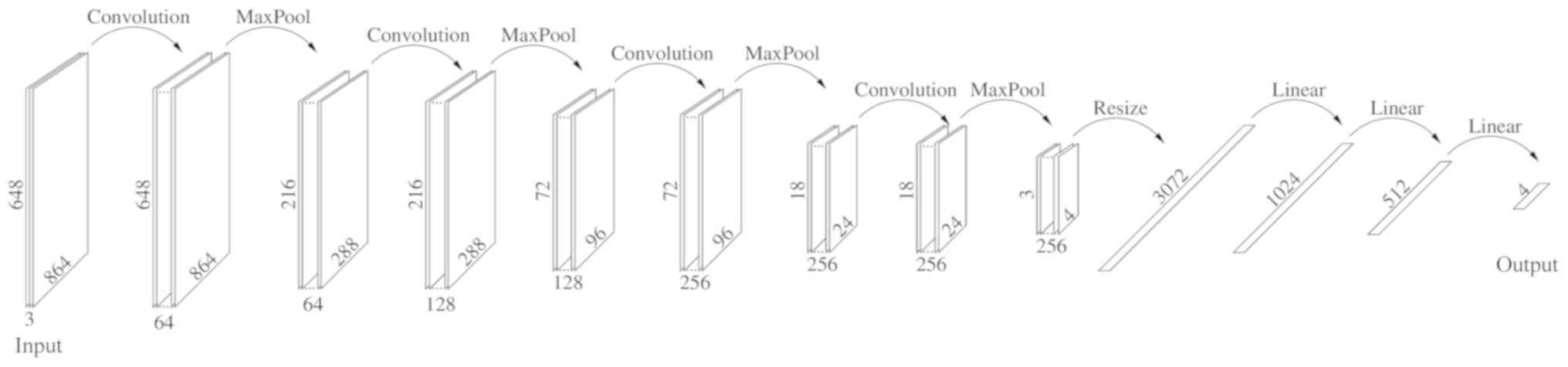
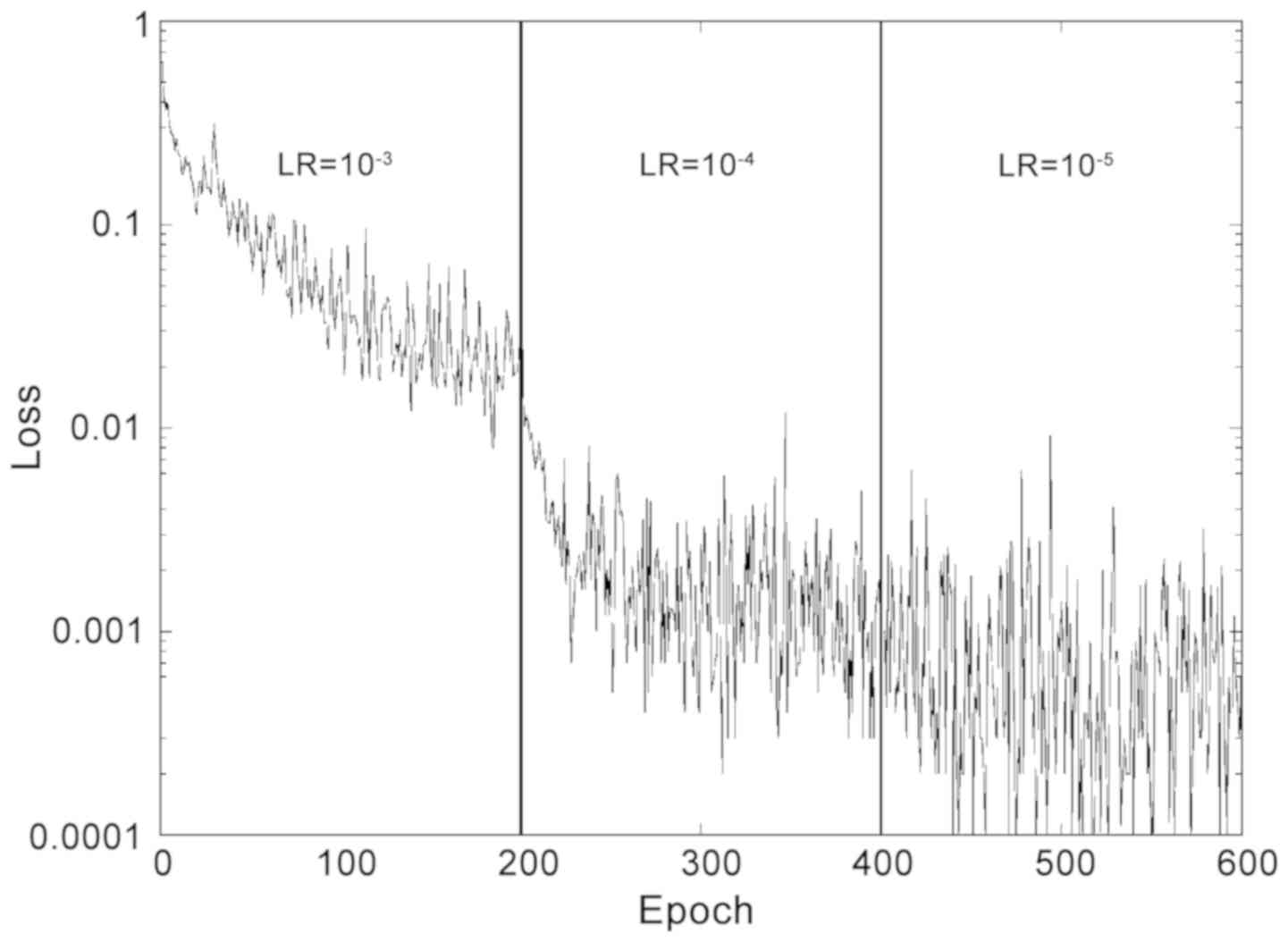
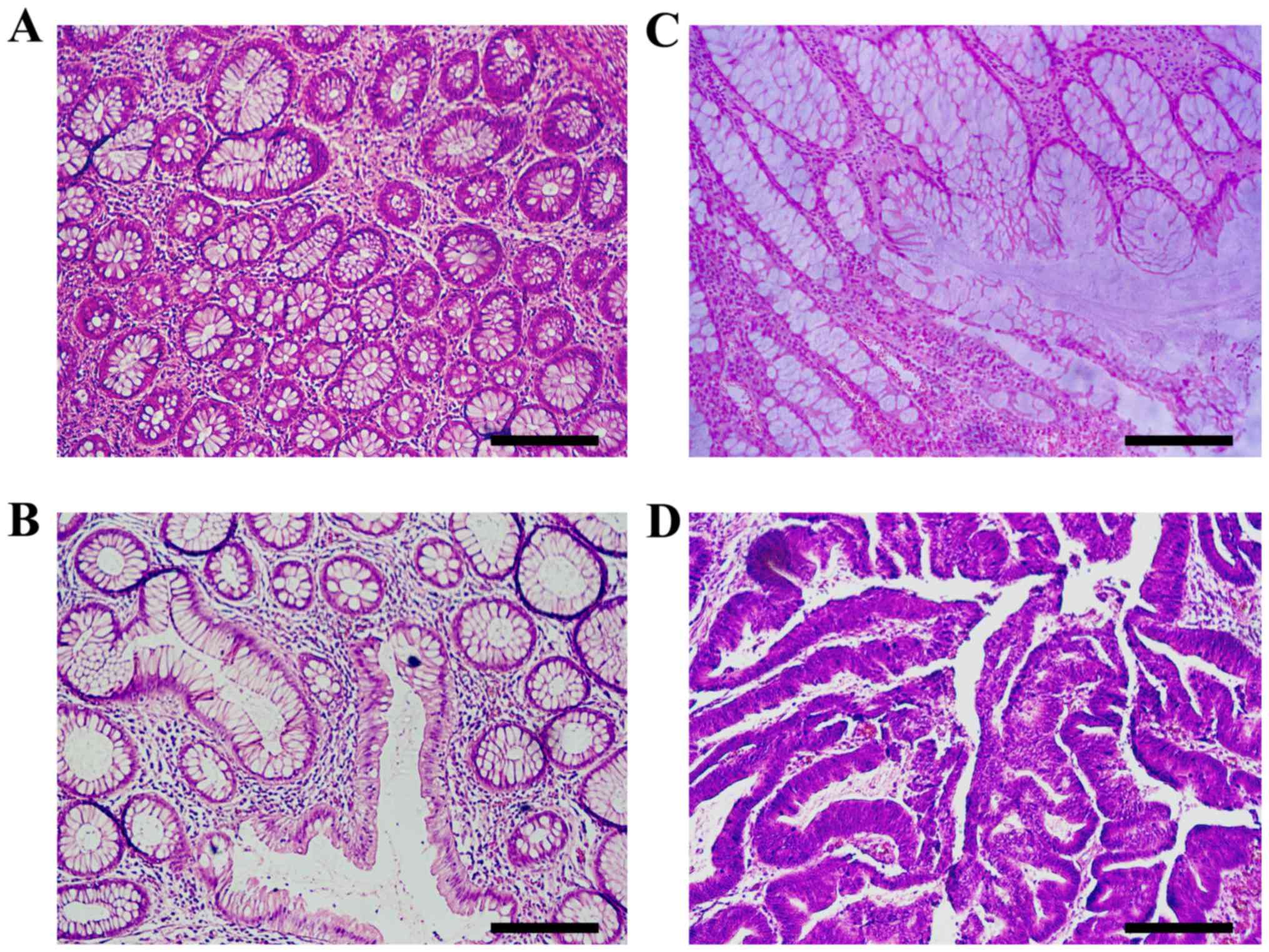
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Das V, Kalita J and Pal M: Predictive and
prognostic biomarkers in colorectal cancer: A systematic review of
recent advances and challenges. Biomed Pharmacother. 87:8–19. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Favoriti P, Carbone G, Greco M, Pirozzi F,
Pirozzi RE and Corcione F: Worldwide burden of colorectal cancer: A
review. Updates Surg. 68:7–11. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Center MM, Jemal A, Smith RA and Ward E:
Worldwide variations in colorectal cancer. CA Cancer J Clin.
59:366–378. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ferlay J, Steliarova-Foucher E,
Lortet-Tieulent J, Rosso S, Coebergh JW, Comber H, Forman D and
Bray F: Cancer incidence and mortality patterns in Europe:
Estimates for 40 countries in 2012. Eur J Cancer. 49:1374–1403.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ahnen DJ, Wade SW, Jones WF, Sifri R,
Mendoza Silveiras J, Greenamyer J, Guiffre S, Axilbund J, Spiegel A
and You YN: The increasing incidence of young-onset colorectal
