Introduction
Macrophages are a class of immune cells residing in
all tissues (1). Macrophages are
broadly classified as M1 pro-inflammatory, or M2 anti-inflammatory
macrophages (2,3). Tumor-associated macrophages (TAMs) were
first identified 30 years ago (4).
Observations indicated that TAMs accumulated around tumors and were
primarily derived from monocytes (5,6). The two
polarization states of macrophages were also observed in TAMs; M1
macrophages were found in the early stages of neoplasia or in
vascularized areas, whilst M2 macrophages were observed during
tumor progression, and were indicative of poor prognosis (2,7–10).
Hypoxia immobilizes macrophages such that they
accumulate in hypoxic regions (11).
As hypoxia is a common feature of most solid tumors, TAMs are
observed in hypoxic regions within a variety of tumor types
(12). High concentrations of
chemokines such as hypoxia-inducible factor (HIF)-1, HIF-2 and
endothelin-2 are secreted from hypoxic tissues, which subsequently
attract macrophages (11,13). Additionally, macrophages in hypoxic
environments express higher levels of growth and angiogenic
factors, including vascular endothelial growth factor (VEGF),
glucose transporter-1 and tumor necrosis factor alpha (TNF-α),
compared with macrophages in normoxic environments (11,14,15).
Overall, hypoxia attracts higher numbers of M2 or M2-like TAMs and
promotes the polarization of M1 to M2 macrophages (13,16).
Lung carcinoma is one of the most common and fatal
carcinomas worldwide, and its incidence is increasing annually
(17,18). The poor prognosis of patients with
lung carcinoma has prompted numerous studies focused either on the
development of therapeutic strategies, or aimed to further the
understanding of lung cancer biology. Hypoxia is an important
factor in the modification of the tumor microenvironment, and thus
plays a pivotal role in all stages of carcinoma development,
including tumorigenesis, progression, angiogenesis and metastasis
(19,20). The effect of hypoxia on lung cancer
has been widely investigated; Meng et al (21) reported that high levels of hypoxia
were positively associated with both higher cancer grades at
diagnosis and poor prognosis. It has also been demonstrated that
the expression level of HIF-1α is positively correlated with poor
prognosis and the expression of various genes in lung cancer,
including epidermal growth factor receptor, matrix
metalloproteinase-9 and p53 (22,23).
However, little is known about the effects of the hypoxia-modified
microenvironment on lung cancer cells, and the subsequent effects
on malignant behaviors in lung cancer.
Considering the influence that hypoxia exerts on
TAMs, the present study was undertaken to assess the hypothesis
that hypoxia influences the malignant behaviors of several types of
cancer cell (including lung cancer cells) through the modification
of the microenvironment.
Materials and methods
Cell culture and macrophage
polarization
Human myeloid leukemia THP-1 cells were obtained
from the American Type Culture Collection and maintained in RPMI
1640 medium (Thermo Fisher Scientific, Inc.) supplemented with 10%
fetal bovine serum (FBS) and 1% of penicillin-streptomycin (Gibco;
Thermo Fisher Scientific, Inc), and cultured at 37°C with 5%
CO2. To obtain macrophages with an Mφ phenotype, THP-1
cells were differentiated by incubation with 10 ng/ml phorbol
12-myristate 13-acetate (PMA, Sigma-Aldrich; Merck KGaA) for 24 h
at 37°C. Polarization towards the M1 phenotype was subsequently
induced by culturing Mφ cells with 100 ng/ml lipopolysaccharide
(LPS, Sigma-Aldrich; Merck KGaA) and 20 ng/ml Interferon-γ (IFN-γ)
for a further 48 h; polarization towards the M2 phenotype was
induced by culturing Mφ cells with 20 ng/ml interleukin-4
(IL-4).
The human non-small cell lung cancer cell line A549
and the human liver cancer cell line HepG2 were cultured in
Dulbecco's Modified Eagles Medium (Gibco; Thermo Fisher Scientific,
Inc.). The HeLa human cervical cancer cell line and the MCF-7
breast cancer cell line (American Type Culture Collection) were
cultured in RPMI-1640 (Gibco; Thermo Fisher Scientific, Inc.). All
cell cultures were supplemented with 10% FBS and 1%
penicillin-streptomycin.
