Introduction
Colorectal cancer (CRC) was the third most common
type of cancer and the fourth leading cause of cancer-associated
mortality worldwide in 2012 (1,2). CRC is
classified into two categories: i) The microsatellite
instability-high (MSI-H) group, which is caused by defects in the
DNA mismatch repair (MMR) system and accounts for ~15% of tumors;
and ii) the microsatellite stable (MSS) group, which exhibits
chromosomal instability and accounts for the remaining 85% of
tumors (3–5). At the protein level, MSI-H is similar
to deficient MMR (dMMR) status, while MSI-low (MSI-L)/MSS are
similar to proficient MMR (pMMR) status (6,7).
Immunotherapy with anti-programmed cell death 1
(PD-1)/programmed death ligand 1 (PD-L1) monoclonal antibodies
(mAbs) is a standard therapeutic strategy in various types of
cancer, including lung cancer, gastric cancer and renal cell
carcinoma (8–10). Although the results of initial trials
of immune checkpoint inhibitors (ICIs) in CRC were not particularly
promising (11,12), dMMR/MSI-H patients with CRC are
currently considered to be candidates for anti-PD-1/PD-L1
immunotherapy (13,14). In a phase II clinical trial,
treatment with the anti-PD-1 mAb pembrolizumab resulted in a
significant clinical response in patients with dMMR-CRC, but not
those with pMMR-CRC (13). Similar
results were observed in dMMR/MSI-H tumors across 12 different
cancer types (15). Furthermore, a
recent phase II trial demonstrated the clinical benefit of the PD-1
mAb nivolumab in patients with dMMR/MSI-H-CRC (14). Therefore, immunotherapy with ICIs
targeting the PD-1/PD-L1 axis may be a promising treatment option
for patients with dMMR/MSI-H solid tumors of varying histological
origins.
In general, dMMR/MSI-H tumors in CRC are thought to
respond to ICIs due to their highly immunogenic nature (14). For example, dMMR/MSI-H results in the
accumulation of several mutations in microsatellites spread along
the genome, and produces neo-antigens that produce a highly
antigenic microenvironment with a high density of tumor
infiltrating lymphocytes (TILs) (3,5,16,17).
Moreover, simultaneous expression of multiple immune checkpoint
molecules, including PD-1 and PD-L1, has been reported in
dMMR/MSI-H-CRC (3,5,16,17).
Consequently, in patients with dMMR/MSI-H-CRC, treatment with ICIs
targeting the PD-1/PD-L1 axis reactivates anti-tumor specific T
cells that subsequently attack tumor cells.
Previous studies revealed that ICIs targeting the
PD-1/PD-L1 axis were ineffective at treating MSS/pMMR-CRC (11,12).
Clinical studies have established MSI status as a putative response
biomarker for PD-1 blockade, with progression free survival rates
of up to 78% reported in patients with MSI-H-CRC, compared with 11%
of patients with MSS-CRC (13,17).
However, other studies have demonstrated that ~10% of patients with
MSS/pMMR-CRC exhibit a response to PD-1/PD-L1 inhibitors (13,17).
Based on the aforementioned clinical evidence, the aim of the
current study was to compare the immune statuses of patients with
MSI-L/MSS or pMMR-CRC with MSI-H or dMMR-CRC, in order to identify
responders with MSI-L/MSS or pMMR-CRC as distinct biomarker-defined
populations. The current study therefore investigated the
infiltrating grade of CD4 and CD8 TILs, the expression levels of
PD-L1 and PD-L2, the interferon-γ (IFN-γ) and CD8 T effector gene
signatures, and the phosphorylated signal transducer and activator
of transcription 1 (p-STAT1) status in patients with MSI-L/MSS or
pMMR- CRC compared with those with MSI-H or dMMR-CRC.
Materials and methods
The Cancer Genome Atlas (TCGA) dataset
analysis
Level 3 Illumina RNA-Seq data (RNA-Seq V2 RSEM
normalized) for colon adenocarcinoma (COAD) and rectal
adenocarcinoma (READ) in TCGA were downloaded through the
cBioPortal (www.cbioportal.org) in July 2018
(18) and consisted of 342 samples.
MSI testing data (MSI-H, MSI-L or MSS) for each TCGA tumor were
obtained from a previous study by Liu et al (19). The current study analyzed multi-gene
expression signatures, including the CD8 T effector gene signature
[CD8A molecule (CD8A), CD8B molecule (CD8B),
eomesodermin (EOMES), granzyme A (GZMA), granzyme B
(GZMB), interferon γ (IFNG) and perforin 1
(PRF1)] and the IFN-γ gene signature [indoleamine
2,3-dioxygenase 1 (IDO1), C-X-C motif chemokine ligand
(CXCL) 9 and 10, major histocompatibility complex class II
DR α (HLA-DRA), STAT1 and IFNG] (20,21). The
IFN-γ gene signature was identified from three clinical studies
investigating pembrolizumab, and focused on genes associated with
antigen presentation and cytotoxic activity in 220 patients with
nine types of cancer (21). The CD8
T effector gene signature was previously established to identify
activated T cells such as cytotoxic T lymphocytes (20). The signature scores were calculated
by averaging the expression levels of the genes included in each
signature (22), excluding
HLA-DRA expression values, as these were not available in
TCGA RNA-seq data.
Patient samples
Formalin-fixed paraffin-embedded tissue samples from
219 patients with primary CRC, who had undergone surgical resection
without preoperative chemotherapy or radiotherapy in Fukushima
Medical University Hospital (Fukushima, Japan) between January 2007
and December 2013 were analyzed in the current study. A total of
138 men and 81 women (mean age, 67.8±12.4 years; age range, 27–94
years), were included (Table I).
Patients with stage 0 CRC were excluded. Clinical and pathological
data were retrospectively obtained from medical records, with the
last follow-up in April 2017 (Table
I).
 | Table I.Clinical features of the patients
(n=219). |
Table I.
Clinical features of the patients
(n=219).
| Clinical
features | Number of
patients |
|---|
| Sex |
|
|
Male | 138 |
|
Female | 81 |
| TNM
stagea |
|
| I | 49 |
| II | 72 |
|
III | 74 |
| IV | 24 |
| Degree of
differentiation |
|
|
Moderate/high
differentiation | 204 |
| Poor
differentiation/undifferentiated | 15 |
| Lymphatic
metastasisa |
|
| N0 | 126 |
|
N1-3 | 93 |
| Degree of
invasiona |
|
| T1 | 25 |
| T2 | 39 |
| T3 | 95 |
| T4 | 60 |
Immunohistochemistry (IHC)
Tissue blocks were cut into 4-µm-thick sections and
were subsequently deparaffinized by three washes of 3 min with
xylene at room temperature and rehydrated by decreasing gradient of
ethanol at room temperature. Sections were incubated with 0.3%
hydrogen peroxide in methanol for 30 min at room temperature to
block endogenous peroxidase activity. For CD4, CD8 and forkhead box
P3 (FOXP3) staining, antigen retrieval was performed by autoclaving
the sections for 5 min at 105°C in target retrieval solution (pH
9.0; Dako; Agilent Technologies, Inc.). For PD-L1 staining, antigen
retrieval was performed by autoclaving the sections for 10 min at
120°C in target retrieval solution (pH 9.0). For p-STAT1 staining,
antigen retrieval was performed by autoclaving the sections for 5
min at 105°C in citrate buffer solution (pH 6.0). Tissue sections
were subsequently incubated with the following primary antibodies
directed against CD4 (clone 4B12; cat. no. M7310; 1:100; Dako;
Agilent Technologies, Inc.), CD8 (clone C8/144B; cat. no. M7103;
1:100; Dako; Agilent Technologies, Inc.), FOXP3 (clone 236A/E7;
cat. no. ab20034; 1:200; Abcam), PD-L1 (clone E1L3N; cat. no.
