Introduction
Colorectal cancer (CRC) ranks among the three most
common types of cancer in terms of both incidence and
cancer-associated mortality in Western industrialized countries
(1). Each year nearly 1.3 million
new cases of CRC are reported, and ~700,000 patients succumb to the
disease worldwide (2). The lifetime
risk of developing CRC may reach 6% of the population living in
developed countries (3,4). CRC ranks highest in incidence rates in
Europe, second only to lung cancer, and it causes ~204,000 deaths
every year (5). The age-specific
incidence of CRC rises sharply after 35 years of age, with ~90% of
cases occurring in persons >50 years of age (6). As in other developed areas, in Italy,
the incidence rate of CRC ranks third highest for men (after
prostate and lung cancer), and second for women (after breast
cancer) (7). The burden of the
disease, however, remains a serious concern in Italy as well as
worldwide, due to the social impact, costs and rates of mortality
(8). According to the theory by
Vogelstein, CRC progresses through three precisely-connected
stages: Initiation, a process that modifies the molecular message
of the normal cell; promotion, in which signal transduction
cascades are altered; and progression, which involves
phenotypically-altered, transformed cells (9). The first morphological changes observed
in the progression of CRC are represented by the formation of
aberrant crypt foci (ACF). The most striking feature of the ACF is
the shape of the gland lumen, which is considerably modified when
compared with the normal mucosa, and strongly dependent on the
histological structure (10).
Furthermore, the phenotypic and genotypic characteristics of ACF
are different from those of normal crypts. These characteristics
were first described by Bird in mice exposed to azoxymethane, and
were subsequently studied extensively in humans by Roncucci
(11,12). Currently, it is impossible to
identify the ACF via routine colonoscopy; however, in humans, the
presence of ACF is identifiable via the use of high-resolution
chromoendoscopy with the aid of particular dyes, such as methylene
blue or indigo carmine (13). A
previous study reported that ACF occurs sporadically between 40 and
45 years of age, when predominantly single foci are observed
(14). After 45 years, the number of
ACF rapidly increases to reach the plateau phase at ~60 years of
age, and slowly decreases thereafter. Other studies have reported
similar incidence rates of ACF in these age brackets (15,16). It
is essential to identify and remove these early lesions for an
adequate CRC prevention strategy (17). Furthermore, accurate tumor grading is
necessary for patient survival and can be achieved most effectively
in stained histopathological sections harvested via biopsy or
during surgery. Medical databases are fundamental in the
development of new techniques for early detection of neoplastic
cells. They are, however, difficult to obtain, since the labeling
of the images is often operator-dependent and requires specialized
skills. The considerable complexity and quantity of the structures
present in the biological tissue represents a fascinating challenge
for pathologists both in manual and automatic analyses of
histopathological slides. Although certain studies have presented a
reasonable consensus among experienced pathologists and
satisfactory results on their intra-observer reliability, other
studies have stated that even experienced pathologists often
disagree on tissue classification. Therefore, the use of expert
scores as a gold standard for histopathological examination could
lead to inappropriate evaluations (18–20). In
addition, quantitative characterization of pathology imagery is
important not only for clinical applications but also for research
applications. Recent studies have proven that the deep learning
(DL) approach is superior for tasks of classification and
segmentation on histological whole-slide images, as compared with
the previous image processing techniques (21–23). As
examples, DL models have been developed to detect metastatic breast
cancer (24), to identify
mitotically active cells (25), to
identify basal cell carcinoma (26)
and to grade brain gliomas (27)
using hematoxylin and eosin (H&E)-stained images.
Histopathological sections of colon tissues, stained with H&E
representative of the adenoma-carcinoma sequence are presented in
Fig. 1.
DL techniques for image recognition have proven
extremely effective in a broad number of applications, often
surpassing human performances. The general idea is that a
sufficiently flexible software network can be trained, i.e., values
can be assigned to its parameters, in order to recognize images by
examining a broad set of labeled images. Once trained, the network
can be used on unlabeled images to assign the correct corresponding
label.
Although the field is still rapidly evolving, some
general features of the network structure required to perform a
given task are sufficiently understood, meaning that the primary
obstacle for new applications is the lack of labeled training
sets.
Several applications of DL techniques to classify
colorectal cells have been published. A summary of the approaches
used is reported in the GLAS challenge contest summary (28). By the very nature of the challenge,
these methods are based on segmentation, i.e., individual pixel
labeling (29), which is an
effective and sensible approach from a computational perspective,
but requires significant investment in preparing a training set
where each individual pixels is assigned by human inspection to a
given part of the cell or the background. In addition, it is not
clear how the trained networks would perform on images obtained
with different instruments and resolutions. As the quality of
colorectal tissue images is instrument-dependent and tends to
improve over time as new microscopes replace older models, this
last point is particularly relevant.
