5061897 | Down |
|
hsa_circ_0005527 | chr11 | − | Exonic | FCHSD2 |
0.02705 |
2.2952095 | Down |
|
hsa_circ_0030042 | chr13 | − | Exonic | FOXO1 |
0.0174801 |
2.1639648 | Down |
|
hsa_circ_0007201 | chr15 | + | Exonic | IQGAP1 |
0.0248831 |
1.7816364 | Down |
| #N/A | chr11 | + | Exonic | DLAT |
0.0309512 |
1.5343122 | Down |
| #N/A | chr4 | − | Exonic | NR3C2 |
0.0476049 |
1.5305334 | Down |
|
hsa_circ_0084137 | chr8 | − | Exonic | RNF170 |
0.0164973 |
2.1286009 | Down |
|
hsa_circ_0013162 | chr1 | − | Exonic | EVI5 |
0.0337218 |
2.6161063 | Down |
|
hsa_circ_0037969 | chr16 | − | Exonic | PARN |
0.0354487 |
1.5344047 | Down |
|
hsa_circ_0077765 | chr6 | + | Exonic | RNF217 |
0.0169759 |
1.5605383 | Down |
| #N/A | chr11 | + | Exonic | TEAD1 |
0.0014024 |
1.81072 | Down |
|
hsa_circ_0035376 | chr15 | + | Exonic | PIGB |
0.0174 |
1.5311146 | Down |
|
hsa_circ_0000389 | chr12 | + | Over-lapping | FGD4 |
0.0319098 |
1.5397315 | Down |
|
hsa_circ_0030281 | chr13 | − | Exonic | DLEU2 |
0.0245441 |
2.2986061 | Down |
| #N/A | chr16 | + | Intronic | MT2A |
0.0025438 |
2.8617763 | Down |
Confirmation of DE circRNAs in PHC
tissues using RT-qPCR
To confirm the differential expression of circRNAs
(indicated by circRNA microarray screening), RT-qPCR was performed
to assess the expression levels of five candidate DE circRNAs in
PHC tissues, compared with adjacent normal tissues, derived from an
additional 11 patients with PHC. The five circRNAs (two upregulated
and three downregulated) were selected according to the following
criteria: i) Fold-change >1.5; ii) P<0.05; iii)
exonic-related circRNAs; iv) raw intensity ≥100; v) included in
circBase; and vi) exhibiting a gene symbol or miRNA binding sites
that were considered to be significantly associated with tumor
occurrence. Only circRNAs that exhibited a ≥1.5-fold expression
level difference were considered to display statistically
significant changes (P<0.05). Moreover, complete and accurate
sequence information, and other information regarding the selected
exonic-related circRNAs, were retrieved from circBase. A group raw
intensity of ≥100 ensured the reliability of the detected
fluorescence values of circRNAs determined using the microassay.
Finally, circRNAs may influence PHC development and progression
through circRNA-miRNA-mRNA networks. Thus, only circRNAs whose
target genes or miRNA binding sites had a predetermined association
with cancer progression were preferentially selected.
As exhibited in Fig.
2A, only hsa_circ_0003056 expression was consistently and
significantly lower in PHC tissues when compared with paired
adjacent non-tumorous tissues. The other three circRNAs exhibited a
fold-change in expression level between 1.5 and 2, which was not
significantly different from the adjacent tissues.
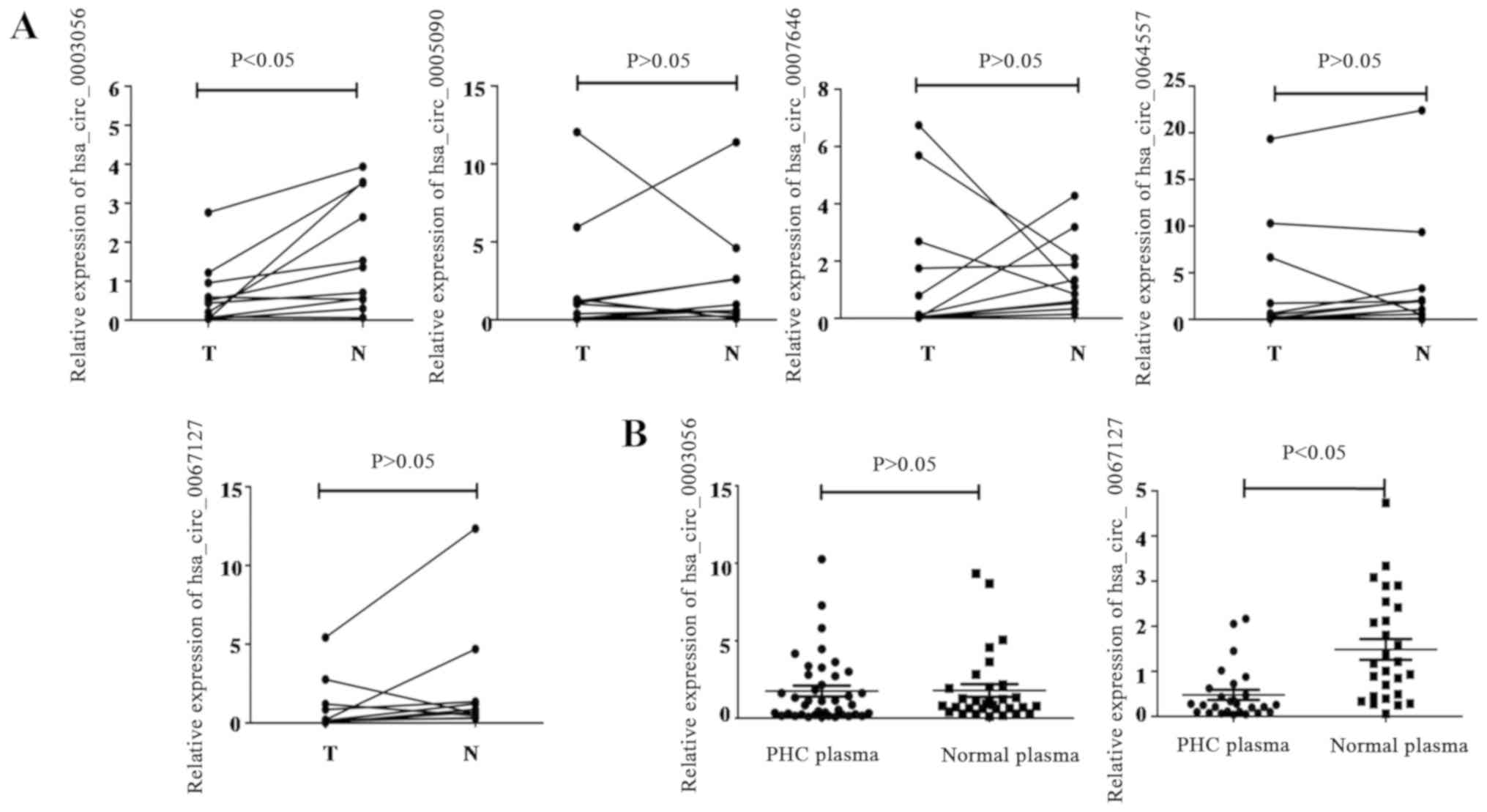 | Figure 2.Confirmation of DE circRNAs in PHC
tissues and plasma by RT-qPCR. (A) Expression levels of