13684S; 1:400; Cell Signaling Technology, Inc.) and p-STAT1 (clone
D3B7; cat. no. 8826S; 1:800; Cell Signaling Technology, Inc.) at
4°C overnight. Following the primary antibody incubation, the
sections were incubated with horseradish peroxidase
(HRP)-conjugated anti-rabbit or anti-mouse secondary antibodies
(Envision + System-HRP; cat. no. K4003 or K4001; ready to use;
Dako; Agilent Technologies, Inc.) for 30 min at room temperature.
The sections were then stained with diaminobenzidine (Dojindo
Molecular Technologies, Inc,) at room temperature for 10 min and
subsequently counterstained with Mayer's hematoxylin solution (cat.
no. 131-09665; Wako Pure Chemical Industries, Ltd.) at room
temperature for 1 min. Sections in which the primary antibodies
were replaced with PBS were used as negative controls. Esophagus
and stomach cancer tissue sections served as positive controls
(23).
Assessment of IHC staining
IHC analysis was performed by four independent
observers (TK, KS, LY and EE) who were blinded to the clinical
data. The TILs at the invasive front region of the tumor were
reviewed in four independent areas using light microscope at a
magnification of ×400. PD-L1 expression was evaluated by assessing
membranous staining without cytoplasmic staining and tumor
specimens were considered to be PD-L1-positive when >1% of the
tumor cells exhibited membranous staining of any intensity
(24). p-STAT1 staining was
evaluated by assessing nuclear staining of the tumor cells.
Positive staining for p-STAT-1 was defined by the presence of
positive nuclear staining in tumor cells. CD4, CD8 and FOXP3
expression was evaluated by counting the number of stained
lymphocytes from four independent areas.
Determination of MMR status
IHC for MMR proteins, including mutL homolog 1, mutS
homolog 2 and 6, and PMS1 homolog 2 mismatch repair system
component, was performed as previously described (25). Loss of a MMR protein was defined as
the absence of nuclear staining of tumor cells in the presence of
positive nuclear staining of normal colon mucosa cells or stromal
cells. Tumors demonstrating the loss of at least one MMR protein
were designated as dMMR, and tumors with intact MMR protein
expression as pMMR.
Statistical analysis
An unpaired Student's t-test was used to compare two
groups and one-way analysis of variance followed by a Tukey's post
hoc test was used to compare multiple groups. The data are
presented as the means ± standard deviation. The correlation of
IFN-γ or CD8 T effector gene signatures with the expression of
PD-L1 and PD-L2, STAT1 and Janus-activated kinase (JAK) 1 or 2, and
the correlation of the IFN-γ gene signature with the CD8 T effector
gene signature were assessed using scatter diagrams and Pearson's
correlation test. Associations between p-STAT1 and PD-L1 expression
were assessed using the χ2 test. All statistical
analyses were conducted using GraphPad Prism software (version 7.0;
GraphPad Software, Inc.). P-values were two-sided and P<0.05 was
considered to indicate a statistically significant difference.
Results
A subset of patients with
MSI-L/MSS-CRC exhibit IFN-γ and CD8 T effector gene signature
upregulation
TCGA COADREAD dataset consisted of 342 samples,
including 51 MSI-H (14.9%) and 291 MSI-L/MSS (85.1%) tumor samples.
The mRNA expression levels of PD-L1 and PD-L2 were significantly
upregulated in MSI-H tumors compared with MSI-L/MSS tumors
(Fig. 1A). The IFN-γ gene signature
consisting of IDO1, CXCL10, CXCL9, STAT1 and IFNG,
and the CD8 T effector gene signature consisting of CD8A, CD8B,
EOMES, GZMA, GZMB, IFNG and PRF1 were subsequently
assessed. The two gene signatures were more prevalent in MSI-H
tumors compared with MSI-L/MSS tumors (Fig. 1A). There was a significant variation
in the IFN-γ gene signature in the MSS/MSI-L group. A subpopulation
with a high IFN-γ gene signature in the MSS/MSI-L group, with
expression levels of the IFN-γ gene signature similar to those in
the MSI-H group was identified (Fig.
1B). Similarly, there was a subset of patients with a high CD8
T effector gene signature in the MSS/MSI-L group, with expression
levels similar to those in the MSI-H group (Fig. 1B). The results obtained in the
current study suggest that there is a subpopulation of patients
with upregulated CD8 T effector and IFN-γ gene signatures in
MSI-L/MSS-CRC.
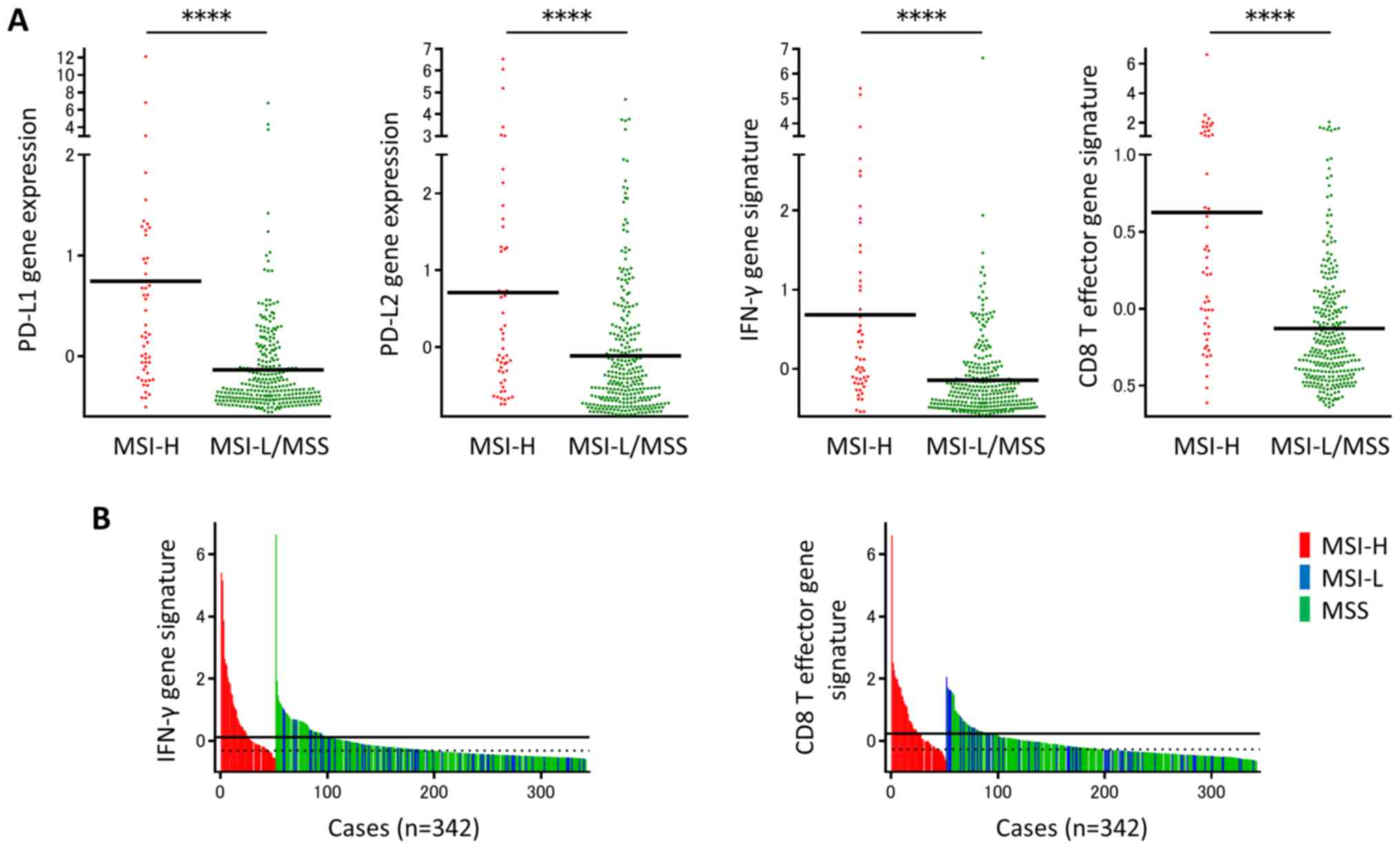 | Figure 1.A subset of patients with
MSI-L/MSS-CRC revealed upregulation of IFN-γ and CD8 T effector
gene signatures. (A) PD-L1 and PD-L2 gene expression levels, and
IFN-γ and CD8 T effector gene signatures in The Cancer Genome Atlas
colorectal adenocarcinoma tumors, according to MSI status (51 MSI-H
and 291 MSI-L/MSS tumors). (B) Subpopulations in MSI-L-/MSS-CRC
showed upregulation of IFN-γ and CD8 T effector gene signatures.