The aim of the present study was to use a DL
algorithm to classify images of colorectal tissues that bypasses
segmentation and labels the raw image directly. Thus, the method
used in the present study did not rely on a segmented training set,
but rather on the readily available labeled images, as routinely
found in a pathologist report. Hence, the network created in the
present study can be easily retrained whenever the quality of the
images changes, with minimal human effort.
In addition, a more articulate classification was
performed in the present study; instead of two labels (benign and
malignant), four categories were employed (normal, preneoplastic,
adenoma and cancer) representing the disease progression. On the
one hand, this implied more uncertainty in the labeling, as images
for neighboring categories may appear similar and a trained
pathologist would rely also on other information and/or a
collection of several images to provide a diagnosis. On the other
hand, the ability to recognize the different stages of cancer
development will prove extremely valuable in providing more
specific diagnoses and eventually early treatments.
In the present study highly accurate and
reproducible results were obtained from biomedical image analysis
(overall accuracy, >95%), with the potential to significantly
improve both the quality and speed of medical diagnoses. The
present study also states the performance level of the DL algorithm
on the GLAS challenge images. Although no patent should be based on
this work without explicit written consent from the authors, the
algorithm used is not proprietary and other researchers are invited
to share it for testing purposes.
Materials and methods
Patients and samples
The database used in the present study consisted of
393 images, divided into the 4 categories or labels: Normal mucosa
(m), preneoplastic lesion (ACF) (p), adenoma (a) and colon cancer
(k). All images were obtained from patients who underwent primary
surgery or colonoscopy at the Modena University Hospital between
1998–2008. Normal mucosa images were obtained from 44 patients,
preneoplastic lesions (ACF) from 45 patients, adenoma images from
76 patients, and colon cancer tissue images from 58 patients
diagnosed with CRC. The present study was granted ethical approval
by the local operative ethics committee of the Policlinico Hospital
of Modena. Written informed consent was not required, since the
present study was retrospective in design and the amount of
specimens obtained was extensive. Tissue images were stained with
haematoxylin and eosin (H&E) and digitized with an optical
microscope Leica IRBE with CCD Rising Tech. Sony CCD Sensor (USB2.0
5.0MP CCD ICX452AQ) camera and Software Image Pro Plus (v4.5).
Images were captured under ×20 magnification and a resolution of
600 dpi. Slides were scanned using a unique study ID and without
identifiable data linked to the patient. In the present study, two
pathologists from the Unit of Pathology of Modena, and Bologna
independently reviewed the whole-slide images in the training and
test datasets in order to identify the type of colorectal lesions
as reference standards. When disagreements regarding the image
classification occurred, the pathologists resolved the issue
through further discussions. When it was not possible to reach a
consensus on a lesion type for an image, that image was discarded
and replaced by a new one in order to maintain accuracy.
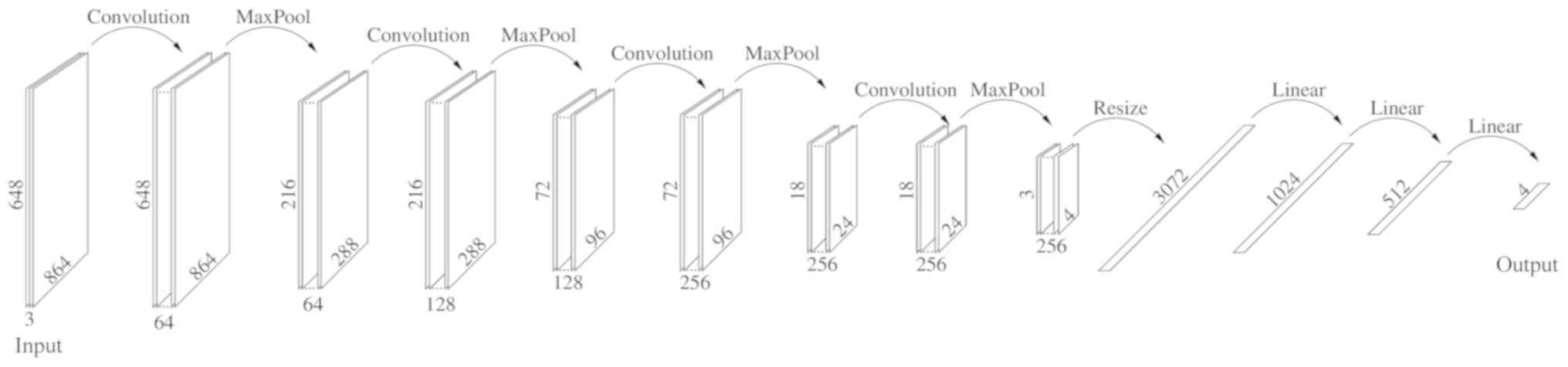
Dataset and the DL algorithm
The present study divided each image in the database
into 9 subimages, each 864×648 pixels, and relabeled each one
individually, discarding those not recognizable, resulting in a
final dataset containing 2,513 images. Disjoint subsets were then
randomly created the: Training (2,012 images), validation (251
images) and test (250 images). Each of the given sets was further
divided into the label categories reported in Table I. This was referred to as the Small
Dataset. A Large Dataset was also created as follows: Each image,
within the corresponding subset, was rotated 180 degrees, and
reflected along both the vertical and horizontal axes, in order to
obtain a dataset four times larger, i.e., comprising 10,052 images
(Table I). The DL algorithm created
in the present study was implemented on the Pytorch platform with
hardware Nvidia Titan XP and Quadro P6000 and consisted of the
sequences reported in Fig. 1.
 | Table I.Small dataset and large dataset. |
Table I.
