|
1
|
Hu T and Xi J: Identification of COX5B as
a novel biomarker in high-grade glioma patients. Onco Targets Ther.
10:5463–5470. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Song X, Zhang N, Han P, Moon BS, Lai RK,
Wang K and Lu W: Circular RNA profile in gliomas revealed by
identification tool UROBORUS. Nucleic Acids Res. 44:e872016.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Louis DN, Perry A, Reifenberger G, von
Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD,
Kleihues P and Ellison DW: The 2016 World Health Organization
classification of tumors of the central nervous system: A summary.
Acta Neuropathol. 131:803–820. 2016. View Article : Google Scholar : PubMed/NCBI
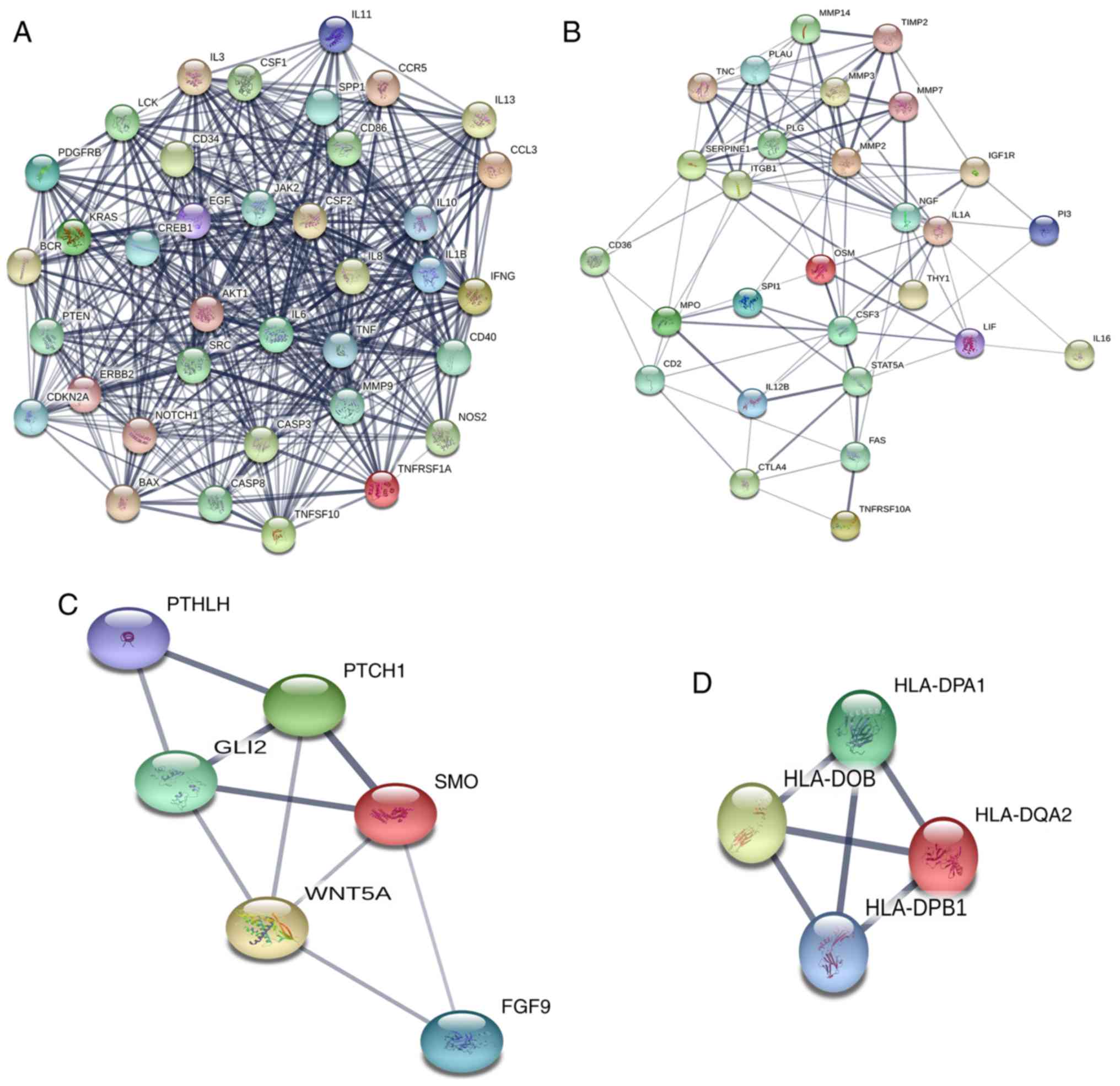
|
|
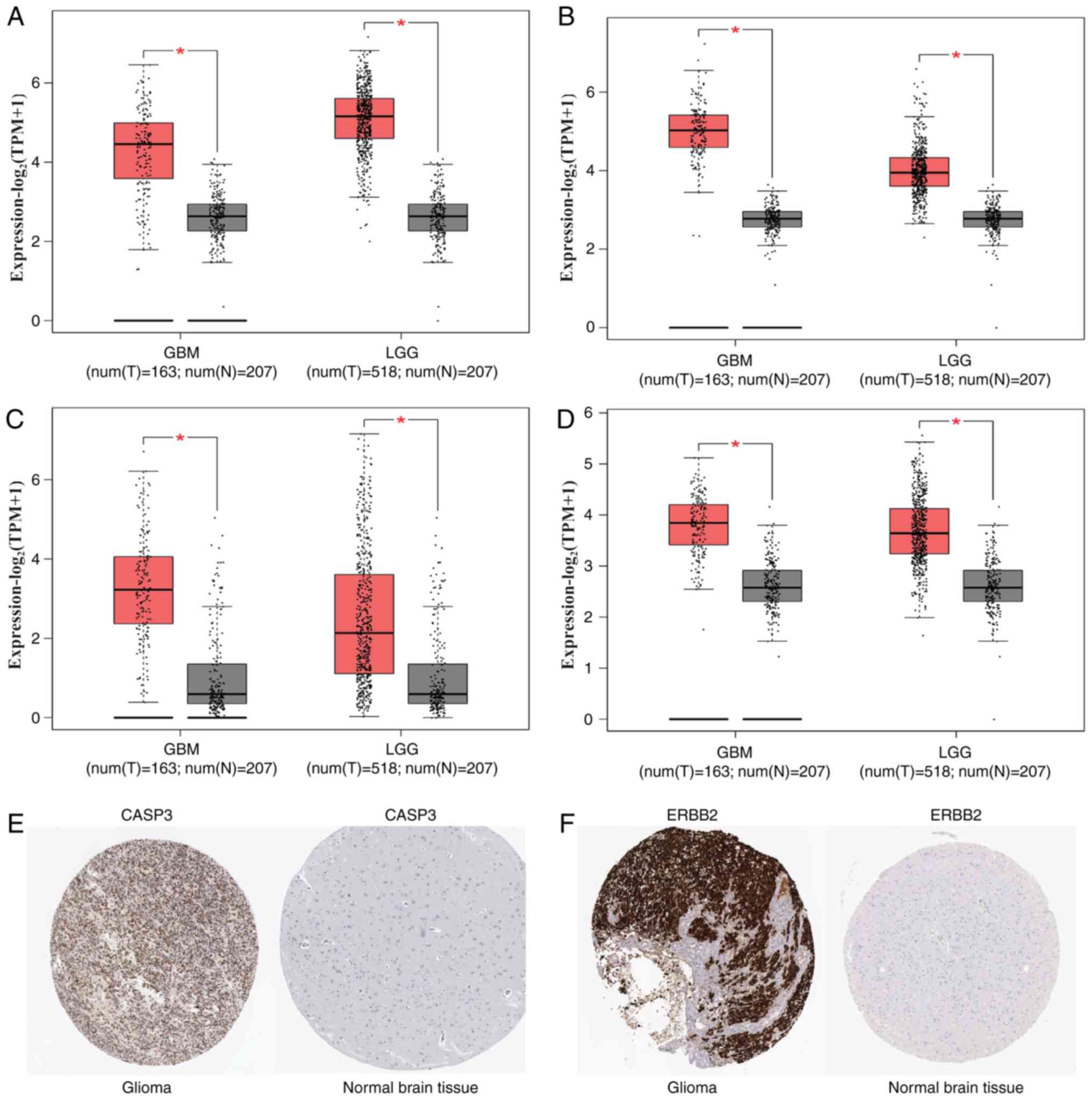
4
|
Komotar RJ, Otten ML, Gaetan M and
Connolly ES Jr: Radiotherapy plus concomitant and adjuvant
temozolomide for glioblastoma-a critical review. Clin Med Oncol.