To identify the effect of hypoxia on the
polarization of macrophages, stimulation was conducted in a Galaxy
14S incubator with oxygen control (New Brunswick Scientific;
Eppendorf) containing 1% O2, 5% CO2 and 94%
N2, with or without 5 µM SB203580, an inhibitor of p38
MAPK. After 24 and 48 h incubation, supernatants and cells were
collected for further analysis.
Western blotting
For each sample, ~1×106 cells were
resuspended in 400 µl RIPA buffer (Sigma-Aldrich; Merck KGaA) and
lysed using the SoniConvert™ sonicator (DocSense, Chengdu, China).
The lysate was quantified using a bicinchoninic acid assay kit
(Sigma Aldrich; Merck KGaA) following the manufacturers'
instructions. A total of 20 µg protein was fractionated on a 4–10%
SDS-PAGE gel and transferred to a PVDF membrane (EMD Millipore).
The membrane was then blocked for 30 min at room temperature in
blocking buffer [5% silk milk, 2.5% normal goat serum
(Sigma-Aldrich; Merck KGaA), 0.025% Tween 20 in PBS] and probed for
the proteins of interest. The primary antibodies used were listed
as follows: Anti-IL-1β (cat. no. ab8320), anti-TNF-α (cat. no.
ab6671), anti-Human Leukocyte Antigen (HLA)-DR (cat. no. 92511),
anti-thymus and activation regulated chemokine (TARC) (cat. no.
ab182793), anti-CD163 (cat no. ab182422), anti-p38 (cat. no.
ab170099), anti-p38 (phospho Y182; cat. no. ab47363), anti-HIF-1α
(cat. no. ab1) and anti-β-actin a (cat. no. ab8227). The goat
anti-rabbit secondary antibody was then employed (cat. no. ab7090).
All antibodies were purchased from Abcam. The primary antibodies
were diluted to 1:2,000 and incubated with the membrane at room
temperature for 1 h. The secondary antibody was diluted to 1:10,000
and the membrane was incubated at room temperature for 1 h. The
blots were then quantified using SuperSignal West Dura ECL
substrate (EMD Millipore).
Flow cytometry
Macrophages were harvested by trypsinization, washed
with PBS and resuspended in staining buffer containing 2% goat
serum (Sigma-Aldrich; Merck KGaA) and 0.5 mM EDTA in PBS. The cells
were then stained with PE-conjugated antibodies against CD86 (cat.
no. 374205; clone, BU63, BioLegend, Inc.) or CD206 (cat. no.
321105; clone, 15-2, BioLegend, Inc.). After 3 min staining in
darkness at room temperature, the cells were washed twice using
staining buffer and analyzed using a 3-laser Navios flow cytometer
(Beckman Coulter, Inc.). FlowJo software was used for data analysis
(version. 1.6.0).
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from the macrophages using
TRIzol® reagent (Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocol; 1 µg total RNA was
employed for reverse transcription using a cDNA Synthesis Kit
(Thermo Fisher Scientific, Inc.) according to the manufacturer's
protocol. qPCR was then performed using a SYBR® Green
PCR Master Mix (Thermo Fisher Scientific, Inc.) on an ABI7500
system (Applied Biosystems; Thermo Fisher Scientific Inc.).
Expression levels were normalized to that of β-actin and the
fold-change in expression was obtained using the 2−ΔΔCq
method (24). The primer sequences
were as follows: β-actin forward, 5′-CATGTACGTTGCTATCCAGGC-3′, and
reverse, 5′-CTCCTTAATGTCACGCACGAT-3′; IL-1β forward,
5′-ATGATGGCTTATTACAGTGGCAA-3′, and reverse,
5′-GTCGGAGATTCGTAGCTGGA-3′; TNF-α forward,
5′-AACAGAGAGGATTTCGTTTCCG-3′, and reverse,
5′-TTTGACCTGAGGGTAAGACTTCT-3′; VEGF forward,
5′-GTCGAGGAAGAGAGAGACGG-3′, and reverse,
5′-GTCTGTCTGTCTGTCCGTCA-3′; HLA-DR forward,
5′-TGGTTTCTATCCAGGCAGCA-3′, and reverse,
5′-TTCAGACCGTGCTCTCCATT-3′; CCL17 forward,
5′-AGGTCTTGAAGCCTCCTCAC-3′, and reverse,
5′-AGTTCAGACAAGGGGATGGG-3′; CD163 forward,
5′-GAGCAGCACATGGGAGATTG-3′, and reverse,
5′-ACCTCCTCCATTTACCAGGC-3′.