Small dataset and large dataset.
| A, Small dataset |
|---|
|
|---|
| Small Dataset | Normal mucosa | Preneoplastic
lesion | Adenoma | Colon cancer | Total |
|---|
| Training | 404 | 407 | 685 | 534 | 2,012 |
| Validation | 50 | 51 | 86 | 64 | 251 |
| Test | 50 | 51 | 85 | 64 | 250 |
|
| B, Large
dataset |
|
|
| Normal
mucosa | Preneoplastic
lesion | Adenoma | Colon
cancer | Total |
|
| Training | 1,616 | 1,628 | 2,736 | 2,068 | 8,048 |
| Validation | 200 | 204 | 344 | 256 | 1,004 |
| Test | 200 | 204 | 340 | 256 | 1,000 |
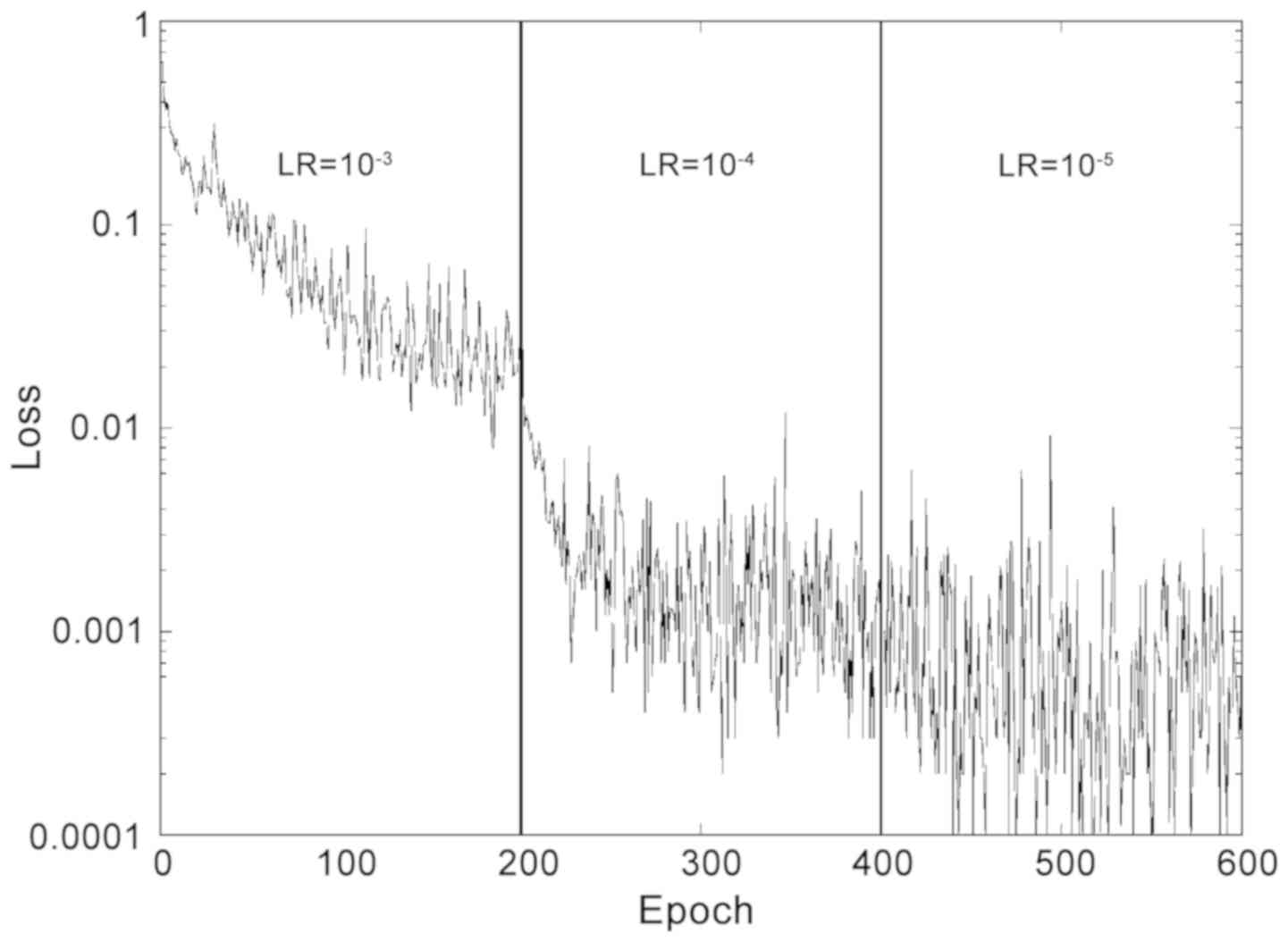
Given the relatively small training set, a small
batch size, consisting of 10 images was selected. During the
validation, performed initially using the Small Dataset, the
optimizer Adam was selected over SGD and Adagrad, which provided
slightly worse levels of accuracy (−5%). For the loss function
calculation, the present study opted for Softmax. The weight decay
was set to 10−5 and cycles of 200 epochs were performed
with different learning rates. A short training of two cycles was
used, with learning rates of 10-3 and 10−4,
respectively, and a long training, which included a third cycle
with a learning rate 10−5. The loss function during
training is reported in Fig. 2.