hsa_circ_0003056, hsa_circ_0005090, hsa_circ_0007646,
hsa_circ_0064557 and hsa_circ_0067127 were assessed in 11 PHC
patients, with comparisons between PHC tissues and adjacent
non-tumor tissues conducted using RT-qPCR; (B) Scatter plots
display the relative expression of hsa_circ_0003056 and
hsa_circ_0067127 in 35 PHC and 32 healthy plasma samples. DE,
differentially expressed; circRNA, circular RNA; PHC, primary
hepatic carcinoma; RT-q, reverse transcription-quantitative; T,
tumor tissue; N, non-tumorous tissues. |
Expression levels and association with
clinicopathological parameters of hsa_circ_0003056 and
hsa_circ_0067127 in PHC patient plasma
Hsa_circ_0003056 expression levels were decreased in
PHC tissues compared with paired, adjacent non-tumor tissues.
Hsa_circ_0067127 also exhibited decreased expression levels in PHC
tissues, but the difference was not significant compared with the
adjacent non-tumorous tissues, which may be due to the small sample
size. To determine whether changes in expression level were also
reflected in patients' plasma, fresh plasma samples were collected
from an additional 35 patients with PHC, and 32 healthy donors.
Subsequently, the expression levels of hsa_circ_0003056 and
hsa_circ_0067127 were detected using RT-qPCR. As indicated in
Fig. 2B, no significant
downregulation or upregulation of hsa_circ_0003056 was identified
in the patients' plasma, when compared with healthy donor plasma.
However, hsa_circ_0067127 was significantly downregulated in the
PHC plasma. Stratified analyses of hsa_circ_0003056 and
hsa_circ_0067127 levels in the plasma samples of patients with PHC
was performed, and is summarized in Table I. Significant downregulation of
hsa_circ_0003056 was observed in patients who were >60 years old
compared those <60 years old. hsa_circ_0067127 levels were also
significantly higher in patients with an AFP level <200 ng/ml.
No association was demonstrated between other clinical and
laboratory characteristics (including sex and HBsAg level) and
hsa_circ_0003056.
Follow-up was carried out for the 35 patients with
PHC and the median overall survival (OS) time was 69 months.
Survival data were analyzed using the log-rank test and survival
curves were generated using the Kaplan-Meier method. Regarding
hsa_circ_0003056 expression, the corresponding 35 patients with PHC
were separated into an hsa_circ_0003056 low-expression group
(<mean value in healthy donors' plasma; n=15; OS=69) and
hsa_circ_0003056 high-expression group (>mean value in healthy
donors' plasma; n=20; OS=72). The difference between OS times
observed between groups was not statistically significant (data not
shown). In terms of hsa_circ_0067127 expression, only 3 out of 35
patients with PHC displayed higher expression in their plasma than
the mean value in the healthy donors. As a result, the survival
curves could not be analyzed.