ELISA
The expression levels of the assayed proteins were
determined using the following corresponding ELISA kits according
to the manufacturer's protocol [Multisciences (Lianke) Biotech Co.,
Ltd]: IL-1β (cat. no. 70-EK101B), TNF-α (cat. no. 70-EK182), VEGF
[including the predominant isoforms, VEGF189,
VEGF165, VEGF145 and VEGF121 (cat.
no. 70-EK183)] and CCL17 (cat. no. 70-ek1115).
Cell counting kit (CCK)-8 assay
To determine the cell proliferation rate, a CCK-8
assay was performed. Originally, 2×104 A549, HeLa, HepG2
and MCF-7 cells were incubated in conditioned medium for 1 to 5
days. Each day, 10 µl CCK-8 solution (Sigma-Aldrich; Merck KGaA)
was added to each well and incubated at 37°C for 2 h in a
humidified incubator. The absorbance value was measured at a
wavelength of 450 nm on a Multiskan spectrum microplate reader
(Thermo Fisher Scientific, Inc.), and the experiment was repeated
three times.
Colony formation assay
Resuspended cells were plated in 6-well plates at a
density of 1×103 cells/well, and incubated for 2 weeks.
The cells were fixed with methanol containing 1% crystal violet for
30 min, and images were captured using a X71 (U-RFL-T) fluorescence
microscope (Olympus Corporation; magnification, ×40).
Tumor formation in soft agar
For each well, 1×104 A549, Hela, HepG2
and MCF-7 cells were suspended in diluted 0.3% low-melt agar medium
and added to pre-set 0.6% low-melt agar medium in 6-well plates.
The plates were incubated for 2 weeks at 37°C and then stained with
crystal violet (0.01% solution), washed with PBS, and imaged under
a X71 (U-RFL-T) fluorescence microscope (Olympus Corporation) at
×40 magnification.
Statistical analysis
SPSS 17.0 (SPSS, Inc.) software was used to conduct
the statistical analyses, and all data are presented as the mean ±
SD. Differences between two groups were compared using the
Student's t-test, whilst the differences between ≥3 groups were
assessed using one-way ANOVA followed by Tukey's post-hoc test.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Stimulation of M1 and M2 polarized
macrophages
To obtain M1 and M2 polarized macrophages, THP-1
cells were first treated with PMA for 24 h to induce Mφ
differentiation. Then, Mφ cells were further stimulated with
IFN-γ/LPS or IL-4 for 48 h, and the cytokine and chemokine
expression profiles of THP-1-derived Mφ, M1 and M2 macrophages were
evaluated by RT-qPCR (Fig. 1A). The
data showed that classically activated (M1-polarized) macrophages
exhibited an upregulation of several proinflammatory mediators,
including IL-1β and TNF-α. By contrast, CCL17 and CD163 were
significantly upregulated in M2-polarized macrophages. The changes
in the expression of these cytokines and chemokines in specific
macrophage subsets were then confirmed by western blotting, and the
protein levels of these mediators showed similar trends (Fig. 1B). The correct macrophage
polarization was also confirmed by detecting cell surface markers
of M1 and M2 polarization. As shown in Fig. 1C, M1-polarized macrophages exhibited
a significantly higher level of CD86 expression, which is reported
to be an M1 macrophage-specific surface marker (25,26).
However, M2-polarized macrophages exhibited a significantly higher
level of CD206 compared with unpolarized M1 macrophages (25,26).