During training, each epoch runs in ~58 sec (Small Dataset) or 3.50
min (Large Dataset), so the complete training is completed in
<10 (Small) or 40 (Large) hours on this system. Evaluation of
the test set took ~10−2 sec/image, making this Neural
Network practical for fast screening of thousands of images in
seconds. In order to estimate the stability of the optimized
network, after the aforementioned training, the present study
performed 10 evaluations of the test set separated by 4 epochs of
further training. The accuracies are reported as the average value
from these 10 evaluations, and the standard deviation was used as
an estimate of the uncertainty. The same procedure was followed for
a number of optimizations starting from scratch and using different
partitions of the original image dataset and different weights for
the Adam optimizer. It was observed that the accuracies were
generally within two standard deviations of each other.
Results
The accuracy of the test set following the Short and
Long trainings is presented in Table
II. The present study also measured the accuracy for the
nearest match. Accuracies, expressed as a percentage of correct
results ± standard deviation, were computed for the test set using
different optimization and evaluation conditions. Short training
indicates 200 epochs with learning rate 10−3 and 200
epochs with learning rate 10−4. Long training adds
another 200 epochs with learning rate 10−5. For the
exact match, predictions were considered correct only if they
matched the target exactly. In the nearest match, neighboring
labels were also regarded as correct, as described later. The Small
Dataset refers to the original 2,513 images divided into training,
validation and test sets. The Large Dataset also includes rotations
and reflections, for a total of 10,052 images. For normal mucosa
(m) the nearest match is the preneoplastic lesion (p); for (p),
they are (m) and (a) the adenoma; for (a), they are (p) and (k)
cancer; while for (k) is just (a). Representative histological
images of the aforementioned categories are presented in Fig. 3, and these four images were all
correctly labeled by the DL algorithm.
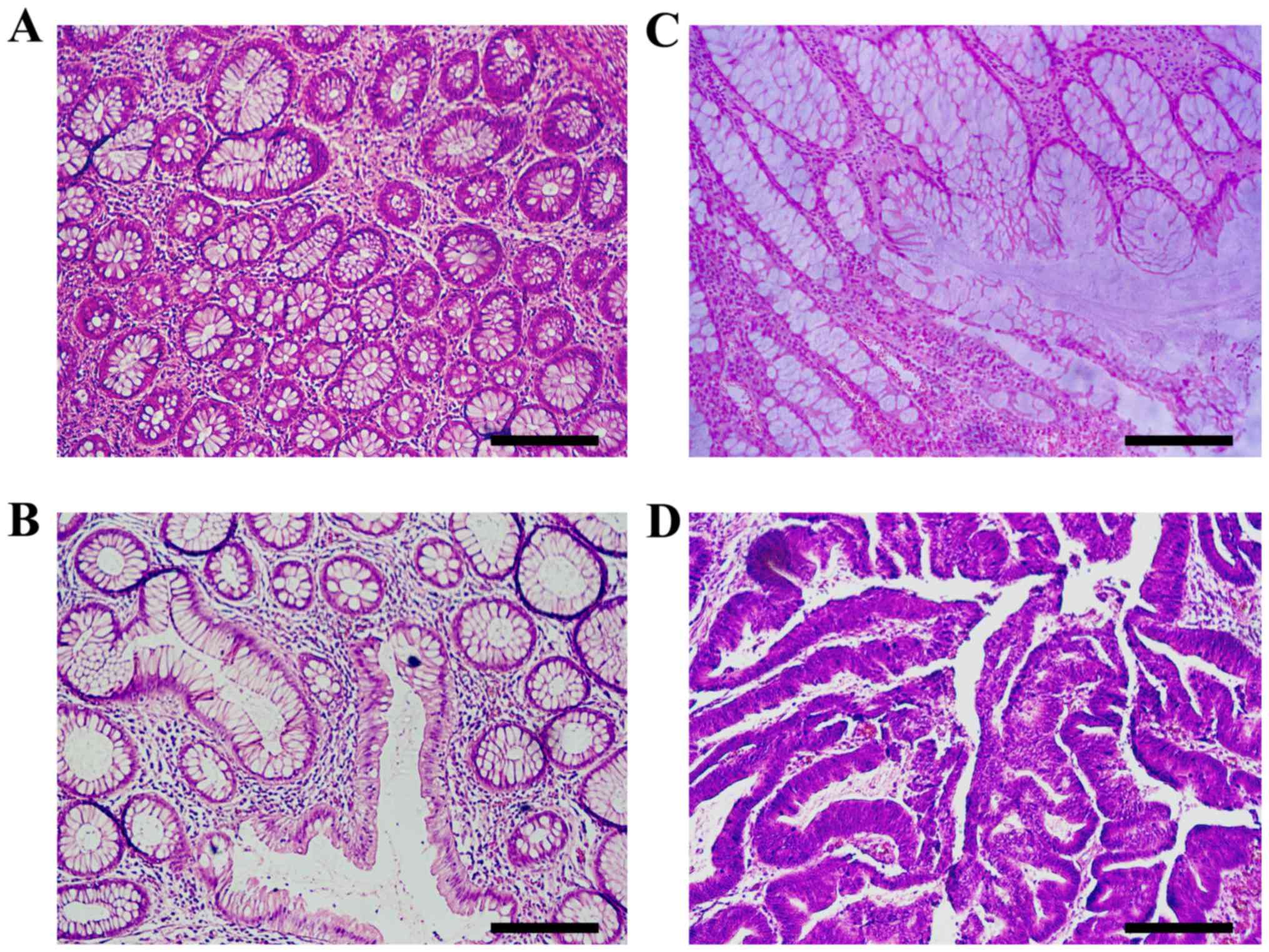 | Figure 3.Histopathological sections of
hematoxylin and eosin stained colon tissues that are representative
of colorectal carcinogenesis. (A) Normal mucosa exhibits benign
glands consisting of a regular circular lumen in cross section. (B)
Pre-neoplastic lesion (aberrant crypt foci) consists of larger
glands with enlarged epithelial nuclei, often stratified and
crowded. (C) Adenoma is characterized by ovoid enlarged nuclei,
vesicular dispersed chromatin and occasional mitoses. Sections