Construction of ceRNA networks of
hsa_circ_0003056 and hsa_circ_0067127, and KEGG/GO enrichment
analyses
The target miRNAs of hsa_circ_0003056 and
hsa_circ_0067127 were identified and ranked based on their mirSVR
scores and TargetScan. This indicated that hsa_circ_0003056 and
hsa_circ_0067127 serve as miRNA sponges, regulating the ceRNA
network. The five top miRNAs were selected to establish
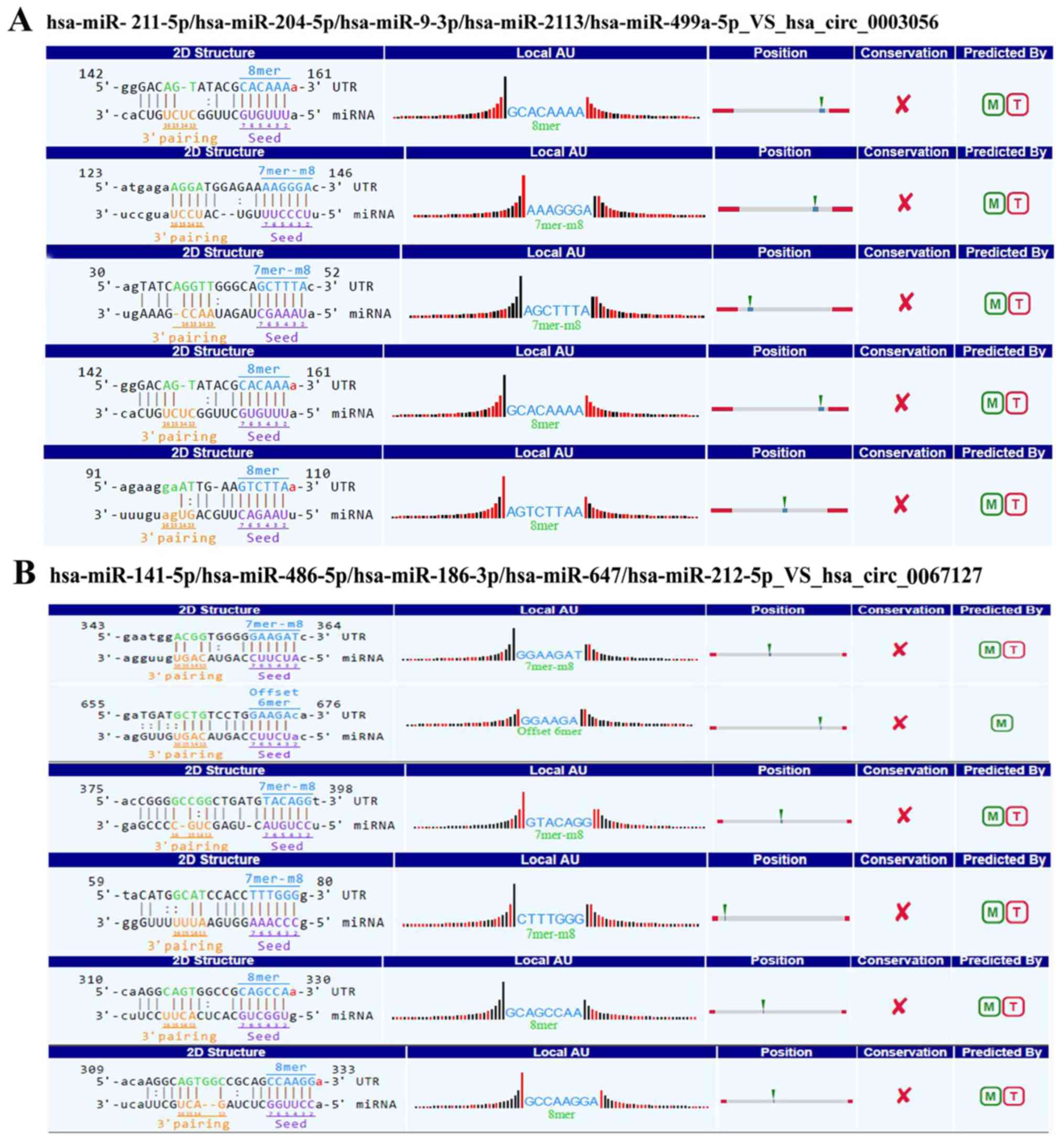
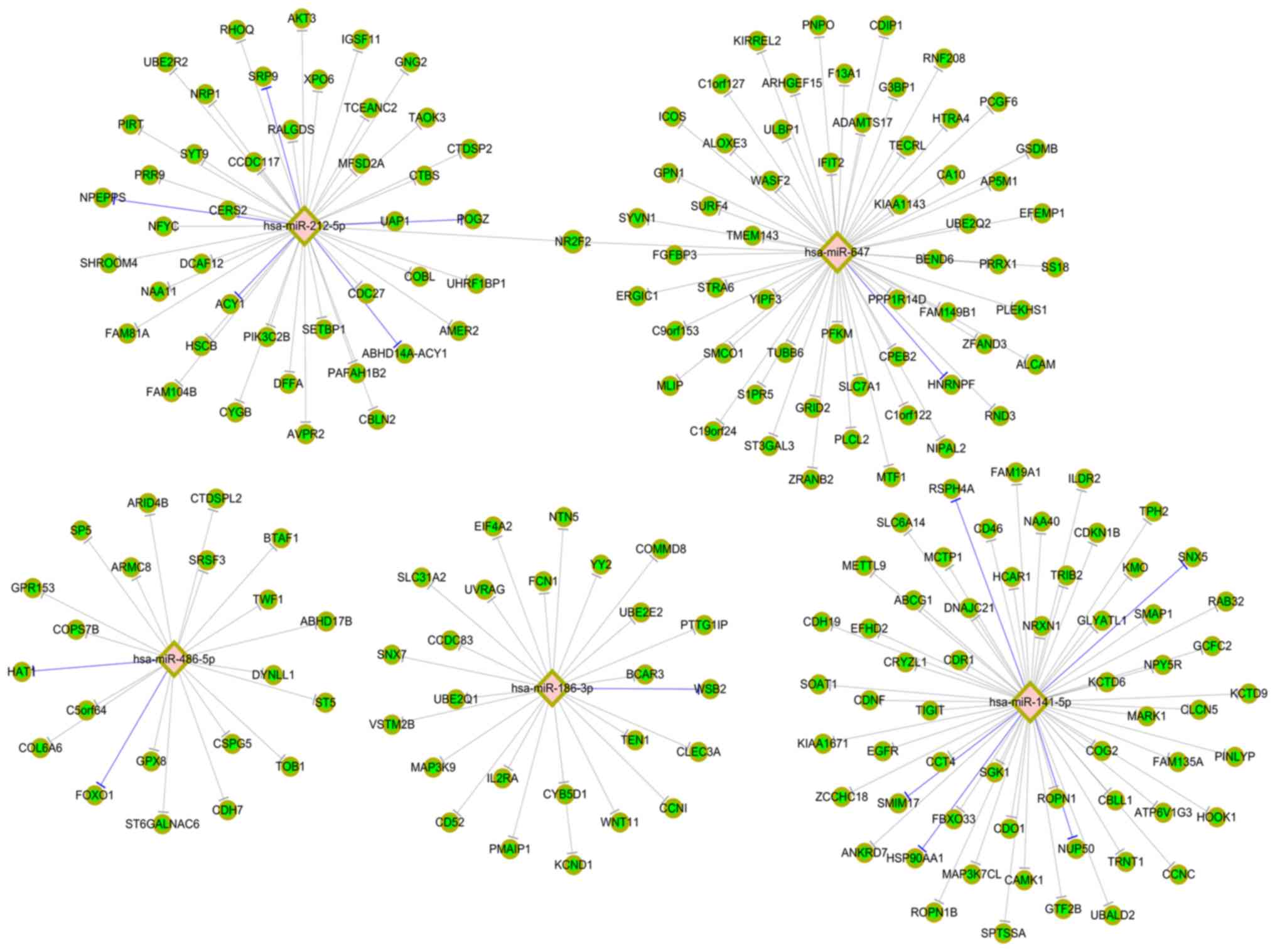
