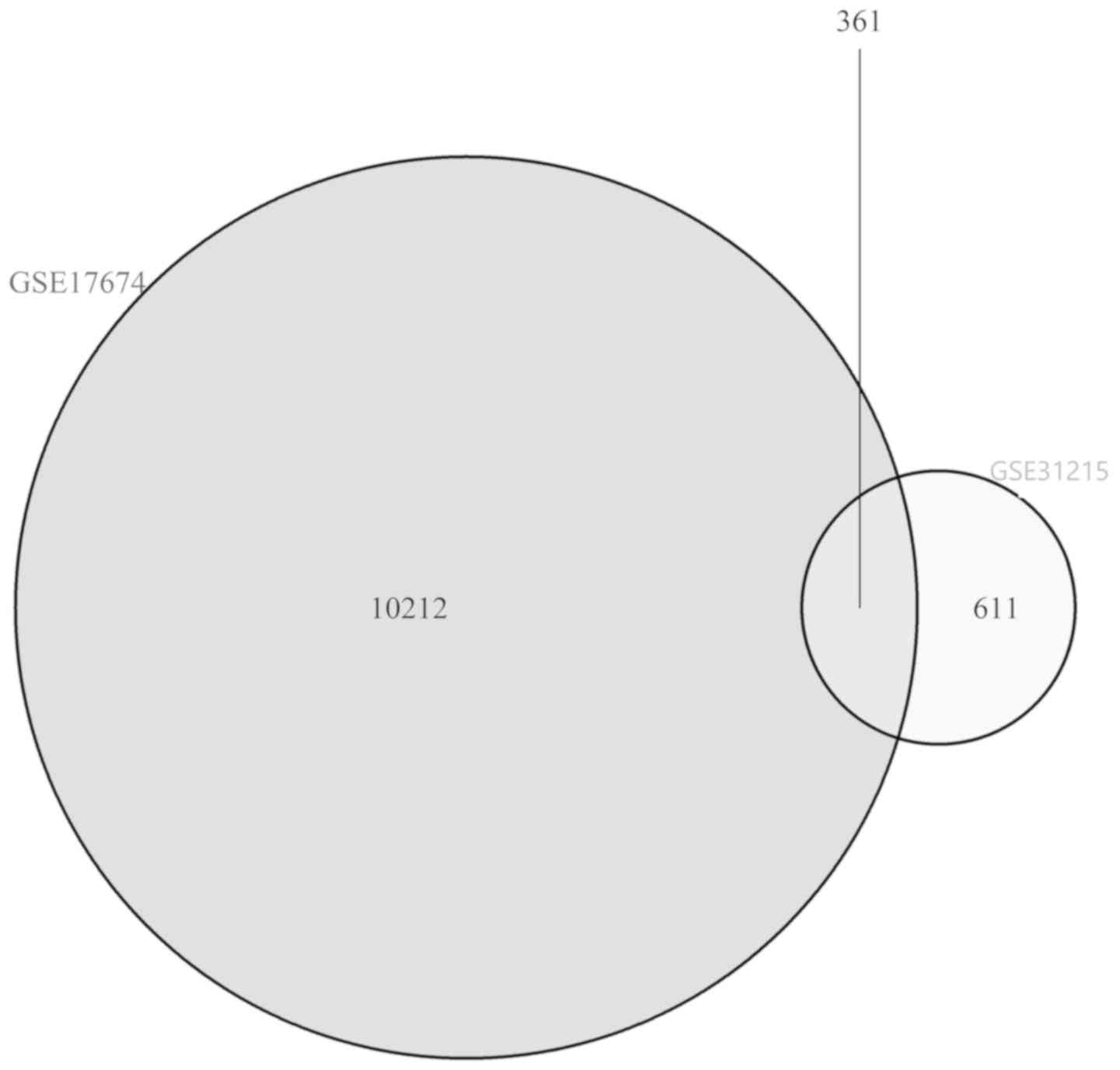
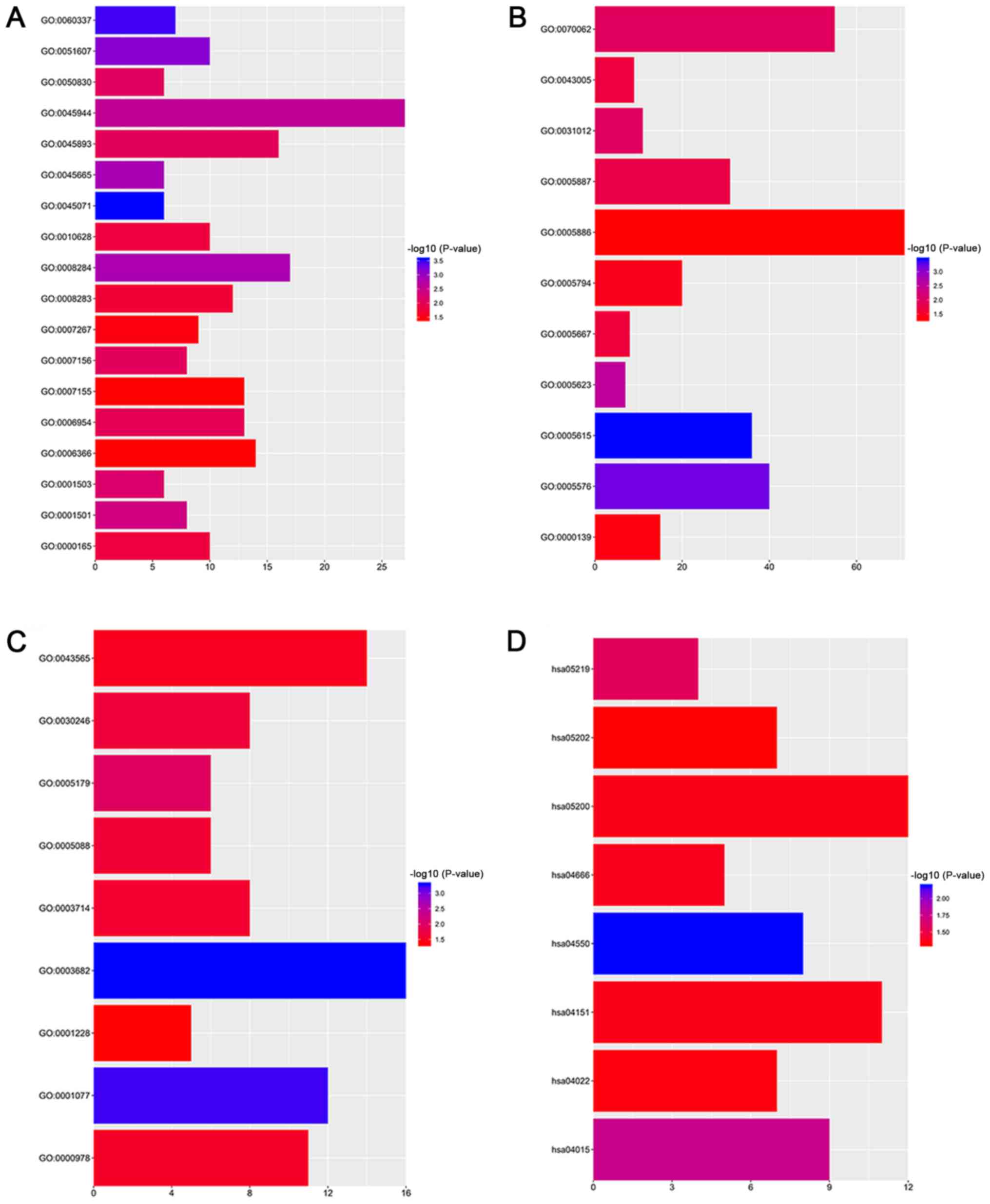
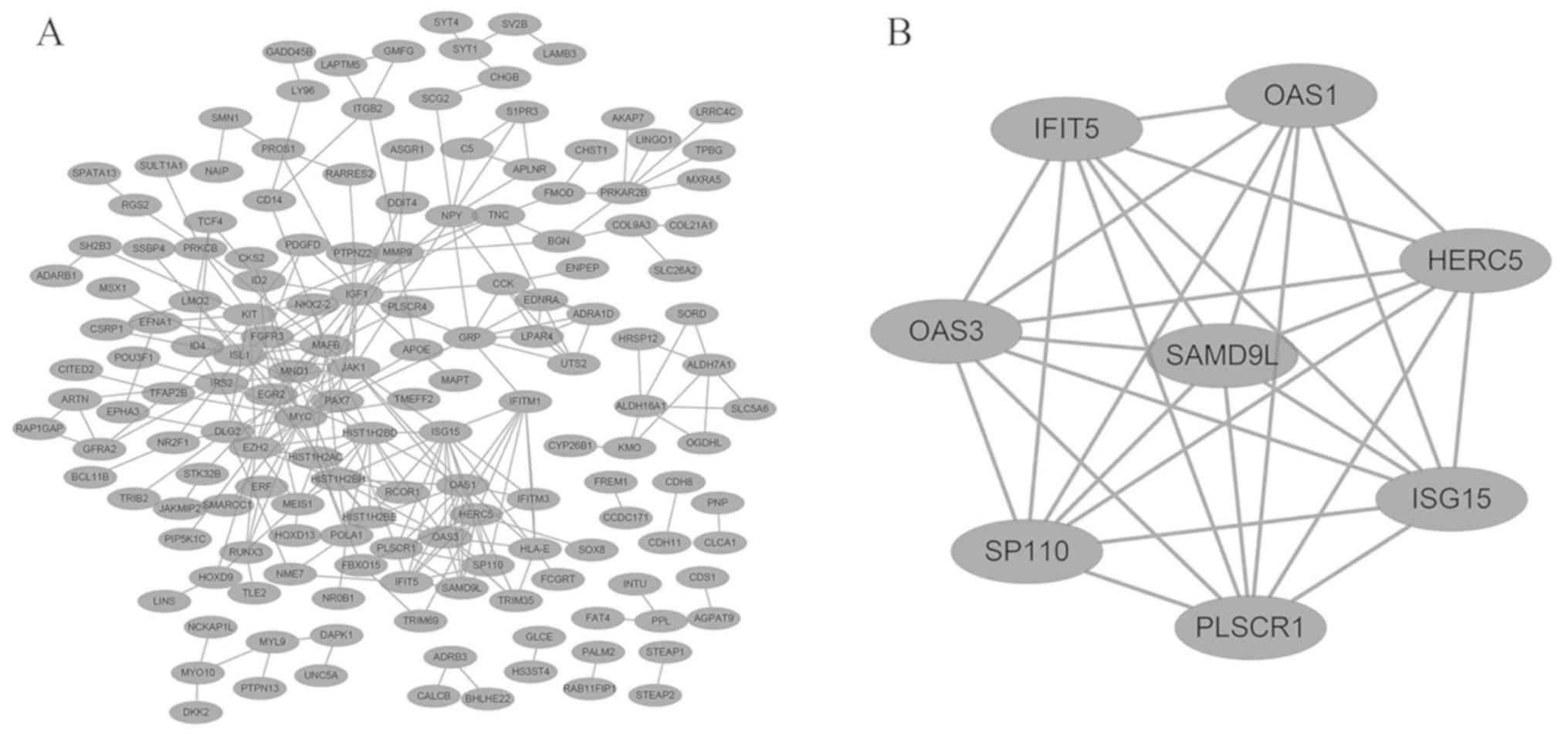
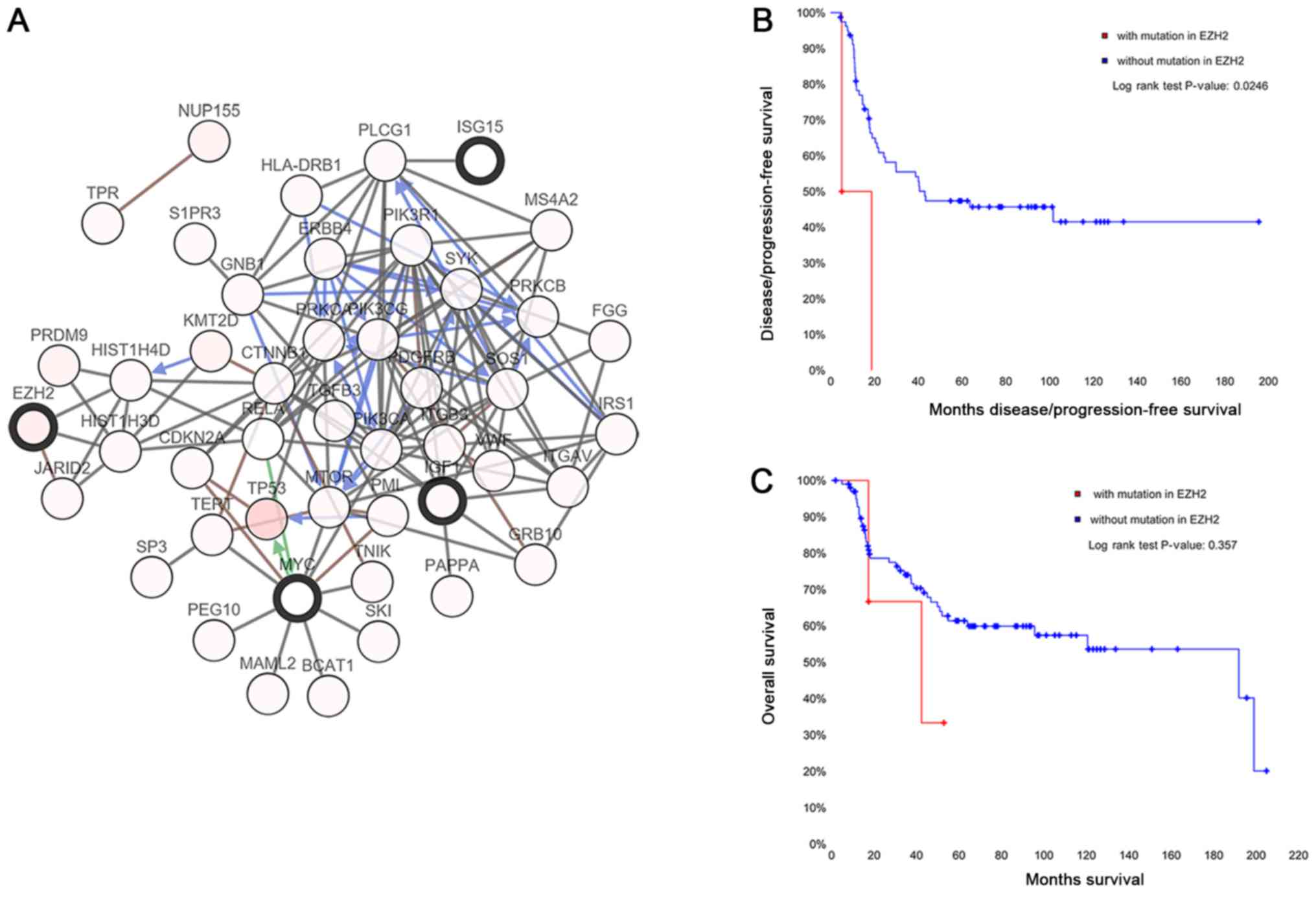
|
1
|
Balamuth NJ and Womer RB: Ewing's sarcoma.
Lancet Oncol. 11:184–192. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Womer RB, West DC, Krailo MD, Dickman PS,
Pawel BR, Grier HE, Marcus K, Sailer S, Healey JH, Dormans JP and
Weiss AR: Randomized controlled trial of interval-compressed
chemotherapy for the treatment of localized Ewing sarcoma: A report
from the Children's Oncology Group. J Clin Oncol. 30:4148–4154.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Shachak A, Ophir R and Rubin E: Applying