IFN-γ and CD8 T effector gene signatures were shown in 342
colorectal tumors in relation to MSI status. Individual samples are
represented as color bars (red, blue or green) to denote MSI-H,
MSI-L or MSS tumors, respectively. Black lines represent the median
of each signature in MSI-H tumors, and dotted lines represent the
median of each signature in MSI-L/MSS tumors. ****P<0.0001. MSI,
microsatellite instability; MSI-L, microsatellite instability-low;
MSS, microsatellite stable; CRC, colorectal cancer; IFN-γ,
interferon γ; CD, cluster of differentiation; PD-L, programmed cell
death ligand; MSI-H, microsatellite instability-high. |
Correlations of PD-L1 and PD-L2 with
the IFN-γ and CD8 T effector gene signatures
There were significant positive correlations between
the IFN-γ gene signature and PD-L1, between the IFN-γ gene
signature and PD-L2, between the CD8 T effector gene signature and
PD-L1, between the CD8 T effector gene signature and PD-L2, and
finally between the CD8 T effector and IFN-γ gene signatures in all
342 CRC cases (Fig. 2A). The current
study demonstrated that a subset of patients with CD8 T effector
and IFN-γ gene signature upregulation in the MSI-L/MSS-CRC group
exhibited upregulation of immune checkpoint molecules, including
PD-L1 and PD-L2.
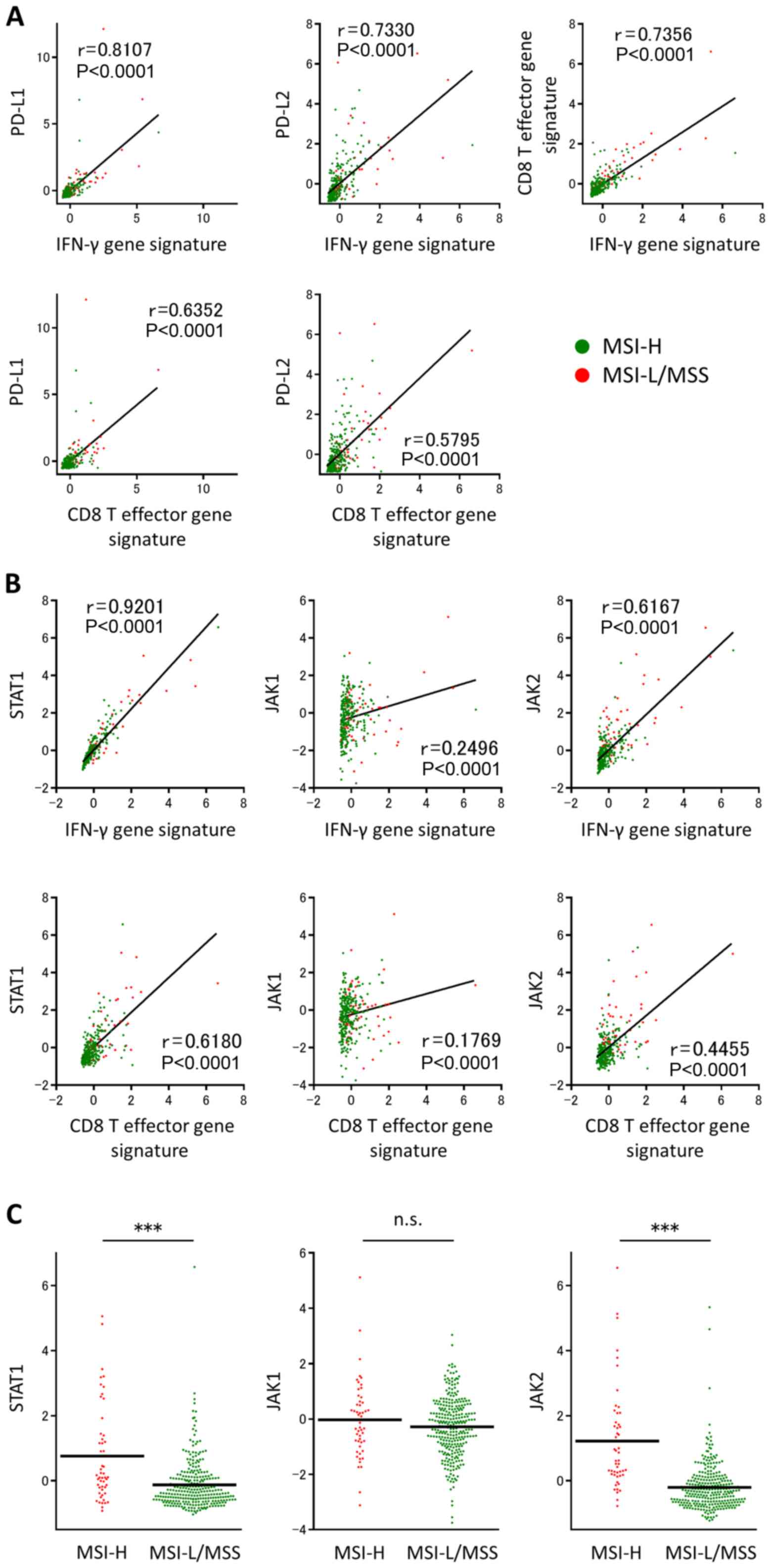 | Figure 2.Association of IFN-γ and CD8 T
effector gene signatures with the expression of PD-L1 or PD-L2 in
TCGA dataset. Individual samples are represented as red or green
dots, indicating MSI-H or MSI-L/MSS, respectively. (A) Significant
positive correlations were revealed between the IFN-γ or CD8 T
effector gene signatures and PD-L1 or PD-L2, and between the IFN-γ
and CD8 T effector gene signatures. (B) There were also significant
positive correlations between the IFN-γ or CD8 T effector gene
signatures and STAT1, JAK1 or JAK2. (C) STAT1, JAK1 and JAK2 gene
expression levels in TCGA colorectal adenocarcinoma tumors,
according to MSI status. ***P<0.001. CD, cluster of
differentiation; IFN-γ, interferon γ; PD-L, programmed cell death
ligand; TCGA, The Cancer Genome Atlas; STAT1, signal transducer and
activator of transcription 1; JAK, Janus-activated kinase; MSI,
microsatellite instability; MSI-H, MSI-high; MSI-L, MSI-low; MSS,
microsatellite stable. |
In the present study, there were significant
positive correlations between the IFN-γ or CD8 T effector gene
signatures and STAT1 or JAK1/2 (Fig.