circRNA-miRNA-mRNA networks: hsa-miR-211-5p, hsa-miR-204-5p,
hsa-miR-9-3p, hsa-miR-2113 and hsa-miR-499a-5p targeting to
hsa_circ_0003056 (Fig. 3A), and
hsa-miR-141-5p, hsa-miR-486-5p, hsa-miR-186-3p, hsa-miR-647 and
hsa-miR-212-5p targeting to hsa_circ_0067127 (Fig. 3B). Regarding hsa_circ_0003056, a Venn
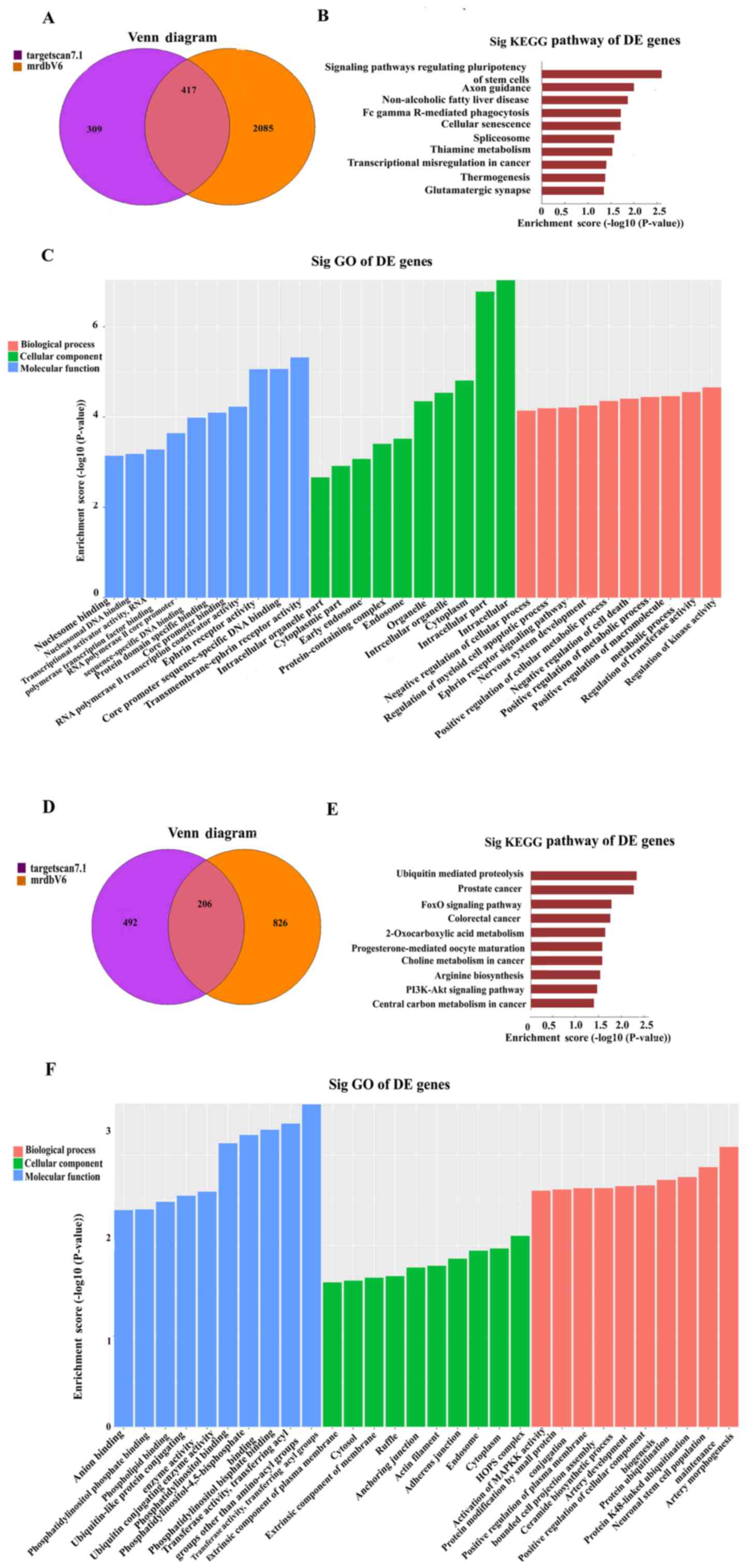
diagram (Fig. 4A) indicated that
overlapping results common to the two databases were accepted.