2:421–422. 2008.PubMed/NCBI
|
|
5
|
Sturm D, Pfister SM and Jones DTW:
Pediatric gliomas: Current concepts on diagnosis, biology, and
clinical management. J Clin Oncol. 35:2370–2377. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Duffau H: Paradoxes of evidence-based
medicine in lower-grade glioma: To treat the tumor or the patient?
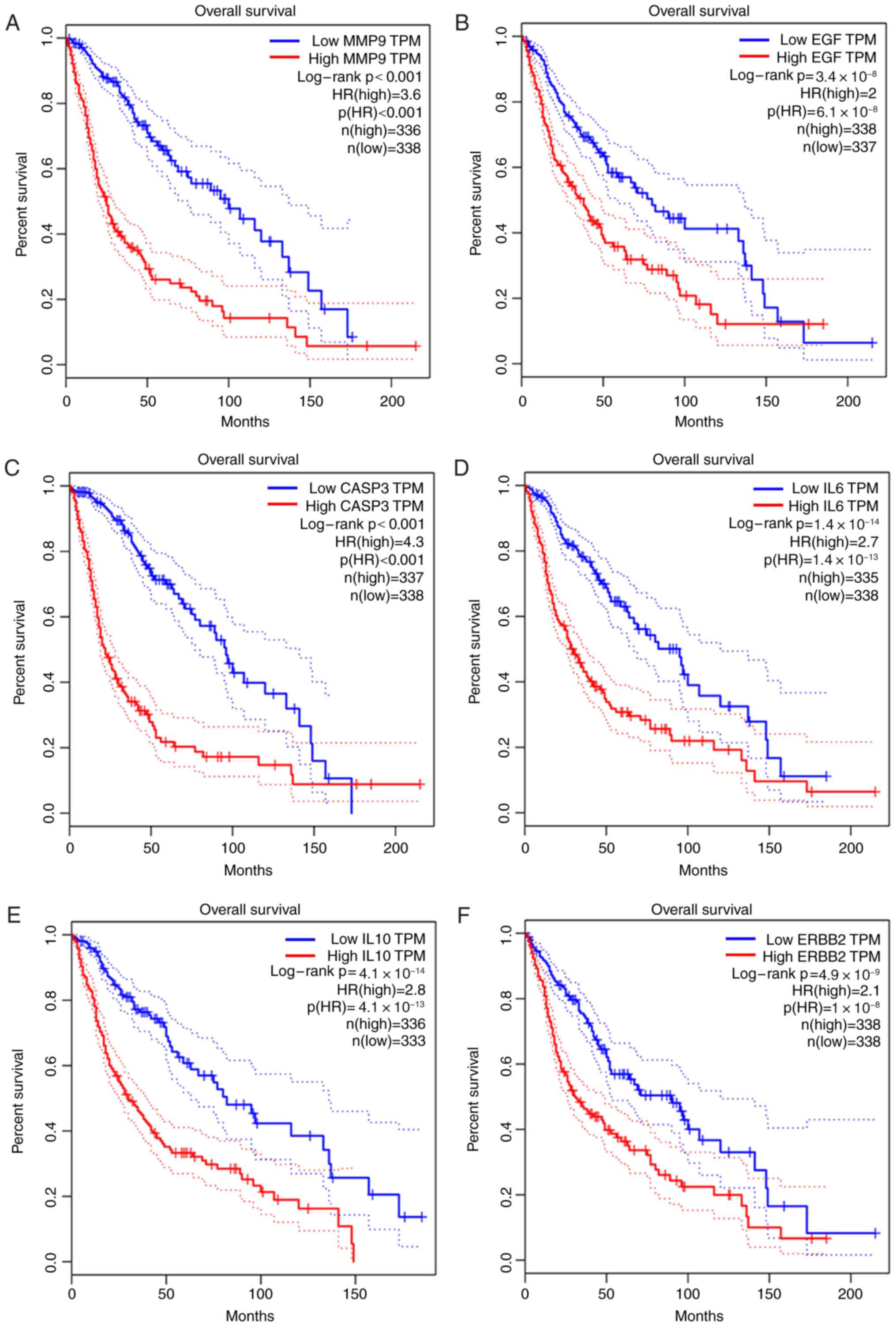
Neurology. 91:657–662. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Demuth T and Berens ME: Molecular
mechanisms of glioma cell migration and invasion. J Neurooncol.