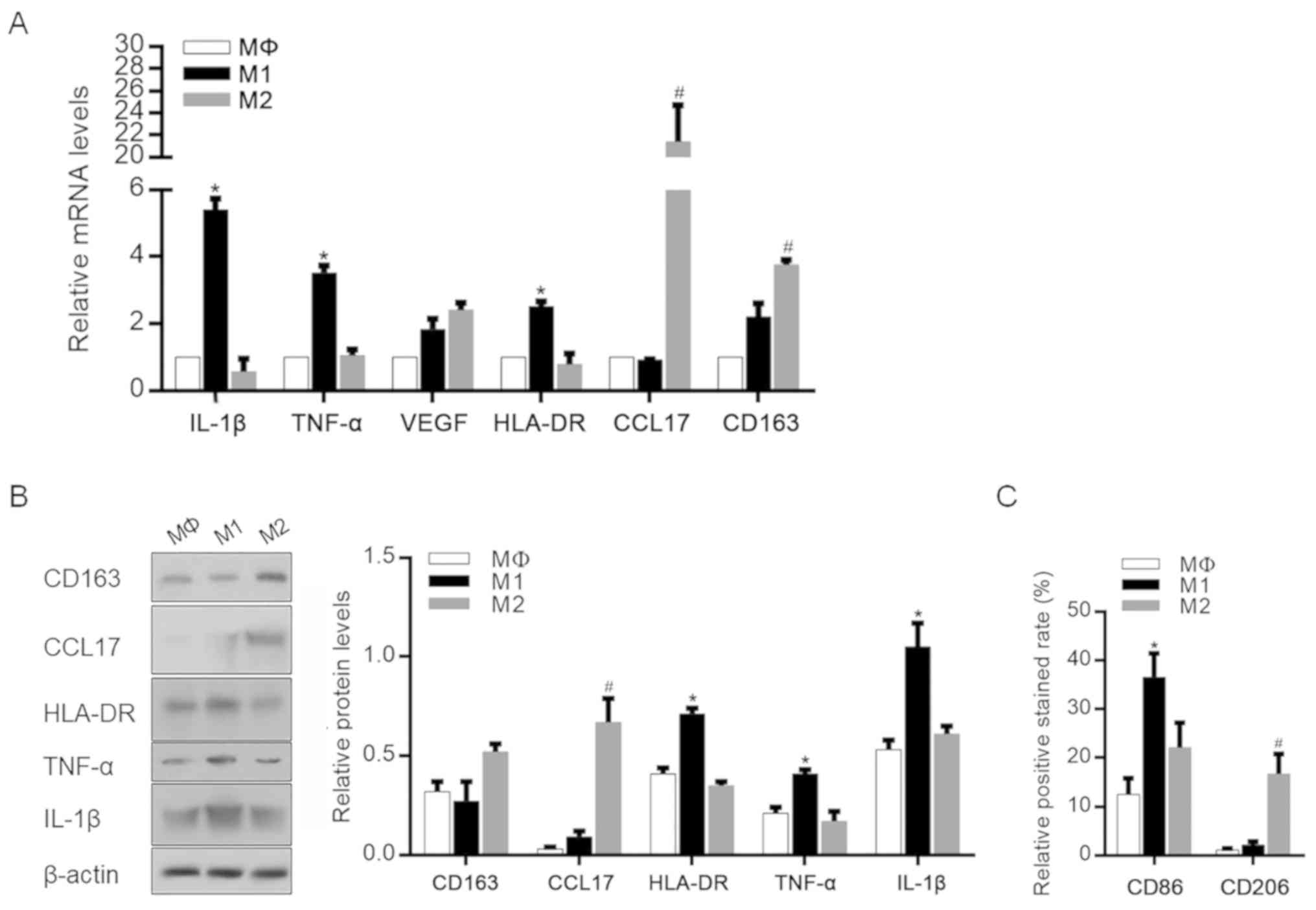 | Figure 1.Identification of M1 and M2
macrophage phenotypes after differentiation. (A) mRNA levels of
IL-1β, TNF-α, VEGF, HLA-DR, CCL17 and CD163 in MΦ, M1 and M2
macrophages after specific stimulation. *P<0.05, vs.
Mφ-stimulated group; #P<0.05, vs. Mφ-stimulated
group. (B) Protein levels of IL-1β, TNF-α, HLA-DR, CCL17 and CD163
in MΦ-, M1- and M2-polarized macrophages. *P<0.05, vs.