TargetScan 7.1 and mirdbV5 are used to forecast the target genes of
miRNAs. The bar plot presents the enrichment scores
[-log10 (P-value)] of the top 10 significant enrichment
pathways. KEGG pathway analysis demonstrated that DE genes were
significantly enriched in pathways regulating the pluripotency of
stem cells (Fig. 4B). The bar plot
presents the enrichment score [-log10 (P-value)] values of the top
10 significant GO enrichments. The hsa_circ_0003056-miRNAs-targets
network exhibited a strong association with ‘regulation of kinase
activity’, ‘intracellular-’ and ‘transmembrane-ephrin receptor’
activity in BP, CC and MF, respectively (Fig. 4C). The overlapping results between
the two databases were also accepted for hsa_circ_0067127 (Fig. 4D). DE genes were significantly
enriched in the pathways associated with ‘ubiquitin-mediated
proteolysis’ and ‘prostate cancer’ (Fig.
4E). The top 10 significant GO enrichments are presented in
Fig. 4F. The
hsa_circ_0067127-miRNAs-targets network exhibited a strong
association with ‘artery morphogenesis activity’, ‘HOPS complex’
and ‘transferase activity, transferring acyl groups’ in BP, CC and
MF, respectively.
Cytoscape analysis of
hsa_circ_0003056- and hsa_circ_0067127-miRNA-target gene
interaction networks
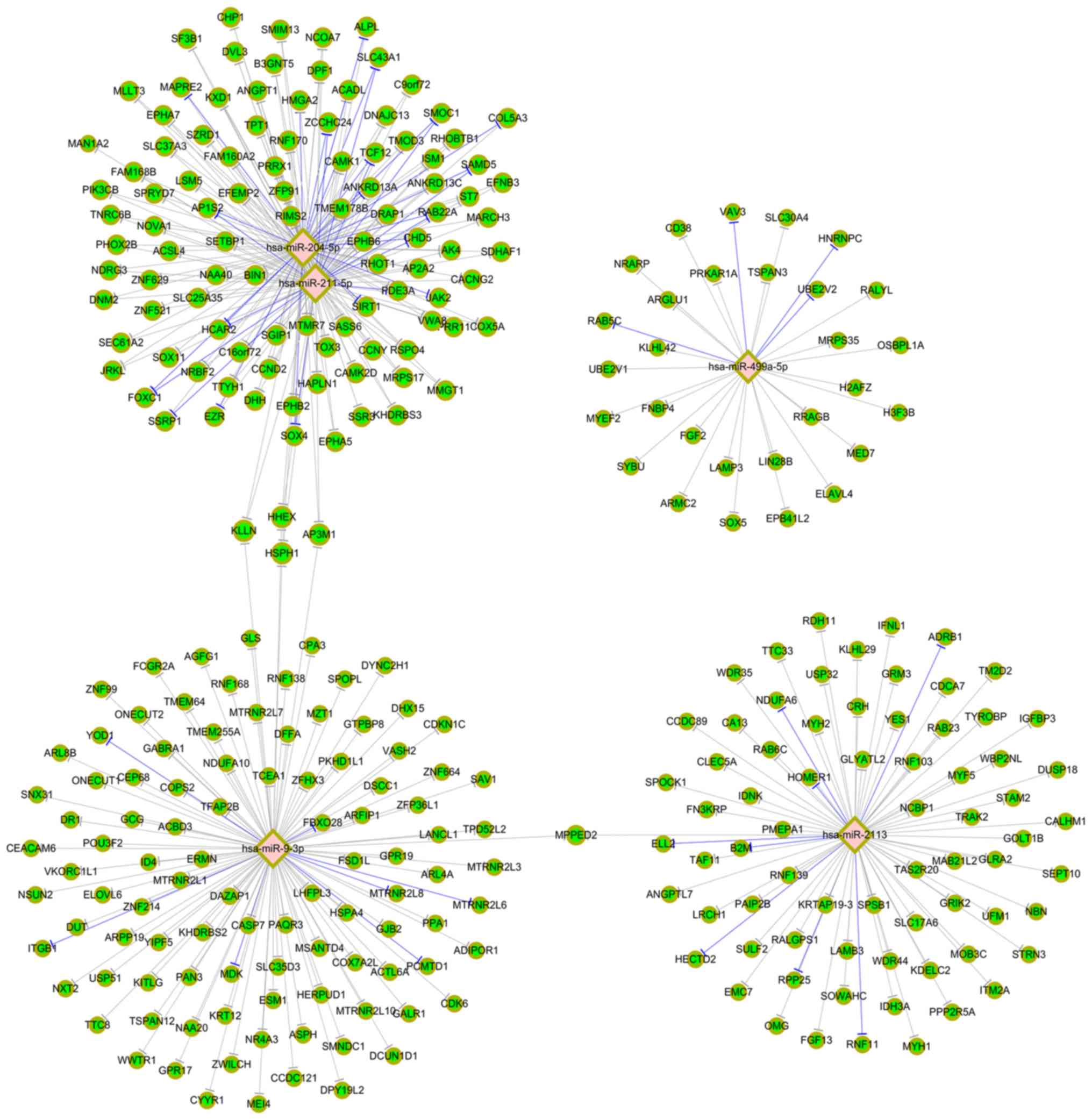
Based on hsa_circ_0003056 and hsa_circ_0067127, the
networks of five miRNAs with their target mRNAs were constructed.