70:217–228. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yang AS, Estecio MR, Garcia-Manero G,
Kantarjian HM and Issa JP: Comment on ‘Chromosomal instability and
tumors promoted by DNA hypomethylation’ and ‘Induction of tumors in
nice by genomic hypomethylation’. Science. 302:11532003. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Manel E: CpG island hypermethylation and
tumor suppressor genes: A booming present, a brighter future.
Oncogene. 21:5427–5440. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Feinberg AP and Vogelstein B:
Hypomethylation of ras oncogenes in primary human cancers. Biochem
Biophys Res Commun. 111:47–54. 1983. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang L, Wang M, Wang W and Mo J:
Incidence and prognostic value of multiple gene promoter
methylations in gliomas. J Neurooncol. 116:349–356. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Esteller M, Garcia-Foncillas J, Andion E,
Goodman SN, Hidalgo OF, Vanaclocha V, Baylin SB and Herman JG:
Inactivation of the DNA-repair gene MGMT and the clinical response
of gliomas to alkylating agents. N Engl J Med. 343:1350–1354. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li X, Pu J, Liu J, Chen Y, Li Y, Hou P,
Shi B and Yang Q: The prognostic value of DAPK1 hypermethylation in
gliomas: A site-specific analysis. Pathol Res Pract. 214:940–948.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Arifin MT, Hama S, Kajiwara Y, Sugiyama K,
Saito T, Matsuura S, Yamasaki F, Arita K and Kurisu K: Cytoplasmic,
but not nuclear, p16 expression may signal poor prognosis in
high-grade astrocytomas. J Neurooncol. 77:273–277. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Watanabe T, Katayama Y, Yoshino A, Yachi
K, Ohta T, Ogino A, Komine C and Fukushima T: Aberrant
hypermethylation of p14ARF and O6-methylguanine-DNA
methyltransferase genes in astrocytoma progression. Brain Pathol.
17:5–10. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Darnay BG, Besse A, Poblenz AT, Lamothe B
and Jacoby JJ: TRAFs in RANK Signaling. Adv Exp Med Biol.
597:152–159. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Berdasco M, Ropero S, Setien F, Fraga MF,
Lapunzina P, Losson R, Alaminos M, Cheung NK, Rahman N and Esteller
M: Epigenetic inactivation of the Sotos overgrowth syndrome gene
histone methyltransferase NSD1 in human neuroblastoma and glioma.
Proc Natl Acad Sci USA. 106:21830–21835. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Pan Q, Shai O, Lee LJ, Frey BJ and
Blencowe BJ: Deep surveying of alternative splicing complexity in
the human transcriptome by high-throughput sequencing. Nat Genet.
40:1413–1415. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
19
|
Fernandez AF, Assenov Y, Martin-Subero JI,
Balint B, Siebert R, Taniguchi H, Yamamoto H, Hidalgo M, Tan AC,
Galm O, et al: A DNA methylation fingerprint of 1628 human samples.
Genome Res. 22:407–419. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Davis S and Meltzer PS: GEOquery: A bridge
between the Gene Expression Omnibus (GEO) and BioConductor.
Bioinformatics. 23:1846–1847. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: Tool for the unification of biology. The Gene
Ontology Consortium. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kanehisa M and Goto S: KEGG: Kyoto
Encyclopaedia of Genes and Genomes. Nucleic Acids Res. 28:27–30.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ono K, Muetze T, Kolishovski G, Shannon P
and Demchak B: CyREST: Turbocharging cytoscape access for external
tools via a RESTful API. F1000Res. 4:4782015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Bader GD and Hogue CW: An automated method
for finding molecular complexes in large protein interaction
networks. BMC Bioinformatics. 4:22003. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res. 45:W98–W102.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ostrom QT, Gittleman H, Farah P, Ondracek
A, Chen Y, Wolinsky Y, Stroup NE, Kruchko C and Barnholtz-Sloan JS:
CBTRUS statistical report: Primary brain and central nervous system
tumors diagnosed in the United States in 2006–2010. Neuro Oncol. 15
(Suppl 2):ii1–ii56. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Alexander BM and Cloughesy TF: Adult
Glioblastoma. J Clin Oncol. 35:2402–2409. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Bralten LB and French PJ: Genetic
alterations in glioma. Cancers (Basel). 3:1129–1140. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Gaudet F, Hodgson JG, Eden A,
Jackson-Grusby L, Dausman J, Gray JW, Leonhardt H and Jaenisch R:
Induction of tumors in mice by genomic hypomethylation. Science.