2B). Furthermore, STAT1 and JAK2 mRNA expression levels were
significantly upregulated in MSI-H tumors compared with MSI-L/MSS
tumors (Fig. 2C). These findings
suggested that the activation of IFN-γ signaling is associated with
IFN-γ production and CD8 T cell infiltration within the tumor
microenvironment, and that IFN-γ signaling is significantly
activated in MSI-H tumors compared with MSI-L/MSS tumors. Since the
STAT1 mRNA expression level was correlated to the highest degree
with the IFN-γ and CD8 T effector gene signatures (Fig. 2B), and the binding of IFN-γ to its
cognate receptor leads to the phosphorylation-dependent activation
of STAT1 (26), the p-STAT1
expression level in clinical samples was analyzed to investigate
the effect of IFN-γ in the tumor microenvironment.
A subset of patients in the pMMR-CRC
group exhibit increased CD8 and CD4 infiltration
In the current study, a total of 219 patients
received colorectal surgery (Table
I). IHC was used to classify patients based on MMR protein
expression, and a total of 18 (8.2%) and 201 (91.8%) patients were
identified to have dMMR- and pMMR-CRC, respectively. Representative
IHC staining for MMR proteins are presented in Fig. 3A. Compared with TCGA COADREAD
database, the prevalence of dMMR-CRC in the current study was low
(14.9 vs. 8.2%). The immune status, including PD-L1 and p-STAT1 in
the tumors, and CD4(+), CD8(+) and FOXP3(+) TILs in the tumor
microenvironments, was evaluated by IHC as presented in Fig. 3B. There were significant associations
between PD-L1 expression and the grade of CD4(+) or CD8(+) TIL
infiltration; however, there was no significant association between
PD-L1 expression and FOXP3(+) TIL infiltration (Fig. 4A-C). Furthermore, there was a
tendency towards a positive association between PD-L1 and p-STAT1
expression levels (Fig. 4D). The
immune parameters of patients with dMMR- and pMMR-CRC were
compared, a subpopulation of PD-L1(+) and p-STAT1(+) patients with
pMMR-CRC (n=26) showed increasing grades of infiltrating CD4(+) or
CD8(+) TILs compared with pMMR/others including pMMR-CRC without a
subpopulation of PD-L1(+) and p-STAT1(+) patients, which were
similar to the levels seen in patients with dMMR-CRC (Fig. 4E and F). Although the number of
FOXP3(+) TILs was increased in patients with pMMR-CRC with PD-L1(+)
and p-STAT1(+) compared with patients with dMMR-CRC, there were no
significant difference between pMMR-CRC/others and dMMR- or
pMMR-CRC with PD-L1(+) and p-STAT1(+) regarding FOXP3 TIL
infiltration (Fig. 4G). The
aforementioned observations in the clinical cohort suggested that
there was a subset of patients with pMMR-CRC with elevated CD8(+)
and CD4(+) TIL infiltration.
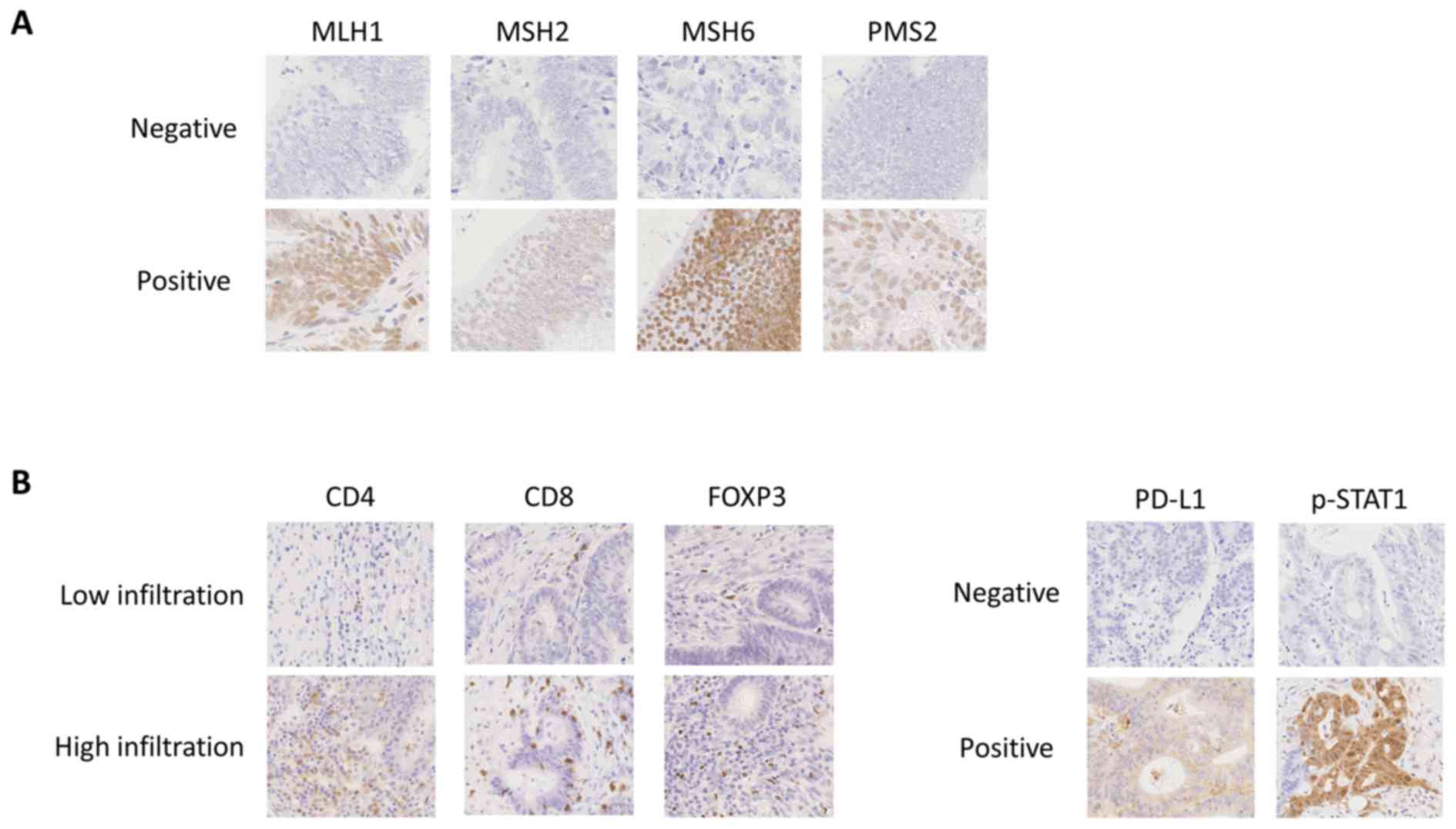 | Figure 3.Representative IHC staining. (A)
Representative IHC staining of MMR proteins including MLH1, MSH2,