Mφ-stimulated group; #P<0.05, vs. Mφ-stimulated
group. (C) CD86 and CD206 expression rates were measured.
*P<0.05, vs. Mφ-stimulated group; #P<0.05, vs.
Mφ-stimulated group. IL-1β, interleukin-β; TNF-α, tumor necrosis
factor-α; VEGF, vascular endothelial growth factor; HLA-DR, human
leukocyte antigen-DR; CCL17, chemokine (C-C motif) ligand 17. |
Effects of hypoxia on the
microenvironment of M1- and M2-polarized macrophages
To identify the effect of hypoxic exposure on the
expression profiles of specific cytokines and chemokines, Mφ-, M1-
and M2-polarized macrophages were induced under normoxic and
hypoxic conditions. As shown in Fig.
2A, hypoxia significantly decreased IL-1β expression in M1
macrophages and increased the VEGF mRNA levels in both M1 and M2
macrophages. The changes in the protein levels of these cytokines
were confirmed via western blot analysis (Fig. 2B). Interestingly, the detection of
M1- and M2-specific cell surface markers revealed that macrophage
differentiation towards M2 polarization was promoted, without
affecting M2 polarization status by incubating with IL-4/hypoxia
conditioned medium (Fig. 2C). The
release of IL-1β, TNF-α, VEGF (including the predominant isoforms
VEGF189, VEGF165, VEGF145 and
VEGF121) and CCL17 in the cell supernatants was then
investigated. Consistent with the aforementioned results, hypoxia
decreased the secretion of proinflammatory mediators in
M1-polarized macrophages (Fig.
2D).