exhibit an uneven distribution of goblet cells within crypts,
luminal serration, budding, branching, crowding and fusion of
glands. (D) In carcinoma, architectural changes increase with
evolution and progression of malignancy. Luminal serration,
budding, branching, crowding and fusion of glands are presented.
Scale bar, 210 µm. |
 | Table II.Test accuracy after short and long
training. |
Table II.
Test accuracy after short and long
training.
| Training | Exact match large
dataset (%) | Nearest match large
dataset (%) | Exact match small
dataset (%) | Nearest match small
dataset (%) |
|---|
| Short |
93.79±0.76 |
99.85±0.11 | 92.92±0.86 | 99.20±0.25 |
| Long |
95.28±0.19 |
99.90±0.00 | 93.08±0.57 | 99.20±0.00 |
The GLAS challenge dataset (29,30) was
used to further validate the approach of the present study against
drastically different images. All images from the GLAS training
were considered, including test A and test B sets combined. These
differ from those of the present study: Both sets refer to ×20
magnification, but the images used in the present study were
collected with a resolution of 0.426 mm/pixel, vs. 0.620 mm/pixel
in the GLAS dataset. They were also different sizes. Hence, each
GLAS image was expanded by bicubic interpolation by a factor of
0.620/0.426 in order to achieve the same nominal resolution as that
in the present study. Of course, this introduces some blurring and
the quality of the resulting images was different from that used
for training. Each of the GLAS images were then cropped to 864×648
pixels. Of the 165 GLAS images, 14 were discarded, as they were too
small. The majority of the remaining 151 were then cropped to focus
on the central part of the image. A few cases comprising
significant portions of non-tissue background were cropped close to
one edge or corner in order to include as much tissue as
possible.
Discussion
Researchers both in the image analysis and pathology
fields have recognized the importance of quantitative analysis of
pathology images. The current pathological diagnosis is based on a
detailed and careful observation of the abnormal morphological
changes, and is based on specific and precise criteria. However, it
is a subjective, though educated opinion. Thus, the need for a
quantitative assessment based on slide images of digital pathology
is urgently required. This quantitative analysis of digital
pathology is important not only from the diagnostic point of view,
but also to understand the reasons that led to a specific diagnosis
(for example, the altered size of the lumen glands of the
Lieberkuhn crypts, indicating a potential malignant hyperplasia).