cancer: A call to action. Mayo Clin Proc. 89:216–224. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
I Numeri Del Cancro in Italia 2016. Il
Pensiero Scientifico Editore, . 2016.
|
|
8
|
Altobelli E, Lattanzi A, Paduano R,
Varassi G and di Orio F: Colorectal cancer prevention in Europe:
Burden of disease and status of screening programs. Prev Med.
62:132–141. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Vogelstein B, Fearon ER, Hamilton SR, Kern
SE, Preisinger AC, Leppert M, Nakamura Y, White R, Smits AM and Bos
JL: Genetic alterations during colorectal-tumor development. N Engl
J Med. 319:525–532. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Di Gregorio C, Losi L, Fante R, Modica S,
Ghidoni M, Pedroni M, Tamassia MG, Gafà L, Ponz de Leon M and
Roncucci L: Histology of aberrant crypt foci in the human colon.
Histopathology. 30:328–334. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bird RP: Observation and quantification of
aberrant crypts in the murine colon treated with a colon
carcinogen: Preliminary findings. Cancer Lett. 37:147–151. 1987.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Losi L, Roncucci L, di Gregorio C, de Leon
MP and Benhattar J: K-ras and p53 mutations in human colorectal
aberrant crypt foci. J Pathol. 178:259–263. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lopez-Ceron M and Pellise M: Biology and
diagnosis of aberrant crypt foci. Colorectal Dis. 14:e157–e164.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Roncucci L, Pedroni M, Vaccina F, Benatti
P, Marzona L and De Pol A: Aberrant crypt foci in colorectal
carcinogenesis. Cell and crypt dynamics. Cell Prolif. 33:1–18.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Roncucci L, Stamp D and Medline AA:
Identification and quantification of aberrant crypt foci and
microadenomas in the human colon. Hum Pathol. 22:287–294. 1991.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kowalczyk M, Siermontowski P, Mucha D,
Ambroży T, Orłowski M, Zinkiewicz K, Kurpiewski W, Paśnik K,
Kowalczyk I and Pedrycz A: Chromoendoscopy with a
standard-resolution colonoscope for evaluation of rectal aberrant
crypt foci. PLoS One. 11:e01482862016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Nascimbeni R, Villanacci V, Mariani PP, Di
Betta E, Ghirardi M, Donato F and Salerni B: Aberrant crypt foci in
the human colon: Frequency and histologic patterns in patients with
colorectal cancer or diverticular disease. Am J Surg Pathol.