In these networks, green circular nodes represent mRNAs, pink
diamond nodes represent miRNAs, gray lines represent those not
experimentally validated, and blue lines represent those
experimentally validated (miRTarBase 7.0.). Fig. 5 indicates the top five miRNAs
targeting to hsa_circ_0003056 and their target mRNAs. The top three
mRNAs corresponding to both hsa-miR-211-5p and hsa-miR-204-5p were
adaptor related protein complex 1 subunit sigma 2, solute carrier
family 37 member 3 and RAB22A, member RAS oncogene family.
Similarly, Fig. 6 presents the top
five miRNAs targeting to hsa_circ_0067127 and their target
mRNAs.
Discussion
It has been determined that circRNAs may influence
the initiation and development of a range of different cancer
types. Certain circRNAs are enriched in exosomes (16), suggesting that they exhibit the
potential to function as tumor biomarkers, and may therefore be
used to support diagnosis. However, few reports have investigated
this with regards to PHC tissues and plasma. The current study
provided a comprehensive circRNA profile in three pairs of PHC
tissues and adjacent tissues prior to treatment, using circRNA chip
screening. A total of five circRNAs were selected for validation of
their respective expression levels in both PHC tissues, and
adjacent tissues from 11 patients, using RT-qPCR analysis. However,
only hsa_circ_0003056 was confirmed to be significantly
downregulated in PHC tissues. According to the initial analyses of
circRNA chip screening, the gene coding for hsa_circ_0003056 is
located on chromosome 22, and the sequence of its best transcript
is NM_012399, with a gene symbol of phosphatidylinositol (Ptdins)
transfer protein beta (PITPNB). The relatively linear PITPNB
stimulates Ptdins-4-phosphate synthesis and signaling in eukaryotic
cells (27). However, whether PITPNB
influences tumor progression is yet to be determined. Therefore,
the current study analyzed the associations between circPITPNB and
certain clinicopathological parameters. However, the results
revealed no significant association, which may be due to the small
sample size of 11 cases. Further functional experiments are
required to determine the mechanism behind the downregulation of
hsa_circ_0003056 expression in PHC tissues.
Using RT-qPCR, hsa_circ_0003056 expression levels
were determined in fresh plasma samples retrieved from 35 PHC
patients and 32 healthy donors. The expression levels of
hsa_circ_0003056 did not significantly decrease in the plasma taken
from patients with PHC. This may be due to the fact that most of
the plasma samples were collected from patients undergoing
interventional therapy, and only a small number were subjected to
specimen collection prior to treatment. It has been suggested that
interventional therapies may influence the expression levels of
hsa_circ_0003056 in the plasma. circRNAs are relatively stable in
plasma, however, the total free nucleic acid content of the
peripheral blood is low (28). There
are no stable, internal or precise quantification methods for the
determination of these levels. β-actin or GAPDH can be used in
plasma or serum (29), but they are
not ideal internal references due to their instability and high
individual variation. Although circRNAs exhibit a degree of tissue
specificity, they are expressed in plasma and can be detected.
These circRNAs may represent novel potential serum biomarkers. For
example, circRNA-284 and miR-221 both exhibit the potential to
become diagnostic biomarkers of carotid plaque ruptures and strokes
(30). In the present study, the
expression level of hsa_circ_0067127 was lower in PHC plasma but
not in PHC tissue (compared to normal plasma and para-tumor
tissues, respectively), and was also associated with AFP level, as
determined by stratified analysis. To further validate the
expression levels of hsa_circ_0003056 and hsa_circ_0067127 in PHC
plasma, the plasmid vector method should be used in future studies
as a standard to quantify the two circRNAs, with an increased
sample size for improved reliability.
Mechanisms describing the roles of circRNAs in
cancer progression have not been clearly elucidated. The primary
function of circRNAs is to act as miRNA sponges by forming
circRNA-miRNA-mRNA axes (22). The
circ_0067934/miR-1324/FZD5/b-catenin signaling complexes may serve
as a promising therapeutic target for HCC intervention (31). Other functions of circRNAs, such as
their interaction with RNA-binding proteins and translating
proteins, have previously been described (32). In the present study, the top five
target miRNAs that were indicated to interact with
hsa_circ_0003056, were selected based on their mirSVR scores and
TargetScan. These were hsa-miR-211-5p, hsa-miR-204-5p,
hsa-miR-9-3p, hsa-miR-2113 and hsa-miR-499a-5p. Hsa-miR-211-5p
levels in HCC tissues were lower than those in normal tissues,
suggesting an inhibitory role in HCC by inhibiting zinc finger
E-box-binding protein (ZEB2) expression (33). Hsa-miR-211-5p is also associated with
renal cell carcinoma, thyroid tumors and triple-negative breast
cancer (34–36). Hsa-miR-204-5p is associated with a
variety of malignancies, including HCC and laryngeal squamous cell
carcinoma (37,38). Whether hsa-miR-204-5p is associated
with PHC progression, through its interaction with
hsa_circ_0003056, remains undetermined. A practical method is
urgently required to identify specific interactions between miRNAs
and circRNAs, currently identified using complicated bioinformatics
prediction networks. KEGG pathway analyses revealed that DE genes
were significantly enriched in pathways that regulates the
pluripotency of stem cells. GO analyses revealed that
hsa_circ_0003056 was strongly associated with ‘regulation of kinase
activity’, and ‘intracellular and transmembrane-ephrin receptor
activity’ in BP, CC and MF, respectively, which is consistent with
its linear form (PITPNB), a gene encoding a cytoplasmic protein
that catalyzes the transfer of phosphatidylinositol and
phosphatidylcholine between membranes.
In the future, functional experiments should be
performed to further explore the mechanism of circRNA in the
regulation of PHC progression. Hepatic carcinoma cell lines with
high or low expression levels of hsa_circ_0003056 should be
constructed, and used to conduct proliferation and migration
experiments. The binding and co-localization of hsa_circ_0003056
with the top five target miRNAs should be confirmed, and the target
genes (mRNAs) corresponding to the miRNAs (as shown using GO
enrichment analyses) should be quantified.