MSH6 and PMS2. (B) Representative IHC staining of CD4, CD8, FOXP3,
PD-L1, and p-STAT1. Magnification, ×400. IHC, immunohistochemistry;
MMR, mismatch repair; CD, cluster of differentiation; MLH, mutL
homolog; MSH6, mutS homolog 6; PMS2, PMS1 homolog 2 mismatch repair
system component; FOXP3, forkhead box P3; PD-L1, programmed cell
death ligand 1; p-STAT1; phosphorylated-signal transducer and
activator of transcription 1. |
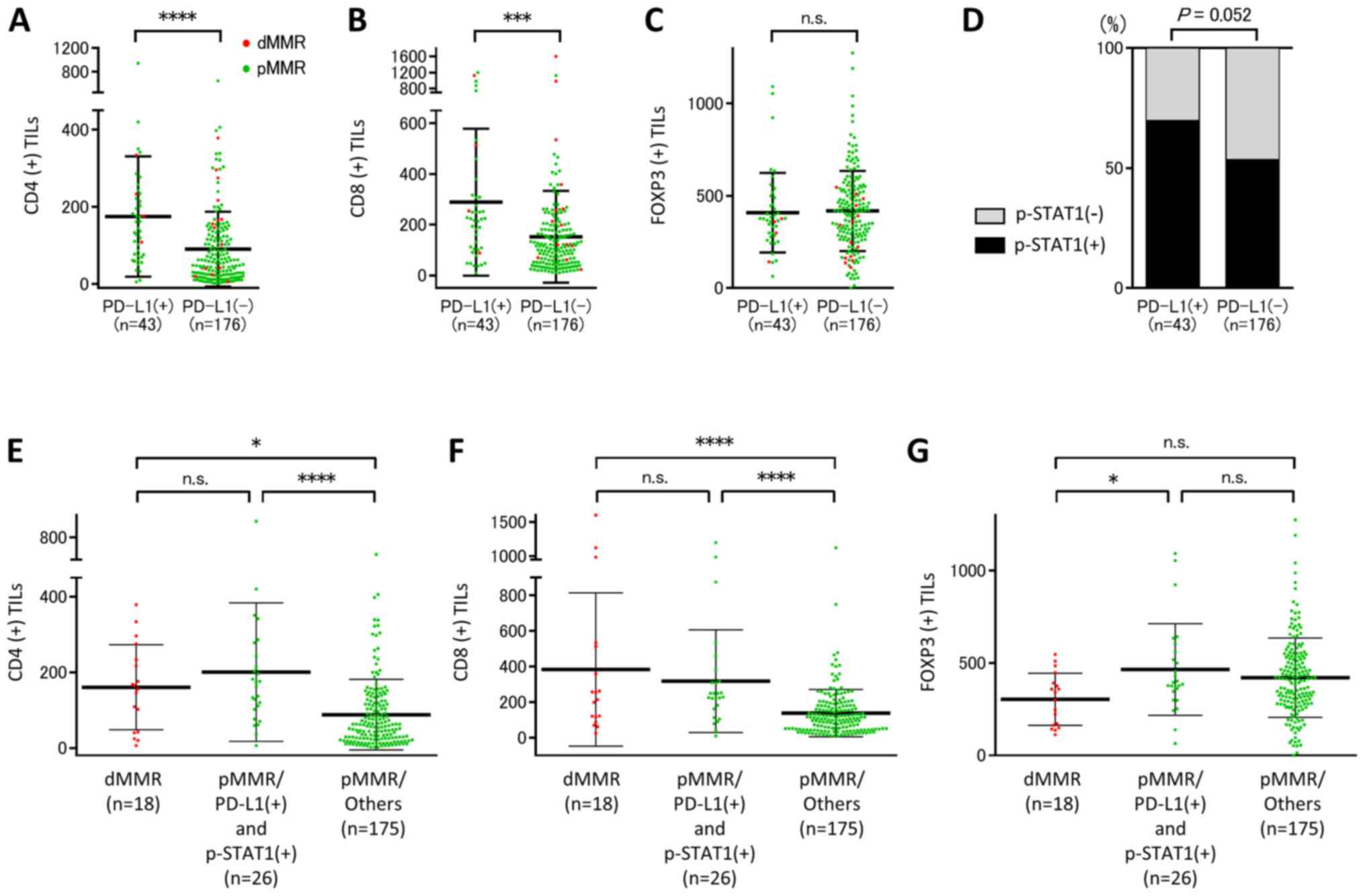 | Figure 4.A subset of patients with PD-L1(+)
and p-STAT1(+) pMMR-CRC exhibited increased CD4(+) and CD8(+) TIL
infiltration as seen in dMMR-CRC. In all patients with CRC, there
were significant positive correlations (A) between the number of
CD4(+) TILs and PD-L1 expression and (B) between the number of
CD8(+) TILs and PD-L1 expression. (C) There was no significant
difference between the number of FOXP3(+) TILs and PD-L1 expression
and (D) between p-STAT1 and PD-L1 expression on tumor cells. A
subpopulation of patients with pMMR-CRC with PD-L1(+) and
p-STAT1(+) exhibited increased (E) CD4(+) and (F) CD8(+) TILs,
which were similar to levels observed in patients with dMMR-CRC,
compared with patients with pMMR-CRC/others. (G) The number of
FOXP3(+) TILs was increased in a subpopulation of patients with
pMMR-CRC with PD-L1(+) and p-STAT1(+) compared with patients with
dMMR-CRC. Error bars represent the means ± SD. *P<0.05,
***P<0.001, ****P<0.0001. PD-L1, programmed cell death ligand
1; p-STAT1, phosphorylated-signal transducer and activator of
transcription 1; MMR, mismatch repair; pMMR, proficient MMR; CRC,
colorectal cancer; CD, cluster of differentiation; TIL, tumor
infiltrating lymphocyte; dMMR, deficient MMR; FOXP3, forkhead box
P3; n.s., not significant. |
Discussion
There are several potential biomarkers to predict
response to ICIs, including tumor mutation burden (TMB), MSI
status, PD-L1 expression, CD8 infiltration and the IFN-γ gene
signature (27–33). Several clinical studies have
established MSI status as a putative response biomarker for PD-1
blockade, and immunotherapy with ICIs against the PD-1/PD-L1 axis
is clinically approved for dMMR/MSI-H solid tumors of varying
histological origin (13,14,17).
dMMR/MSI-H tumors are thought to respond to ICIs for a number of
reasons. dMMR/MSI-H tumors have an antigenic microenvironment with
a high density of TILs and express multiple immune checkpoint
molecules, including PD-1 and PD-L1, due to mutation-induced
neo-antigens (3,5,16,17). The
present study used clinical samples and TCGA database to reveal
that dMMR/MSI-H tumors have immunogenic tumor microenvironments and
exhibit upregulated PD-L1 and PD-L2 expression levels, and an
increase in p-STAT1, and increased infiltration of CD8 T cells
compared with pMMR/MSS tumors, and that these factors are closely
associated with IFN-γ production within the tumor microenvironment.