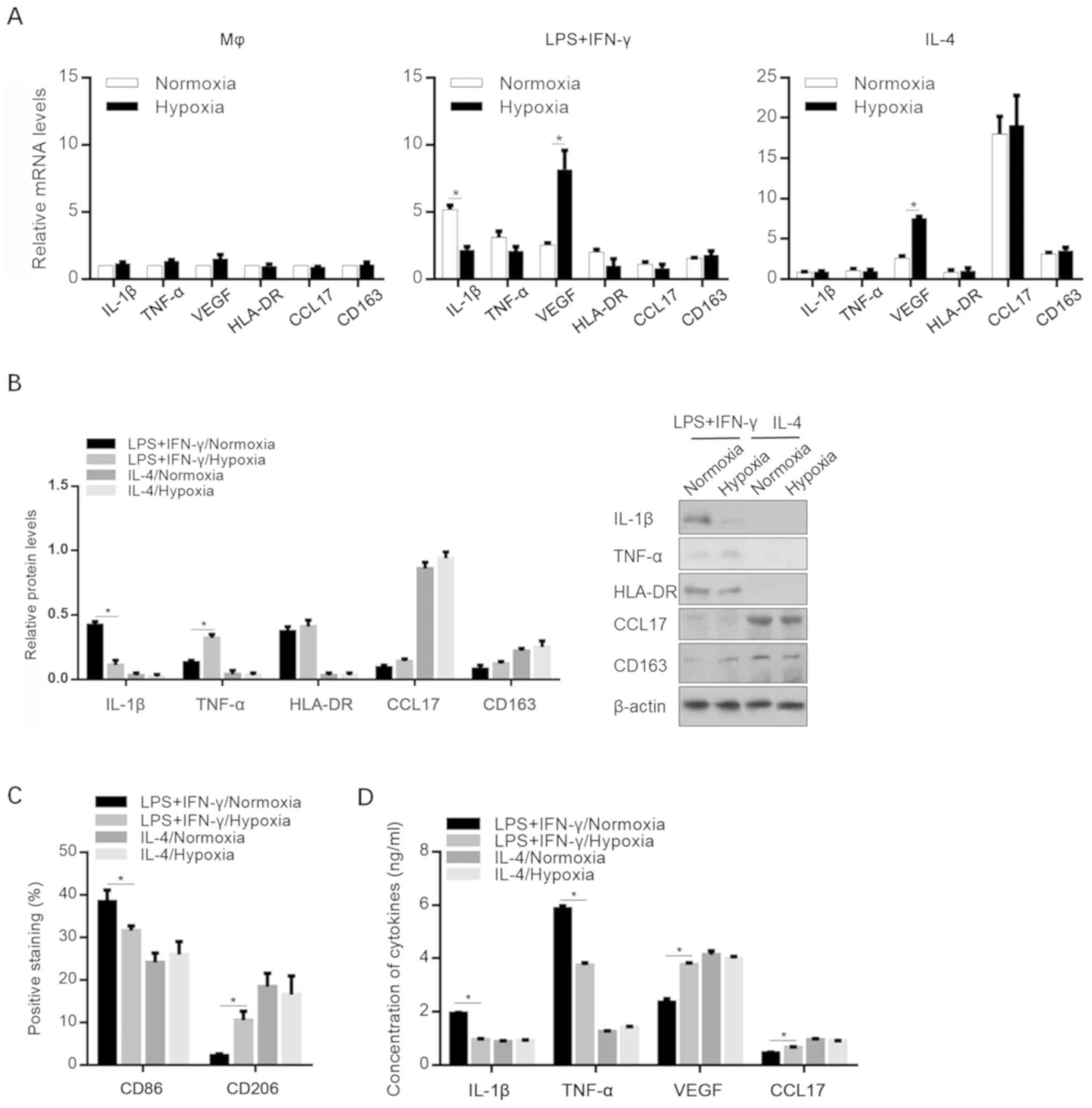 | Figure 2.Effects of hypoxia on the expression
levels of cytokines and chemokines. (A) mRNA levels of IL-1β,
TNF-α, VEGF, HLA-DR, CCL17 and CD163 in MΦ, M1 (LPS+IFN-γ) and M2
(IL-4) macrophages after specific stimulation under normoxic or
hypoxic conditions. *P<0.05, vs. Normoxic group. (B) Protein
levels of IL-1β, TNF-, HLA-DR, CCL17 and CD163 in MΦ-, M1- and
M2-polarized macrophages under normoxic or hypoxic conditions.
*P<0.05, vs. LPS+IFN-γ/Normoxic group. (C) CD86 and CD206
expression levels. *P<0.05, vs. LPS+IFN-γ/Normoxic group. (D)
Secretion of IL-1β, TNF-α, VEGF, and CCL17. *P<0.05, vs.
LPS+IFN-γ/Normoxic group. IL-1β, interleukin-β; TNF-α, tumor
necrosis factor-α; VEGF, vascular endothelial growth factor; CCL17,
chemokine (C-C motif) ligand 17; LPS, lipopolysaccharide; IFN-γ,
interferon-γ. |
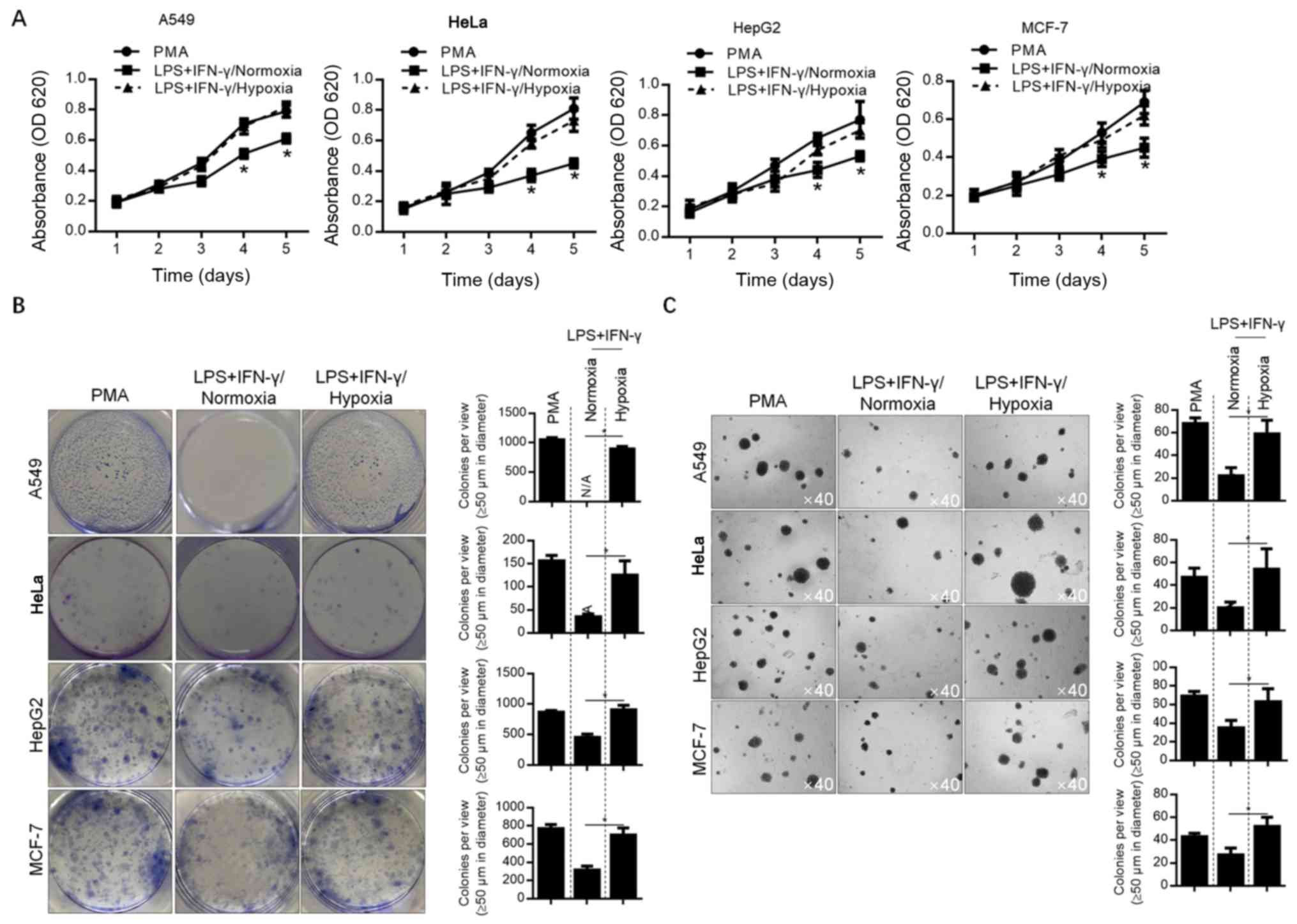
Hypoxia-modified microenvironments
promote malignant behavior in A549 cells
To detect the effect of hypoxia-modified
microenvironments on A549 lung cancer cells, as well as MCF-7,
HepG2 and HeLa cells, conditioned-medium was used to culture each
cell line. Considering the less pronounced effect of hypoxia on
M2-polarized macrophages, the conditioned medium of M1-polarized
macrophages was employed for further analyses. The malignant
behaviors of A549, HeLa, HepG2 and MCF-7 cells (including
proliferation, colony formation and soft-agar tumor formation) were