Furthermore, the quantitative characterization of pathological
images is important both for clinical applications (e.g., to
decrease/eliminate inter- and intra-observer variations in
diagnosis) and for research applications (e.g., to understand the
biological mechanisms underlying the pathological process). From a
histopathological perspective, the crypts of the Lieberkuhn have
different characterizing components, including the lumen,
epithelial cells and stroma (connective tissue, blood vessels,
nervous tissue, etc.). The epithelial cells form the outline of the
gland, which encloses the cytoplasm and the lumen, while the stroma
is not considered part of the gland. If only non-cancerous (benign)
glands are considered, the DL algorithms must actually be able to
manage and recognize a significant variability of shape, size,
position, consistency and staining of the glands. Obviously, when
the analysis is performed on the cancerous tissue, the glands
appear significantly different from the benign glands, and the
presence of artefacts or corrupted areas further aggravates the
problem. Therefore, machine learning approaches are primarily used
to develop extremely precise models trained with labelled examples
that can address the problematic tissue variability. An effective
algorithm for medical diagnoses requires large training datasets,
which, in general, are extremely difficult to obtain, in order to
correctly classify the different features of benign and malignant
gland types (Fig. 3). Colorectal
carcinogenesis is a multi-step process characterized by marked
morphological changes. The normal epithelium becomes a
hyperproliferative mucosa and subsequently gives rise to a benign
adenoma, which can then progress into carcinoma and metastases
(9). Furthermore, CRC presents an
intratumor heterogeneity highlighted by genetic analysis (31). From the reported accuracies, it was
observed in the present study that the Large Dataset generally
provided better results than the Small Dataset. Thus, as previously
reported (28,29), including the same image with
different orientations is a viable strategy to improve the dataset.
In the present study, the transformed images were placed in the
same set (training, validation or test) as the original so as to
not bias the results by including the same image in the test set as
was used in training, with a different orientation. The effect of a
total randomization of the Large Dataset was also tested, and
although the performance was slightly improved, the differences
were within two standard deviations. The reported results also
indicated that the last 200 epochs in the Long training time had a
small effect on the loss. This seemed to improve the accuracy for
the exact match, but not for the neighboring match. Although from
Table I it appears that the standard
deviation was significantly smaller after the Long training, this
effect was largely due to the 10 samples used to compute the
average and standard deviation being separated by 4 epochs of
training. The learning rate for this extra training was
10−4 after the Short optimization, but it was 10 times
smaller after the Long one. Since the parameters change was tied to
the learning rate, the sets of parameters used for the 10 cases
after the Long optimization were more similar to each other than
those after the Short optimization, hence they provided more
similar classifications. Thus, a direct comparison of standard
deviations is only meaningful when the same learning rate is used.
In the present study, this effect was tested by increasing the
learning rate or the number of epochs between evaluations after the
Long training, and revealed that this was indeed the case. This
also indicates that there is an association between the 10
evaluations after the Long training, so the present study
recommends using the standard deviation from the Short training to
estimate the uncertainty. The results for the Long training and
Large Dataset are detailed in the confusion matrix in Table III, where percentages are presented
as averages over the 10 measures. In this case, only 1 of the 1,006
test cases falls outside the nearest neighbor range. When repeating
the optimization it was typically observed that this number was
between 0 and 2. The accuracy for exact matches was close to 95%,
making the procedure attractive for clinical use. In addition, when
the nearest cases were included, it surpassed 99.8%, indicating
that most mislabeling refer to one of the nearest neighbors. As
mentioned in the ‘Introduction’ of the present study, when using
four labels, this kind of mislabeling would not be unusual even for
trained pathologists. Finally, the reported DL network structure
derives from several attempts where different numbers and sizes of
hidden layers were used. Although efforts were made to maximize the
accuracy on the validation set, it is still possible that
modifications of the network used in the present study may improve
the performance. To the best of our knowledge, there are currently
no alternative methods for automatic classification into the four
key categories of colorectal tissue images based on the approach
that is proposed in the present study. A comparison can be
attempted with the GLAS Challenge contest (28), although the aim there was
segmentation and classification based on only two categories.
Furthermore, accuracies were based on individual pixels and
segmented objects, and were between 80–90%, significantly lower
than the ones obtained in the present study. To this end, the
present study processed the GLAS dataset as described in the
previous section, obtaining images that are comparable to those in
the present study, but with a different resolution. As the GLAS
challenge only has two categories (benign and malignant), three of
the four categories (preneoplastic, adenoma and cancer) were
grouped as malignant, leaving the fourth (normal mucosa) as benign.