instructional design theories to bioinformatics education in
microarray analysis and primer design workshops. Cell Biol Educ.
4:199–206. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Vogelstein B, Papadopoulos N, Velculescu
VE, Zhou S, Diaz LA Jr and Kinzler KW: Cancer genome landscapes.
Science. 339:1546–1558. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Edgar R, Domrachev M and Lash AE: Gene
Expression Omnibus: NCBI gene expression and hybridization array
data repository. Nucleic Acids Res. 30:207–210. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: Tool for the unification of biology. The Gene
Ontology Consortium. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kanehisa M and Goto S: KEGG: Kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Savola S, Klami A, Myllykangas S, Manara
C, Scotlandi K, Picci P, Knuutila S and Vakkila J: High expression
of complement component 5 (C5) at tumor site associates with
superior survival in Ewing's sarcoma family of tumour patients.
ISRN Oncol. 2011:1687122015.
|
|
9
|
Riggi N, Suvà M, De Vito C, Provero P,
Stehle J, Baumer K, Cironi L, Janiszewska M, Petricevic T, Suvà D,
et al: EWS-FLI-1 modulates miRNA145 and SOX2 expression to initiate
mesenchymal stem cell reprogramming toward Ewing sarcoma cancer
stem cells. Genes Dev. 24:916–932. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Irizarry RA, Hobbs B, Collin F,
Beazer-Barclay YD, Antonellis KJ, Scherf U and Speed TP:
Exploration, normalization, and summaries of high density
oligonucleotide array probe level data. Biostatistics. 4:249–264.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Gautier L, Cope L, Bolstad BM and Irizarry
RA: Affy-analysis of Affymetrix GeneChip data at the probe level.