Since the binding of IFN-γ to its cognate receptor leads to the
phosphorylation-dependent activation of JAK1/2 and STAT1, and that
p-STAT1 forms subsequently homodimers that translocate to the
nucleus and activate the transcription of IFN-γ-stimulated genes
(26), p-STAT1 expression was
investigated in the present study. These observations are in line
with previous studies investigating gastric cancer, which reported
that the upregulation of PD-L1 and PD-L2 is significantly
associated with IFN-γ production and CD8 T cell infiltration within
the tumor microenvironment (34,35).
However, previous studies revealed that in
pMMR/MSS-CRC, a small subset of patients may still benefit from
immunotherapy with anti-PD-1/PD-L1 antibodies (13–15).
Therefore, the identification of distinct biomarker-defined
populations with pMMR/MSS-CRC may improve patient outcome. The
present study revealed a subset of patients with MSI-L/MSS-CRC with
upregulated CD8 T effector and IFN-γ gene signatures, as seen in
MSI-H-CRC. Previous studies reported that the number of
CD8+ TILs was increased in MSI-CRC compared with MSS-CRC
(36–38). Also, De Smedt et al (36) used IHC to demonstrate that there were
no significant differences in the number of CD4(+) TILs in MSI- or
MSS-CRC; however, Boissière-Michot et al (37) reported that the density of CD4(+)
TILs was increased in MSI-CRC compared with MSS-CRC. Pagès et
al (38) revealed that 21% of
patients with MSS tumors had high immunoscore densities of
CD3+ and CD8+ TILs, similar to MSI tumors.
The present study identified a subset of patients with pMMR-CRC
with increased CD8(+) and CD4(+) TIL infiltration, as seen in
dMMR-CRC. The aforementioned patients may benefit from
immunotherapy with anti-PD-1/PD-L1 mAbs.
It is well known that regulatory T (Treg) cells
express FOXP3 and suppress antitumor immune responses (39–43). In
patients with CRC, the number of Treg cells (which suppress the
activity of tumor antigen-specific T cells) is increased in the
tumor microenvironment (44–46). Although several studies have
evaluated the association between Treg cells and MSI status, the
results were controversial (37,39–41,47).
Differences in these studies may be attributed to the use of
different methodologies, such as PCR and IHC, the types of lesions
analyzed, including invasive front and surface of tumor, and the
phenotypic diversity of Treg cells (48). Further investigation is required to
clarify the association between Treg cells and MSI or MMR status in
CRC.
A recent study demonstrated that in a large cohort
of patients with CRC, those with high TMB accounted for 3% of
MSS-CRC cases (49), indicating that
a subset of patients with MSS-CRC with high TMB may benefit from
treatment with ICIs. As the TMB status was not evaluated in the
present study, it is currently unclear whether the subpopulation of
patients with MSI-L/MSS-CRC with upregulated CD8 T effector and
IFN-γ gene signatures corresponds with high mutation burden groups.
Furthermore, conclusions about the responsiveness to ICI therapies
cannot be drawn in the current study. Further investigations to
clarify the association between TMB and immune profiling in
MSI-L/MSS-CRC, as well as the clinical response in the identified
subpopulation, are required.
While the MSS/TMB-high group may benefit from
treatment with ICIs, there are several unresolved issues to
accurately identify patients with MSS/TMB-high status. TMB analysis
from whole exome sequencing is not widely available, as it is a
time and cost intensive method (50,51).
More importantly, it has recently been shown that patients with
metastatic MSI-H/dMMR-CCR who exhibit primary resistance to ICIs
may have been misdiagnosed (52). In
other words, patients who had been initially diagnosed as MSI-H
were actually MSS, indicating that MSI-testing is currently not
accurate enough and not fully established. Furthermore, Wang et
al (53) reported that patients
with a polymerase ε-mutation present a hyper-mutated tumor
phenotype caused by high frequent base substitution mutations,
without necessarily giving rise to the short tandem repeat
signature identified through MSI-H testing. In light of the
aforementioned complexities, multiple predictive biomarkers are
required to accurately select ICI responders with MSS/pMMR-CRC. The
evaluation of CD8 T effector and IFN-γ gene signatures described in
the current study may be used as part of a combination testing
strategy for ICI responder identification.
The present study demonstrated that a subset of
patients with upregulated CD8 T effector and IFN-γ gene signatures
MSI-L/MSS- or pMMR-CRC may benefit from immunotherapy with
anti-PD-1/PD-L1 mAbs.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets generated and/or analyzed during the
current study are available in The Cancer Genome Atlas and were
downloaded via cBioPortal (www.cbioportal.org).
Authors' contributions
KM and KK conceived and designed the study. HO and
YN analyzed TCGA dataset. KS, LY, EE, WS, SF, HE, MS, TM, ZS and SO
contributed to the acquisition of the patient samples. TK, KS, LY
and EE performed and evaluated the IHC staining. TK, KM, HO, YN,
WS, SF, HE, MS, TM, ZS and SO analyzed the patient data. TK, HO, KM
and KK drafted the manuscript. All authors have read and approved
the final manuscript.
Ethics approval and consent to
participate
All procedures were conducted in accordance with the
Helsinki Declaration and were approved by the responsible committee
on human experimentation at Fukushima Medical University (Reference
2847). Written informed consent was obtained from all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
CRC
|
colorectal cancer
|
|
dMMR
|
deficient mismatch repair
|
|
ICIs
|
immune checkpoint inhibitors
|
|
IHC
|
Immunohistochemistry
|
|
IFN-γ
|
interferon-γ
|
|
JAK
|
Janus-activated kinase
|
|
MSI-H
|
microsatellite instability-high
|
|
MSI-L
|
MSI-low
|
|
MSS
|
microsatellite stable
|
|
mAbs
|
monoclonal antibodies
|
|
p-STAT1
|
phosphorylated signal transducer and
activator of transcription 1
|
|
pMMR
|
proficient MMR
|
|
PD-1
|
programmed cell death 1
|
|
PD-L1
|
programmed death ligand 1
|
|
Treg
|
regulatory T
|
|
TCGA
|
The Cancer Genome Atlas
|
|
TILs
|
tumor infiltrating lymphocytes
|
|
TMB
|
tumor mutation burden
|
References
|
1
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Brenner H, Kloor M and Pox CP: Colorectal
cancer. Lancet. 383:1490–1502. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Cancer Genome Atlas Network, .
Comprehensive molecular characterization of human colon and rectal
cancer. Nature. 487:330–337. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kloor M and von Knebel Doeberitz M: The
immune biology of microsatellite-unstable cancer. Trends Cancer.