In summary, hsa_circ_0003056 expression is
significantly decreased in PHC tissues, but is relatively stable in
plasma. Conversely, hsa_circ_0067127 is downregulated in PHC plasma
but not in PHC tissue. The detailed molecular mechanisms by which
hsa_circ_0003056 and hsa_circ_0067127 function as miRNA sponges to
regulate PHC occurrence and development require further
investigation.
Acknowledgements
Not applicable.
Funding
The present study was supported by grants from the
Natural Science Fund of Education Department of Anhui Province
(grant no. KJ2016A380 and KJ2018A0797), the Science and Technology
Project of Clinical Medicine Foundation of Jiangsu Province (grant
no. BL2014005), the Key Development Project for Medical Science and
Technology of Jiangsu Province (grant no. ZKX12038) and the Key
Project of Nanjing Health Bureau (grant no. 201208038). The
sponsors had no role in the study design, data collection and
analysis, preparation of the manuscript or the decision to submit
the article for publication.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
YY and CS designed and supervised the research. YD,
AF and JY performed the experiments and data analyses. JD and NW
collected the specimens. YD and CS wrote the manuscript. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of the Second Hospital of Nanjing (2017-LY-Kt007).
Written informed consent was obtained from each patient and healthy
donor enrolled. All procedures performed in the study involving
human participants were in accordance with the ethical standards of
the Institutional and/or National Research Committee and with the
1964 Helsinki declaration and its later amendments or comparable
ethical standards.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
circRNAs
|
circular RNAs
|
|
PHC
|
Primary hepatic carcinoma
|
|
HCC
|
Hepatocellular carcinoma
|
|
RT-qPCR
|
reverse transcription-quantitative
PCR
|
|
miRNAs
|
microRNAs
|
|
GO
|
Gene ontology
|
|
KEGG
|
Kyoto Encyclopedia of Genes and
Genomes
|
|
ncRNA
|
non-coding RNA
|
|
DE
|
differential expression
|
References
|
1
|
Ma L, Wang B, Long Y and Li H: Effect of
traditional Chinese medicine combined with Western therapy on
primary hepatic carcinoma: A systematic review with meta-analysis.
Front Med. 11:191–202. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wu X, Li J, Wang C, Zhang G, Zheng N and
Wang X: Application of different imaging methods in the early
diagnosis of primary hepatic carcinoma. Gastroenterol Res Pract.
2016:87632052016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Berretta M, Cavaliere C, Alessandrini L,
Stanzione B, Facchini G, Balestreri L, Perin T and Canzonieri V:
Serum and tissue markers in hepatocellular carcinoma and
cholangiocarcinoma: Clinical and prognostic implications.
Oncotarget. 8:14192–14220. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Peng L, Chen G, Zhu Z, Shen Z, Du C, Zang
R, Su Y, Xie H, Li H, Xu X, et al: Circular RNA ZNF609 functions as
a competitive endogenous RNA to regulate AKT3 expression by
sponging miR-150-5p in Hirschsprung's disease. Oncotarget.
8:808–818. 2017.PubMed/NCBI
|
|
6
|
Memczak S, Jens M, Elefsinioti A, Torti F,
Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer
M, et al: Circular RNAs are a large class of animal RNAs with
regulatory potency. Nature. 495:333–338. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Salzman J: Circular RNA Expression: Its
potential regulation and function. Trends Genet. 32:309–316. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Memczak S, Papavasileiou P, Peters O and
Rajewsky N: Identification and characterization of circular RNAs as
a new class of putative biomarkers in human blood. PLoS One.
10:e1412142015. View Article : Google Scholar
|
|
9
|
Li F, Zhang L, Li W, Deng J, Zheng J, An
M, Lu J and Zhou Y: Circular RNA ITCH has inhibitory effect on ESCC
by suppressing the Wnt/β-catenin pathway. Oncotarget. 6:6001–6013.
2015.PubMed/NCBI
|
|
10
|
Liu Y, Cui H, Wang W, Li L, Wang Z, Yang S
and Zhang X: Construction of circular miRNA sponges targeting
miR-21 or miR-221 and demonstration of their excellent anticancer
effects on malignant melanoma cells. Int J Biochem Cell Biol.
45:2643–2650. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sand M, Bechara FG, Sand D, Gambichler T,
Hahn SA, Bromba M, Stockfleth E and Hessam S: Circular RNA
expression in basal cell carcinoma. Epigenomics. 8:619–632. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang C, Wu H, Wang Y, Zhao Y, Fang X,
Chen C and Chen H: Expression patterns of circular RNAs from
primary kinase transcripts in the mammary glands of lactating rats.
J Breast Cancer. 18:235–241. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bachmayr-Heyda A, Reiner AT, Auer K,
Sukhbaatar N, Aust S, Bachleitner-Hofmann T, Mesteri I, Grunt TW,
Zeillinger R and Pils D: Correlation of circular RNA abundance with
proliferation-Exemplified with colorectal and ovarian cancer,
idiopathic lung fibrosis, and normal human tissues. Sci Rep.
5:80572015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Li P, Chen S, Chen H, Mo X, Li T, Shao Y,
Xiao B and Guo J: Using circular RNA as a novel type of biomarker
in the screening of gastric cancer. Clin Chim Acta. 444:132–136.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Qu S, Song W, Yang X, Wang J, Zhang R,
Zhang Z, Zhang H and Li H: Microarray expression profile of
circular RNAs in human pancreatic ductal adenocarcinoma. Genomics
Data. 5:385–387. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J,
Chen D, Gu J, He X and Huang S: Circular RNA is enriched and stable
in exosomes: A promising biomarker for cancer diagnosis. Cell Res.