300:489–492. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Győrffy B, Bottai G, Fleischer T, Munkácsy
G, Budczies J, Paladini L, Børresen-Dale AL, Kristensen VN and
Santarpia L: Aberrant DNA methylation impacts gene expression and
prognosis in breast cancer subtypes. Int J Cancer. 38:87–97. 2016.
View Article : Google Scholar
|
|
31
|
Dai D, Zhou B, Xu W, Jin H and Wang X:
CHFR promoter hypermethylation is associated with gastric cancer
and plays a protective role in gastric cancer process. J Cancer.
10:949–956. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wang ZL, Zhang CB, Cai JQ, Li QB, Wang Z
and Jiang T: Integrated analysis of genome-wide DNA methylation,
gene expression and protein expression profiles in molecular
subtypes of WHO II–IV gliomas. J Exp Clin Cancer Res. 34:1272015.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Johnstone RW, Ruefli AA and Lowe SW:
Apoptosis: A link between cancer genetics and chemotherapy. Cell.
108:153–164. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Schlabach MR, Luo J, Solimini NL, Hu G, Xu
Q, Li MZ, Zhao Z, Smogorzewska A, Sowa ME, Ang XL, et al: Cancer
proliferation gene discovery through functional genomics. Science.
319:620–624. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Lu P, Weaver VM and Werb Z: The
extracellular matrix: A dynamic niche in cancer progression. J Cell
Biol. 196:395–406. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Chen DS and Mellman I: Oncology meets
immunology: The cancer-immunity cycle. Immunity. 39:1–10. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Lippitz BE: Cytokine patterns in patients
with cancer: A systematic review. Lancet Oncol. 14:e218–e228. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Fresno Vara JA, Casado E, de Castro J,
Cejas P, Belda-Iniesta C and González-Barón M: PI3K/Akt signalling
pathway and cancer. Cancer Treat Rev. 30:193–204. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Majewska E and Szeliga M: AKT/GSK3β
signaling in glioblastoma. Neurochem Res. 42:918–924. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Burdon T, Smith A and Savatier P:
Signalling, cell cycle and pluripotency in embryonic stem cells.
Trends Cell Biol. 12:432–438. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Doi A, Park IH, Wen B, Murakami P, Aryee
MJ, Irizarry R, Herb B, Ladd-Acosta C, Rho J, Loewer S, et al:
Differential methylation of tissue- and cancer-specific CpG island
shores distinguishes human induced pluripotent stem cells,
embryonic stem cells and fibroblasts. Nat Genet. 41:1350–1353.
2009. View
Article : Google Scholar : PubMed/NCBI
|
|
42
|
Sood AK, Coffin JE, Schneider GB, Fletcher
MS, DeYoung BR, Gruman LM, Gershenson DM, Schaller MD and Hendrix
MJ: Biological significance of focal adhesion kinase in ovarian
cancer: Role in migration and invasion. Am J Pathol. 165:1087–1095.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Luo M and Guan JL: Focal adhesion kinase:
A prominent determinant in breast cancer initiation, progression
and metastasis. Cancer Lett. 289:127–139. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Cao F, Zhang Q, Chen W, Han C, He Y, Ran Q
and Yao S: IL-6 increases SDCBP expression, cell proliferation, and
cell invasion by activating JAK2/STAT3 in human glioma cells. Am J
Transl Res. 9:4617–4626. 2017.PubMed/NCBI
|
|
45
|
Samaras V, Piperi C, Korkolopoulou P,
Zisakis A, Levidou G, Themistocleous MS, Boviatsis EI, Sakas DE,
Lea RW, Kalofoutis A and Patsouris E: Application of the ELISPOT
method for comparative analysis of interleukin (IL)-6 and IL-10
secretion in peripheral blood of patients with astroglial tumors.
Mol Cell Biochem. 304:343–351. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Xue Q, Cao L, Chen XY, Zhao J, Gao L, Li
SZ and Fei Z: High expression of MMP9 in glioma affects cell
proliferation and is associated with patient survival rates. Oncol
Lett. 13:1325–1330. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Carpten JD, Faber AL, Horn C, Donoho GP,
Briggs SL, Robbins CM, Hostetter G, Boguslawski S, Moses TY, Savage
S, et al: A transforming mutation in the pleckstrin homology domain
of AKT1 in cancer. Nature. 448:439–444. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Holland EC, Celestino J, Dai C, Schaefer
L, Sawaya RE and Fuller GN: Combined activation of Ras and Akt in
neural progenitors induces glioblastoma formation in mice. Nat
Genet. 25:55–57. 2000. View
Article : Google Scholar : PubMed/NCBI
|
|
49
|
Kjær IM, Olsen DA, Alnor A, Brandslund I,
Bechmann T and Madsen JS: EGFR and EGFR ligands in serum in healthy
women; reference intervals and age dependency. Clin Chem Lab Med.