2:121–133. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Koopman M, Kortman GA, Mekenkamp L,
Ligtenberg MJ, Hoogerbrugge N, Antonini NF, Punt CJ and van Krieken
JH: Deficient mismatch repair system in patients with sporadic
advanced colorectal cancer. Br J Cancer. 100:266–273. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sargent DJ, Marsoni S, Monges G, Thibodeau
SN, Labianca R, Hamilton SR, French AJ, Kabat B, Foster NR, Torri
V, et al: Defective mismatch repair as a predictive marker for lack
of efficacy of fluorouracil-based adjuvant therapy in colon cancer.
J Clin Oncol. 28:3219–3226. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Borghaei H, Paz-Ares L, Horn L, Spigel DR,
Steins M, Ready NE, Chow LQ, Vokes EE, Felip E, Holgado E, et al:
Nivolumab versus docetaxel in advanced nonsquamous non-small-cell
lung cancer. N Engl J Med. 373:1627–1639. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kang YK, Boku N, Satoh T, Ryu MH, Chao Y,
Kato K, Chung HC, Chen JS, Muro K, Kang WK, et al: Nivolumab in
patients with advanced gastric or gastro-oesophageal junction
cancer refractory to, or intolerant of, at least two previous
chemotherapy regimens (ONO-4538-12, ATTRACTION-2): A randomised,
double-blind, placebo-controlled, phase 3 trial. Lancet.
390:2461–2471. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Escudier B, Motzer RJ, Sharma P, Wagstaff
J, Plimack ER, Hammers HJ, Donskov F, Gurney H, Sosman JA, Zalewski
PG, et al: Treatment beyond progression in patients with advanced
renal cell carcinoma treated with nivolumab in CheckMate 025. Eur
Urol. 72:368–376. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Brahmer JR, Tykodi SS, Chow LQ, Hwu WJ,
Topalian SL, Hwu P, Drake CG, Camacho LH, Kauh J, Odunsi K, et al:
Safety and activity of anti-PD-L1 antibody in patients with
advanced cancer. N Engl J Med. 366:2455–2465. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Topalian SL, Hodi FS, Brahmer JR,
Gettinger SN, Smith DC, McDermott DF, Powderly JD, Carvajal RD,
Sosman JA, Atkins MB, et al: Safety, activity, and immune
correlates of anti-PD-1 antibody in cancer. N Engl J Med.
366:2443–2454. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Le DT, Uram JN, Wang H, Bartlett BR,
Kemberling H, Eyring AD, Skora AD, Luber BS, Azad NS, Laheru D, et
al: PD-1 blockade in tumors with mismatch-repair deficiency. N Engl
J Med. 372:2509–2520. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Overman MJ, McDermott R, Leach JL, Lonardi
S, Lenz HJ, Morse MA, Desai J, Hill A, Axelson M, Moss RA, et al:
Nivolumab in patients with metastatic DNA mismatch repair-deficient
or microsatellite instability-high colorectal cancer (CheckMate
142): An open-label, multicentre, phase 2 study. Lancet Oncol.
18:1182–1191. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Le DT, Durham JN, Smith KN, Wang H,
Bartlett BR, Aulakh LK, Lu S, Kemberling H, Wilt C, Luber BS, et
al: Mismatch repair deficiency predicts response of solid tumors to
PD-1 blockade. Science. 357:409–413. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Bauer K, Nelius N, Reuschenbach M, Koch M,
Weitz J, Steinert G, Kopitz J, Beckhove P, Tariverdian M, von
Knebel Doeberitz M and Kloor M: T cell responses against
microsatellite instability-induced frameshift peptides and
influence of regulatory T cells in colorectal cancer. Cancer
Immunol Immunother. 62:27–37. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Dudley JC, Lin MT, Le DT and Eshleman JR:
Microsatellite instability as a biomarker for PD-1 blockade. Clin
Cancer Res. 22:813–820. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Gao J, Aksoy BA, Dogrusoz U, Dresdner G,
Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al:
Integrative analysis of complex cancer genomics and clinical
profiles using the cBioPortal. Sci Signal. 6:pl12013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liu Y, Sethi NS, Hinoue T, Schneider BG,
Cherniack AD, Sanchez-Vega F, Seoane JA, Farshidfar F, Bowlby R,
Islam M, et al: Comparative molecular analysis of gastrointestinal
adenocarcinomas. Cancer Cell. 33:721–735.e8. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wallin JJ, Bendell JC, Funke R, Sznol M,
Korski K, Jones S, Hernandez G, Mier J, He X, Hodi FS, et al:
Atezolizumab in combination with bevacizumab enhances
antigen-specific T-cell migration in metastatic renal cell
carcinoma. Nat Commun. 7:126242016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ayers M, Lunceford J, Nebozhyn M, Murphy
E, Loboda A, Kaufman DR, Albright A, Cheng JD, Kang SP, Shankaran
V, et al: IFN-γ-related mRNA profile predicts clinical response to
PD-1 blockade. J Clin Invest. 127:2930–2940. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Nakayama Y, Mimura K, Tamaki T, Shiraishi
K, Kua LF, Koh V, Ohmori M, Kimura A, Inoue S, Okayama H, et al:
Phospho-STAT1 expression as a potential biomarker for
anti-PD-1/anti-PD-L1 immunotherapy for breast cancer. Int J Oncol.
54:2030–2038. 2019.PubMed/NCBI
|
|
23
|
Ashizawa M, Okayama H, Ishigame T, Thar
Min AK, Saito K, Ujiie D, Murakami Y, Kikuchi T, Nakayama Y, Noda
M, et al: miRNA-148a-3p regulates immunosuppression in DNA mismatch
repair-deficient colorectal cancer by targeting PD-L1. Mol Cancer
Res. 17:1403–1413. 2019.PubMed/NCBI
|
|
24
|
Herbst RS, Baas P, Kim DW, Felip E,
Pérez-Gracia JL, Han JY, Molina J, Kim JH, Arvis CD, Ahn MJ, et al:
Pembrolizumab versus docetaxel for previously treated,
PD-L1-positive, advanced non-small-cell lung cancer (KEYNOTE-010):
A randomised controlled trial. Lancet. 387:1540–1550. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Noda M, Okayama H, Tachibana K, Sakamoto
W, Saito K, Thar Min AK, Ashizawa M, Nakajima T, Aoto K, Momma T,
et al: Glycosyltransferase gene expression identifies a poor
prognostic colorectal cancer subtype associated with mismatch
repair deficiency and incomplete glycan synthesis. Clin Cancer Res.
24:4468–4481. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Khodarev NN, Roizman B and Weichselbaum
RR: Molecular pathways: Interferon/stat1 pathway: Role in the tumor
resistance to genotoxic stress and aggressive growth. Clin Cancer
Res. 18:3015–3021. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
McGranahan N, Furness AJ, Rosenthal R,
Ramskov S, Lyngaa R, Saini SK, Jamal-Hanjani M, Wilson GA, Birkbak
NJ, Hiley CT, et al: Clonal neoantigens elicit T cell
immunoreactivity and sensitivity to immune checkpoint blockade.
Science. 351:1463–1469. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Postow MA, Callahan MK and Wolchok JD:
Immune checkpoint blockade in cancer therapy. J Clin Oncol.
33:1974–1982. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
McLaughlin J, Han G, Schalper KA,
Carvajal-Hausdorf D, Pelekanou V, Rehman J, Velcheti V, Herbst R,
LoRusso P and Rimm DL: Quantitative assessment of the heterogeneity
of PD-L1 expression in non-small-cell lung cancer. JAMA Oncol.