Bioinformatics. 20:307–315. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ihaka R and Gentleman R: R: A language for
data analysis and graphics. J Comput Graph Stat. 5:299–314. 1996.
View Article : Google Scholar
|
|
13
|
Troyanskaya O, Cantor M, Sherlock G, Brown
P, Hastie T, Tibshirani R, Botstein D and Altman RB: Missing value
estimation methods for DNA microarrays. Bioinformatics. 17:520–525.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Smyth GK: Limma: Linear models for
microarray dataBioinformatics Comput Biol Sol Using R Bioconductor.
Gentleman R, Carey V, Dudoit S, Irizarry R and Huber W: Springer;
NY: pp. 397–420. 2005
|
|
15
|
Huang DW, Sherman BT, Tan Q, Collins JR,
Alvord WG, Roayaei J, Stephens R, Baseler MW, Lane HC and Lempicki
RA: The DAVID Gene Functional Classification Tool: A novel
biological module-centric algorithm to functionally analyze large
gene lists. Genome Biol. 8:R1832007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kanehisa M: The KEGG database. Novartis
Found Symp. 247:91–101. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Szklarczyk D, Franceschini A, Wyder S,
Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos
A, Tsafou KP, et al: STRING v10: Protein-protein interaction
networks, integrated over the tree of life. Nucleic Acids Res.
43:D447–D452. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Smoot ME, Ono K, Ruscheinski J, Wang PL
and Ideker T: Cytoscape 2.8: New features for data integration and
network visualization. Bioinformatics. 27:431–432. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bader GD and Hogue CW: An automated method
for finding molecular complexes in large protein interaction
networks. BMC Bioinformatics. 4:22003. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chin CH, Chen SH, Wu HH, Ho CW, Ko MT and
Lin CY: cytoHubba: Identifying hub objects and sub-networks from
complex interactome. BMC Syst Biol. 8 (Suppl 4):S112014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gao J, Aksoy BA, Dogrusoz U, Dresdner G,
Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al:
Integrative analysis of complex cancer genomics and clinical
profiles using the cBioPortal. Sci Signal. 6:pl12013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Nakamura F, Nakamura Y, Maki K, Sato Y and
Mitani K: Cloning and characterization of the novel chimeric gene
TEL/PTPRR in acute myelogenous leukemia with inv(12)(p13q13).
Cancer Res. 65:6612–6621. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zheng T, Wang A, Hu D and Wang Y:
Molecular mechanisms of breast cancer metastasis by gene expression
profile analysis. Mol Med Rep. 16:4671–4677. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
He R, Wu JX, Zhang Y, Che H and Yang L:
LncRNA LINC00628 overexpression inhibits the growth and invasion
through regulating PI3K/Akt signaling pathway in osteosarcoma. Eur
Rev Med Pharmacol Sci. 22:5857–5866. 2018.PubMed/NCBI
|
|
25
|
Kawano M, Tanaka K, Itonaga I, Iwasaki T
and Tsumura H: MicroRNA-20b promotes cell proliferation via
targeting of TGF-β receptor II and upregulates MYC expression in
Ewing's sarcoma cells. Int J Oncol. 51:1842–1850. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Poli V, Fagnocchi L, Fasciani A, Cherubini
A, Mazzoleni S, Ferrillo S, Miluzio A, Gaudioso G, Vaira V, Turdo
A, et al: MYC-driven epigenetic reprogramming favors the onset of
tumorigenesis by inducing a stem cell-like state. Nat Commun.
9:10242018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Naab TJ, Gautam A, Ricks-Santi L, Esnakula
AK, Kanaan YM, DeWitty RL, Asgedom G, Makambi KH, Abawi M and
Blancato JK: MYC amplification in subtypes of breast cancers in
African American women. BMC Cancer. 18:2742018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Little CD, Nau MM, Carney DN, Gazdar AF
and Minna JD: Amplification and expression of the c-myc oncogene in
human lung cancer cell lines. Nature. 306:194–196. 1983. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yang X, Ye H, He M, Zhou X, Sun N, Guo W,
Lin X, Huang H, Lin Y, Yao R and Wang H: LncRNA PDIA3P interacts
with c-Myc to regulate cell proliferation via induction of pentose
phosphate pathway in multiple myeloma. Biochem Biophys Res Commun.
498:207–213. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Soucek L, Lawlor ER, Soto D, Shchors K,
Swigart LB and Evan GI: Mast cells are required for angiogenesis
and macroscopic expansion of Myc-induced pancreatic islet tumors.