The same evaluation as in the test of the present study, i.e., 10
evaluations separated by 10 training epochs, yields an accuracy of
81.7 with standard deviation 1.1. This accuracy is smaller than
those obtained by some GLAS challenge participants (29). However, considering that the network
training is based on images obtained with different instruments and
resolution, and that the classification scheme is not the same, it
demonstrates that the approach used in the present study is viable
and could be exported to broader datasets, provided enough
diversity is included in the training set. This result also
indicates that networks trained on images from a given microscope
model may not perform as well on images from another one, making
the unsegmented approach of the present study more appealing for
easy adjustments. The fact that our approach is fundamentally
different from those used in the GLAS challenge should be noted, as
it is not based on segmentation and recognition of individual
features, but rather on direct classification from raw images.
 | Table III.Confusion matrix for long training
and large dataset. Rows correspond to predictions, columns to true
labels. |
Table III.
Confusion matrix for long training
and large dataset. Rows correspond to predictions, columns to true
labels.
| True→
Predicted↓ | M | P | A | K | Precision False
positive |
|---|
| M | 18.2 | 0.3 | 0.0 | 0.0 | 98.2 1.8 |
| P | 1.9 | 18.3 | 0.8 | 0.0 | 87.3 12.7 |
| A | 0.0 | 1.5 | 33.1 | 0.0 | 95.6 4.4 |
| K | 0.0 | 0.1 | 0.8 | 25.6 | 99.3 0.7 |
| Recall False
negative | 90.7 9.3 | 90.3 9.7 | 97.4 2.6 | 100.0 0.0 | 95.34.7 |
The results from the present study suggest the
potential for this method to become of practical assistance to
pathologists.
In conclusion, the present study collected a
microscope image database of human colorectal tissue, including
normal mucosa, preneoplastic lesion, adenoma and carcinoma. The DL
algorithm used in the present study, trained on part of the
dataset, was able to correctly assign >95% of the test cases,
and the majority of the mislabeled images were assigned to
neighboring categories. The results from the present study suggest
that DL techniques may provide a valuable tool to assist human
operators for histological classification of the different steps of
the adenoma-carcinoma sequence, typical of colorectal tumors.
Acknowledgements
The authors would like to Dr Alessandro Achille and
Dr Patrik Chaudhari (UCLA) for their helpful comments.
Funding
NVIDIA Corporation (Santa Clara, USA) provided Titan
XP and a Quadro P6000 graphic processing unit through their GPU
grant for research purposes. Marie Slodova Curie Action funded the
current study via the GHAIA Geometric Harmonic Analysis for
Intersciplinary Application (grant no. GA 777822).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
PS, RF and FF conceived and designed the present
study. PS, LL and LR acquired the data. PS, LL, LR and RF analyzed
and interpreted the data. RF, FF and GF implemented the Neural
network. LL and LR critically revised the manuscript. PS, RF and FF
wrote the manuscript.
Ethics approval and consent to
participate
The present study was granted approved by the local
Operative Ethics Committee of The Policlinico Hospital of
Modena.
Patient consent for publication
All patients provided written informed consent for
the publication of any associated data and accompanying images.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Das V, Kalita J and Pal M: Predictive and
prognostic biomarkers in colorectal cancer: A systematic review of
recent advances and challenges. Biomed Pharmacother. 87:8–19. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Favoriti P, Carbone G, Greco M, Pirozzi F,
Pirozzi RE and Corcione F: Worldwide burden of colorectal cancer: A
review. Updates Surg. 68:7–11. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Center MM, Jemal A, Smith RA and Ward E:
Worldwide variations in colorectal cancer. CA Cancer J Clin.
59:366–378. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ferlay J, Steliarova-Foucher E,
Lortet-Tieulent J, Rosso S, Coebergh JW, Comber H, Forman D and
Bray F: Cancer incidence and mortality patterns in Europe:
Estimates for 40 countries in 2012. Eur J Cancer. 49:1374–1403.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ahnen DJ, Wade SW, Jones WF, Sifri R,
Mendoza Silveiras J, Greenamyer J, Guiffre S, Axilbund J, Spiegel A
and You YN: The increasing incidence of young-onset colorectal
cancer: A call to action. Mayo Clin Proc. 89:216–224. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
I Numeri Del Cancro in Italia 2016. Il
Pensiero Scientifico Editore, . 2016.
|
|
8
|
Altobelli E, Lattanzi A, Paduano R,
Varassi G and di Orio F: Colorectal cancer prevention in Europe:
Burden of disease and status of screening programs. Prev Med.
62:132–141. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Vogelstein B, Fearon ER, Hamilton SR, Kern
SE, Preisinger AC, Leppert M, Nakamura Y, White R, Smits AM and Bos
JL: Genetic alterations during colorectal-tumor development. N Engl
J Med. 319:525–532. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Di Gregorio C, Losi L, Fante R, Modica S,
Ghidoni M, Pedroni M, Tamassia MG, Gafà L, Ponz de Leon M and
Roncucci L: Histology of aberrant crypt foci in the human colon.