Jul 16–2019.doi: 10.1515/cclm-2019-0376 (Epub ahead of print).
View Article : Google Scholar
|
|
50
|
Thorne AH, Zanca C and Furnari F:
Epidermal growth factor receptor targeting and challenges in
glioblastoma. Neuro Oncol. 18:914–918. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Hai L, Zhang C, Li T, Zhou X, Liu B, Li S,
Zhu M, Lin Y, Yu S, Zhang K, et al: Notch1 is a prognostic factor
that is distinctly activated in the classical and proneural subtype
of glioblastoma and that promotes glioma cell survival via the
NF-κB(p65) pathway. Cell Death Dis. 9:1582018. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Sarkar S, Mirzaei R, Zemp FJ, Wei W,
Senger DL, Robbins SM and Yong VW: Activation of NOTCH signaling by
Tenascin-C promotes growth of human brain tumor-initiating cells.
Cancer Res. 77:3231–3243. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Li MY, Peng WH, Wu CH, Chang YM, Lin YL,
Chang GD, Wu HC and Chen GC: PTPN3 suppresses lung cancer cell
invasiveness by counteracting Src-mediated DAAM1 activation and
actin polymerization. Oncogene. Aug 12–2019.doi:
10.1038/s41388-019-0948-6 (Epub ahead of print). View Article : Google Scholar
|
|
54
|
Guryanova OA, Wu Q, Cheng L, Lathia JD,
Huang Z, Yang J, MacSwords J, Eyler CE, McLendon RE, Heddleston JM,
et al: Nonreceptor tyrosine kinase BMX maintains self-renewal and
tumorigenic potential of glioblastoma stem cells by activating
STAT3. Cancer Cell. 19:498–511. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Tong L, Wang Y, Ao Y and Sun X: CREB1
induced lncRNA HAS2-AS1 promotes epithelial ovarian cancer
proliferation and invasion via the miR-466/RUNX2 axis. Biomed
Pharmacother. 115:1088912019. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Dong H, Cao W and Xue J: Long noncoding
FOXD2-AS1 is activated by CREB1 and promotes cell proliferation and
metastasis in glioma by sponging miR-185 through targeting AKT1.
Biochem Biophys Res Commun. 508:1074–1081. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Mukherjee S, Tucker-Burden C, Kaissi E,
Newsam A, Duggireddy H, Chau M, Zhang C, Diwedi B, Rupji M, Seby S,
et al: CDK5 Inhibition resolves PKA/cAMP-independent activation of
CREB1 signaling in glioma stem cells. Cell Rep. 23:1651–1664. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Brennan PJ, Kumagai T, Berezov A, Murali R
and Greene MI: HER2/Neu: Mechanisms of
dimerization/oligomerization. Oncogene. 19:6093–6101. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Kim YH, Tran TA, Lee HJ, Jung SI, Lee JJ,
Jang WY, Moon KS, Kim IY, Jung S and Jung TY: Branched multipeptide
immunotherapy for glioblastoma using human leukocyte
antigen-A*0201-restricted cytotoxic T-lymphocyte epitopes from
ERBB2, BIRC5 and CD99. Oncotarget. 7:50535–50547. 2016.PubMed/NCBI
|
|
60
|
Bagnjuk K, Kast VJ, Tiefenbacher A,
Kaseder M, Yanase T, Burges A, Kunz L, Mayr D and Mayerhofer A:
Inhibitor of apoptosis proteins are potential targets for treatment
of granulosa cell tumors-implications from studies in KGN. J
Ovarian Res. 12:762019. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Arman K, Ergün S, Temiz E and Öztuzcu S:
The interrelationship between HER2 and CASP3/8 with apoptosis in
different cancer cell lines. Mol Biol Rep. 41:8031–8036. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Michaelsen SR, Urup T, Olsen LR, Broholm
H, Lassen U and Poulsen HS: Molecular profiling of short-term and
long-term surviving patients identifies CD34 mRNA level as
prognostic for glioblastoma survival. J Neurooncol. 137:533–542.
2018. View Article : Google Scholar : PubMed/NCBI
|