25:981–984. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Qin M, Liu G, Huo X, Tao X, Sun X, Ge Z,
Yang J, Fan J, Liu L and Qin W: Hsa_circ_0001649: A circular RNA
and potential novel biomarker for hepatocellular carcinoma. Cancer
Biomark. 16:161–169. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Shang X, Li G, Liu H, Li T, Liu J, Zhao Q
and Wang C: Comprehensive circular RNA profiling reveals that
hsa_circ_0005075, a new circular RNA biomarker, is involved in
hepatocellular carcinoma development. Medicine (Baltimore).
95:e38112016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Huang XY, Huang ZL, Xu YH, Zheng Q, Chen
Z, Song W, Zhou J, Tang ZY and Huang XY: Comprehensive circular RNA
profiling reveals the regulatory role of the
circRNA-100338/miR-141-3p pathway in hepatitis B-related
hepatocellular carcinoma. Sci Rep. 7:54282017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Fu L, Yao T, Chen Q, Mo X, Hu Y and Guo J:
Screening differential circular RNA expression profiles reveals
hsa_circ_0004018 is associated with hepatocellular carcinoma.
Oncotarget. 8:58405–58416. 2017.PubMed/NCBI
|
|
21
|
Yu J, Xu QG, Wang ZG, Yang Y, Zhang L, Ma
JZ, Sun SH, Yang F and Zhou WP: Circular RNA cSMARCA5 inhibits
growth and metastasis in hepatocellular carcinoma. J Hepatol.
68:1214–1227. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hansen TB, Jensen TI, Clausen BH, Bramsen
JB, Finsen B, Damgaard CK and Kjems J: Natural RNA circles function
as efficient microRNA sponges. Nature. 495:384–388. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Enright AJ, John B, Gaul U, Tuschl T,
Sander C and Marks DS: MicroRNA targets in Drosophila. Genome Biol.
5:R12003. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Pasquinelli AE: MicroRNAs and their
targets: Recognition, regulation and an emerging reciprocal
relationship. Nat Rev Genet. 13:271–282. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Xie Z, Hur SK, Zhao L, Abrams CS and
Bankaitis VA: A Golgi lipid signaling pathway controls apical Golgi
distribution and cell polarity during Neurogenesis. Dev Cell.
44:725–740. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Sorber L, Zwaenepoel K, Jacobs J, De Winne
K, Goethals S, Reclusa P, Van Casteren K, Augustus E, Lardon F,
Roeyen G, et al: Circulating cell-free DNA and RNA analysis as
liquid biopsy: Optimal centrifugation protocol. Cancers (Basel).
11(pii): E 4582019. View Article : Google Scholar
|
|
29
|
Lu R, Shao Y, Ye G, Xiao B and Guo J: Low
expression of hsa_circ_0006633 in human gastric cancer and its
clinical significances. Tumor Biol. 39:1–7. 2017. View Article : Google Scholar
|
|
30
|
Bazan HA, Hatfield SA, Brug A, Brooks AJ,
Lightell DJ Jr and Woods TC: Carotid plaque rupture is accompanied
by an increase in the ratio of serum circR-284 to miR-221 levels.
Circ-Cardiovasc Gene. 10(pii): e00172042017.
|
|
31
|
Zhu Q, Lu G, Luo Z, Gui F, Wu J, Zhang D
and Ni Y: CircRNA circ_0067934 promotes tumor growth and metastasis
in hepatocellular carcinoma through regulation of
miR-1324/FZD5/Wnt/β-catenin axis. Biochem Bioph Res Co.
497:626–632. 2018. View Article : Google Scholar
|
|
32
|
Meng S, Zhou H, Feng Z, Xu Z, Tang Y, Li P
and Wu M: CircRNA: Functions and properties of a novel potential
biomarker for cancer. Mol Cancer. 16:94–102. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Jiang G, Wen L, Deng W, Jian Z and Zheng
H: Regulatory role of miR-211-5p in hepatocellular carcinoma
metastasis by targeting ZEB2. Biomed Pharmacother. 90:806–812.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Quan J, Pan X, He T, Lin C and Lai Y, Chen
P, Zhang Z, Yang S, Wang T and Lai Y: Tumor suppressor miR-211-5p
is associated with cellular migration, proliferation and apoptosis
in renal cell carcinoma. Exp Ther Med. 15:4019–4028.
2018.PubMed/NCBI
|
|
35
|
Chen LL, Zhang ZJ, Yi ZB and Li JJ:
MicroRNA-211-5p suppresses tumour cell proliferation, invasion,
migration and metastasis in triple-negative breast cancer by
directly targeting SETBP1. Brit J Cancer. 117:78–88. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wang L, Shen YF, Shi ZM, Shang XJ, Jin DL
and Xi F: Overexpression miR-211-5p hinders the proliferation,
migration, and invasion of thyroid tumor cells by downregulating
SOX11. J Clin Lab Anal. 32:2018. View Article : Google Scholar
|
|
37
|
Jiang G, Wen L, Zheng H, Jian Z and Deng
W: miR-204-5p targeting SIRT1 regulates hepatocellular carcinoma
progression. Cell Biochem Funct. 34:505–510. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Gao W, Wu Y, He X, Zhang C, Zhu M, Chen B,
Liu Q, Qu X, Li W, Wen S and Wang B: MicroRNA-204-5p inhibits
invasion and metastasis of laryngeal squamous cell carcinoma by
suppressing forkhead box C1. J Cancer. 8:2356–2368. 2017.
View Article : Google Scholar : PubMed/NCBI
|