Histopathology. 30:328–334. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bird RP: Observation and quantification of
aberrant crypts in the murine colon treated with a colon
carcinogen: Preliminary findings. Cancer Lett. 37:147–151. 1987.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Losi L, Roncucci L, di Gregorio C, de Leon
MP and Benhattar J: K-ras and p53 mutations in human colorectal
aberrant crypt foci. J Pathol. 178:259–263. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lopez-Ceron M and Pellise M: Biology and
diagnosis of aberrant crypt foci. Colorectal Dis. 14:e157–e164.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Roncucci L, Pedroni M, Vaccina F, Benatti
P, Marzona L and De Pol A: Aberrant crypt foci in colorectal
carcinogenesis. Cell and crypt dynamics. Cell Prolif. 33:1–18.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Roncucci L, Stamp D and Medline AA:
Identification and quantification of aberrant crypt foci and
microadenomas in the human colon. Hum Pathol. 22:287–294. 1991.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kowalczyk M, Siermontowski P, Mucha D,
Ambroży T, Orłowski M, Zinkiewicz K, Kurpiewski W, Paśnik K,
Kowalczyk I and Pedrycz A: Chromoendoscopy with a
standard-resolution colonoscope for evaluation of rectal aberrant
crypt foci. PLoS One. 11:e01482862016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Nascimbeni R, Villanacci V, Mariani PP, Di
Betta E, Ghirardi M, Donato F and Salerni B: Aberrant crypt foci in
the human colon: Frequency and histologic patterns in patients with
colorectal cancer or diverticular disease. Am J Surg Pathol.
23:1256–1263. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Gupta AK, Pinsky P, Rall C, Mutch M, Dry
S, Seligson D and Schoen RE: Reliability and accuracy of the
endoscopic appearance in the identification of aberrant crypt foci.
Gastrointest Endosc. 70:322–330. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Andrion A, Magnani C, Betta PG, Donna A,
Mollo F, Scelsi M, Bernardi P, Botta M and Terracini B: Malignant
mesothelioma of the pleura: Interobserver variability. J Clin
Pathol. 48:856–860. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Van Putten PG, Hol L, Van Dekken H, Han
van Krieken J, van Ballegooijen M, Kuipers EJ and van Leerdam ME:
Inter-observer variation in the histological diagnosis of polyps in
colorectal cancer screening. Histopathology. 58:974–981. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hadsell R, Sermanet P, Ben J, Erkan A,
Scoffier M, Kavukcuoglu K, Muller U and Lecun Y: Learning
long-range vision for autonomous off-road driving. J Field Robot.
26:120–144. 2009. View Article : Google Scholar
|
|
22
|
Sirinukunwattana K, Ahmed Raza SE, Yee-Wah
Tsang, Snead DR, Cree IA and Rajpoot NM: Locality sensitive deep
learning for detection and classification of nuclei in routine
colon cancer histology images. IEEE Trans Med Imaging.
35:1196–1206. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Janowczyk A and Madabhushi A: Deep
learning for digital pathology image analysis: A comprehensive
tutorial with selected use cases. J Pathol Inform. 7:292016.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Cruz-Roa AA, Ovalle JE, Madabhushi A and
Osorio FAG: International conference on medical image computing and
computer-assisted intervention. A deep learning architecture for
image representation, visual interpretability and automated
basal-cell carcinoma cancer detectionSpringer; pp. 403–410. 2013,
PubMed/NCBI
|
|
25
|
Ertosun MG and Rubin DL: Automated grading
of gliomas using deep learning in digital pathology images: A
modular approach with ensemble of convolutional neural networks.
AMIA Annu Symp Proc. 2015:1899–1908. 2015.PubMed/NCBI
|
|
26
|
Malon CD and Cosatto E: Classification of
mitotic figures with convolutional neural networks and seeded blob
features. J Pathol Inform. 4:92013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wang H, Cruz-Roa A, Basavanhally A,
Gilmore H, Shih N, Feldman M, Tomaszewski J, Gonzalez F and
Madabhushi A: Cascaded ensemble of convolutional neural networks
and handcrafted features for mitosis detection. SPIE Med Imaging.
941:90410B2014.
|
|
28
|
Sirinukunwattana K, Pluim JPW, Chen H, Qi
X, Heng PA, Guo YB, Wang LY, Matuszewski BJ, Bruni E, Sanchez U, et
al: Gland segmentation in colon histology images: The glas
challenge contest. Med Image Anal. 35:489–502. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kainz P, Pfeiffer M and Urschler M:
Segmentation and classification of colon glands with deep
convolutional neural networks and total variation regularization.
PeerJ. 5:e38742017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Sirinukunwattana K, Snead DR and Rajpoot
NM: A stochastic polygons model for glandular structures in colon
histology images. IEEE Trans Med Imaging. 34:2366–2378. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Losi L, Baisse B, Bouzourene H and
Benhattar J: Evolution of intratumoral genetic heterogeneity during
colorectal cancer progression. Carcinogenesis. 26:916–922. 2005.
View Article : Google Scholar : PubMed/NCBI
|