2:46–54. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Bhaijee F and Anders RA: PD-L1 expression
as a predictive biomarker: Is absence of proof the same as proof of
absence? JAMA Oncol. 2:54–55. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Schalper KA, Kaftan E and Herbst RS:
Predictive biomarkers for PD-1 axis therapies: The hidden treasure
or a call for research. Clin Cancer Res. 22:2102–2104. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Nishino M, Ramaiya NH, Hatabu H and Hodi
FS: Monitoring immune-checkpoint blockade: Response evaluation and
biomarker development. Nat Rev Clin Oncol. 14:655–668. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Gibney GT, Weiner LM and Atkins MB:
Predictive biomarkers for checkpoint inhibitor-based immunotherapy.
Lancet Oncol. 17:e542–e551. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Mimura K, Kua LF, Shiraishi K, Kee Siang
L, Shabbir A, Komachi M, Suzuki Y, Nakano T, Yong WP, So J and Kono
K: Inhibition of mitogen-activated protein kinase pathway can
induce upregulation of human leukocyte antigen class I without
PD-L1-upregulation in contrast to interferon-gamma treatment.
Cancer Sci. 105:1236–1244. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Mimura K, Teh JL, Okayama H, Shiraishi K,
Kua LF, Koh V, Smoot DT, Ashktorab H, Oike T, Suzuki Y, et al:
PD-L1 expression is mainly regulated by interferon gamma associated
with JAK-STAT pathway in gastric cancer. Cancer Sci. 109:43–53.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
De Smedt L, Lemahieu J, Palmans S, Govaere
O, Tousseyn T, Van Cutsem E, Prenen H, Tejpar S, Spaepen M,
Matthijs G, et al: Microsatellite instable vs. stable colon
carcinomas: Analysis of tumour heterogeneity, inflammation and
angiogenesis. Br J Cancer. 113:500–509. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Boissière-Michot F, Lazennec G, Frugier H,
Jarlier M, Roca L, Duffour J, Du Paty E, Laune D, Blanchard F, Le
Pessot F, et al: Characterization of an adaptive immune response in
microsatellite-instable colorectal cancer. Oncoimmunology.
3:e292562014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Pagès F, Mlecnik B, Marliot F, Bindea G,
Ou FS, Bifulco C, Lugli A, Zlobec I, Rau TT, Berger MD, et al:
International validation of the consensus Immunoscore for the
classification of colon cancer: A prognostic and accuracy study.
Lancet. 391:2128–2139. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Le Gouvello S, Bastuji-Garin S, Aloulou N,
Mansour H, Chaumette MT, Berrehar F, Seikour A, Charachon A, Karoui
M, Leroy K, et al: High prevalence of Foxp3 and IL17 in
MMR-proficient colorectal carcinomas. Gut. 57:772–779. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Michel S, Benner A, Tariverdian M,
Wentzensen N, Hoefler P, Pommerencke T, Grabe N, von Knebel
Doeberitz M and Kloor M: High density of FOXP3-positive T cells
infiltrating colorectal cancers with microsatellite instability. Br
J Cancer. 99:1867–1873. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Salama P, Phillips M, Grieu F, Morris M,
Zeps N, Joseph D, Platell C and Iacopetta B: Tumor-infiltrating
FOXP3+ T regulatory cells show strong prognostic significance in
colorectal cancer. J Clin Oncol. 27:186–192. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Blatner NR, Bonertz A, Beckhove P, Cheon
EC, Krantz SB, Strouch M, Weitz J, Koch M, Halverson AL, Bentrem DJ
and Khazaie K: In colorectal cancer mast cells contribute to
systemic regulatory T-cell dysfunction. Proc Natl Acad Sci USA.
107:6430–6435. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Frey DM, Droeser RA, Viehl CT, Zlobec I,
Lugli A, Zingg U, Oertli D, Kettelhack C, Terracciano L and
Tornillo L: High frequency of tumor-infiltrating FOXP3(+)
regulatory T cells predicts improved survival in mismatch
repair-proficient colorectal cancer patients. Int J Cancer.
126:2635–2643. 2010.PubMed/NCBI
|
|
44
|
Clarke SL, Betts GJ, Plant A, Wright KL,
El-Shanawany TM, Harrop R, Torkington J, Rees BI, Williams GT,
Gallimore AM and Godkin AJ: CD4+CD25+FOXP3+ regulatory T cells
suppress anti-tumor immune responses in patients with colorectal
cancer. PLoS One. 1:e1292006. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Loddenkemper C, Schernus M, Noutsias M,
Stein H, Thiel E and Nagorsen D: In situ analysis of FOXP3+
regulatory T cells in human colorectal cancer. J Transl Med.
4:522006. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Ling KL, Pratap SE, Bates GJ, Singh B,
Mortensen NJ, George BD, Warren BF, Piris J, Roncador G, Fox SB, et
al: Increased frequency of regulatory T cells in peripheral blood
and tumour infiltrating lymphocytes in colorectal cancer patients.
Cancer Immun. 7:72007.PubMed/NCBI
|
|
47
|
Masugi Y, Nishihara R, Yang J, Mima K, da
Silva A, Shi Y, Inamura K, Cao Y, Song M, Nowak JA, et al: Tumour
CD274 (PD-L1) expression and T cells in colorectal cancer. Gut.
66:1463–1473. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Liston A and Gray DH: Homeostatic control
of regulatory T cell diversity. Nat Rev Immunol. 14:154–165. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Fabrizio DA, George TJ Jr, Dunne RF,
Frampton G, Sun J, Gowen K, Kennedy M, Greenbowe J, Schrock AB,
Hezel AF, et al: Beyond microsatellite testing: Assessment of tumor
mutational burden identifies subsets of colorectal cancer who may
respond to immune checkpoint inhibition. J Gastrointest Oncol.
9:610–617. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Wang Z, Duan J, Cai S, Han M, Dong H, Zhao
J, Zhu B, Wang S, Zhuo M, Sun J, et al: Assessment of blood tumor
mutational burden as a potential biomarker for immunotherapy in
patients with non-small cell lung cancer with use of a
next-generation sequencing cancer gene panel. JAMA Oncol.
5:696–702. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Chan TA, Yarchoan M, Jaffee E, Swanton C,
Quezada SA, Stenzinger A and Peters S: Development of tumor
mutation burden as an immunotherapy biomarker: Utility for the
oncology clinic. Ann Oncol. 30:44–56. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Cohen R, Hain E, Buhard O, Guilloux A,
Bardier A, Kaci R, Bertheau P, Renaud F, Bibeau F, Fléjou JF, et
al: Association of primary resistance to immune checkpoint
inhibitors in metastatic colorectal cancer with misdiagnosis of
microsatellite instability or mismatch repair deficiency status.
JAMA Oncol. 5:551–555. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Wang C, Gong J, Tu TY, Lee PP and Fakih M:
Immune profiling of microsatellite instability-high and polymerase
epsilon (POLE)-mutated metastatic colorectal tumors identifies
predictors of response to anti-PD-1 therapy. J Gastrointest Oncol.
9:404–415. 2018. View Article : Google Scholar : PubMed/NCBI
|


