Nat Med. 13:1211–1218. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
31
|
Jones JI and Clemmons DR: Insulin-like
growth factors and their binding proteins: Biological actions.
Endocr Rev. 16:3–34. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Olmos D, Martins AS, Jones RL, Alam S,
Scurr M and Judson IR: Targeting the insulin-like growth factor 1
receptor in Ewing's sarcoma: Reality and expectations. Sarcoma.
2011:4025082011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
de Groot S, Gelderblom H, Fiocco M, Bovée
JV, van der Hoeven JJ, Pijl H and Kroep JR: Serum levels of IGF-1
and IGF-BP3 are associated with event-free survival in adult Ewing
sarcoma patients treated with chemotherapy. Onco Targets Ther.
10:2963–2970. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kasprzak A, Szaflarski W, Szmeja J,
Andrzejewska M, Przybyszewska W, Kaczmarek E, Koczorowska M,
Kościński T, Zabel M and Drews M: Differential expression of IGF-1
mRNA isoforms in colorectal carcinoma and normal colon tissue. Int
J Oncol. 42:305–316. 2013.PubMed/NCBI
|
|
35
|
Kakuta S, Shibata S and Iwakura Y: Genomic
structure of the mouse 2′,5′-oligoadenylate synthetase gene family.
J Interferon Cytokine Res. 22:981–993. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Lee WB, Choi WY, Lee DH, Shim H, Kim-Ha J
and Kim YJ: OAS1 and OAS3 negatively regulate the expression of
chemokines and interferon-responsive genes in human macrophages.
BMB Rep. 52:133–138. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Herviou L, Cavalli G and Moreaux J: EZH2
is therapeutic target for personalized treatment in multiple
myeloma. Bull Cancer. 105:804–819. 2018.(In French). View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Richter GH, Plehm S, Fasan A, Rössler S,
Unland R, Bennani-Baiti IM, Hotfilder M, Löwel D, von Luettichau I,
Mossbrugger I, et al: EZH2 is a mediator of EWS/FLI1 driven tumor
growth and metastasis blocking endothelial and neuro-ectodermal
differentiation. Proc Natl Acad Sci USA. 106:5324–5329. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Gulati N, Béguelin W and Giulino-Roth L:
Enhancer of zeste homolog 2 (EZH2) inhibitors. Leuk Lymphoma.
59:1574–1585. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wang D, Quiros J, Mahuron K, Pai CC,
Ranzani V, Young A, Silveria S, Harwin T, Abnousian A, Pagani M, et
al: Targeting EZH2 reprograms intratumoral regulatory T cells to
enhance cancer immunity. Cell Rep. 23:3262–3274. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Osiak A, Utermöhlen O, Niendorf S, Horak I
and Knobeloch KP: ISG15, an interferon-stimulated ubiquitin-like
protein, is not essential for STAT1 signaling and responses against
vesicular stomatitis and lymphocytic choriomeningitis virus. Mol
Cell Biol. 25:6338–6345. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Burks J, Reed RE and Desai SD: Free ISG15
triggers an antitumor immune response against breast cancer: A new
perspective. Oncotarget. 6:7221–7231. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Zhao C, Hsiang TY, Kuo RL and Krug RM:
ISG15 conjugation system targets the viral NS1 protein in influenza
A virus-infected cells. Proc Natl Acad Sci USA. 107:2253–2258.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Jeon YJ, Jo MG, Yoo HM, Hong SH, Park JM,
Ka SH, Oh KH, Seol JH, Jung YK and Chung CH: Chemosensitivity is
controlled by p63 modification with ubiquitin-like protein ISG15. J
Clin Invest. 122:2622–2636. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Zuo C, Sheng X, Ma M, Xia M and Ouyang L:
ISG15 in the tumorigenesis and treatment of cancer: An emerging
role in malignancies of the digestive system. Oncotarget.
7:74393–74409. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Tecalco-Cruz AC and Cruz-Ramos E: Protein
ISGylation and free ISG15 levels are increased by interferon gamma
in breast cancer cells. Biochem Biophys Res Commun. 499:973–978.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Yoo L, Yoon AR, Yun CO and Chung KC:
Covalent ISG15 conjugation to CHIP promotes its ubiquitin E3 ligase
activity and inhibits lung cancer cell growth in response to type I
interferon. Cell Death Dis. 9:972018. View Article : Google Scholar : PubMed/NCBI
|