23:1256–1263. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Gupta AK, Pinsky P, Rall C, Mutch M, Dry
S, Seligson D and Schoen RE: Reliability and accuracy of the
endoscopic appearance in the identification of aberrant crypt foci.
Gastrointest Endosc. 70:322–330. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Andrion A, Magnani C, Betta PG, Donna A,
Mollo F, Scelsi M, Bernardi P, Botta M and Terracini B: Malignant
mesothelioma of the pleura: Interobserver variability. J Clin
Pathol. 48:856–860. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Van Putten PG, Hol L, Van Dekken H, Han
van Krieken J, van Ballegooijen M, Kuipers EJ and van Leerdam ME:
Inter-observer variation in the histological diagnosis of polyps in
colorectal cancer screening. Histopathology. 58:974–981. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hadsell R, Sermanet P, Ben J, Erkan A,
Scoffier M, Kavukcuoglu K, Muller U and Lecun Y: Learning
long-range vision for autonomous off-road driving. J Field Robot.
26:120–144. 2009. View Article : Google Scholar
|
|
22
|
Sirinukunwattana K, Ahmed Raza SE, Yee-Wah
Tsang, Snead DR, Cree IA and Rajpoot NM: Locality sensitive deep
learning for detection and classification of nuclei in routine
colon cancer histology images. IEEE Trans Med Imaging.
35:1196–1206. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Janowczyk A and Madabhushi A: Deep
learning for digital pathology image analysis: A comprehensive
tutorial with selected use cases. J Pathol Inform. 7:292016.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Cruz-Roa AA, Ovalle JE, Madabhushi A and
Osorio FAG: International conference on medical image computing and
computer-assisted intervention. A deep learning architecture for
image representation, visual interpretability and automated
basal-cell carcinoma cancer detectionSpringer; pp. 403–410. 2013,
PubMed/NCBI
|
|
25
|
Ertosun MG and Rubin DL: Automated grading
of gliomas using deep learning in digital pathology images: A
modular approach with ensemble of convolutional neural networks.
AMIA Annu Symp Proc. 2015:1899–1908. 2015.PubMed/NCBI
|
|
26
|
Malon CD and Cosatto E: Classification of
mitotic figures with convolutional neural networks and seeded blob
features. J Pathol Inform. 4:92013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wang H, Cruz-Roa A, Basavanhally A,
Gilmore H, Shih N, Feldman M, Tomaszewski J, Gonzalez F and
Madabhushi A: Cascaded ensemble of convolutional neural networks
and handcrafted features for mitosis detection. SPIE Med Imaging.
941:90410B2014.
|
|
28
|
Sirinukunwattana K, Pluim JPW, Chen H, Qi
X, Heng PA, Guo YB, Wang LY, Matuszewski BJ, Bruni E, Sanchez U, et
al: Gland segmentation in colon histology images: The glas
challenge contest. Med Image Anal. 35:489–502. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kainz P, Pfeiffer M and Urschler M:
Segmentation and classification of colon glands with deep
convolutional neural networks and total variation regularization.
PeerJ. 5:e38742017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Sirinukunwattana K, Snead DR and Rajpoot
NM: A stochastic polygons model for glandular structures in colon
histology images. IEEE Trans Med Imaging. 34:2366–2378. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Losi L, Baisse B, Bouzourene H and
Benhattar J: Evolution of intratumoral genetic heterogeneity during
colorectal cancer progression. Carcinogenesis. 26:916–922. 2005.
View Article : Google Scholar : PubMed/NCBI
|