analyzed. The results showed that all of these malignant behaviors
were decreased by normoxia-conditioned medium, compared with
Mφ-conditioned medium (P<0.05). Moreover, hypoxia-conditioned
medium exerted no detectable effects on these malignant behaviors
(Fig. 3A-C). This indicates that the
promotion of cancer cell malignant behavior is inhibited by the
M1-modified microenvironment, which can subsequently be reversed by
hypoxia.
Hypoxia-modified polarization of
macrophages is independent of the expression of HIF-1α, and
partially dependent on the activation of p38
As a key transcriptional regulator, HIF-1α serves a
critical role in the adaptation of tumors to hypoxia via the
regulation of multiple cytokines, including VEGF (27,28).
Following hypoxia or CoCl2 treatment, HIF-1α expression
in macrophages was determined; as expected, HIF-1α was upregulated
in both M1- and M2-polarized macrophages (Fig. 4A and B). The Addition of
CoCl2 to culture medium is commonly used to accumulate
HIF-1α, and to activate its transcriptional activity (29). However, the
CoCl2-conditioned medium of M1 macrophages failed to
modify the release of cytokines and chemokines affected by the
hypoxia-conditioned medium of M1 polarization (Fig. 4C and D). The detection of cell
surface markers also indicated that CoCl2 treatment
failed to exert the same effect with hypoxia, indicating that the
effect of hypoxic exposure is independent of the presence of HIF-1α
(Fig. 4E).
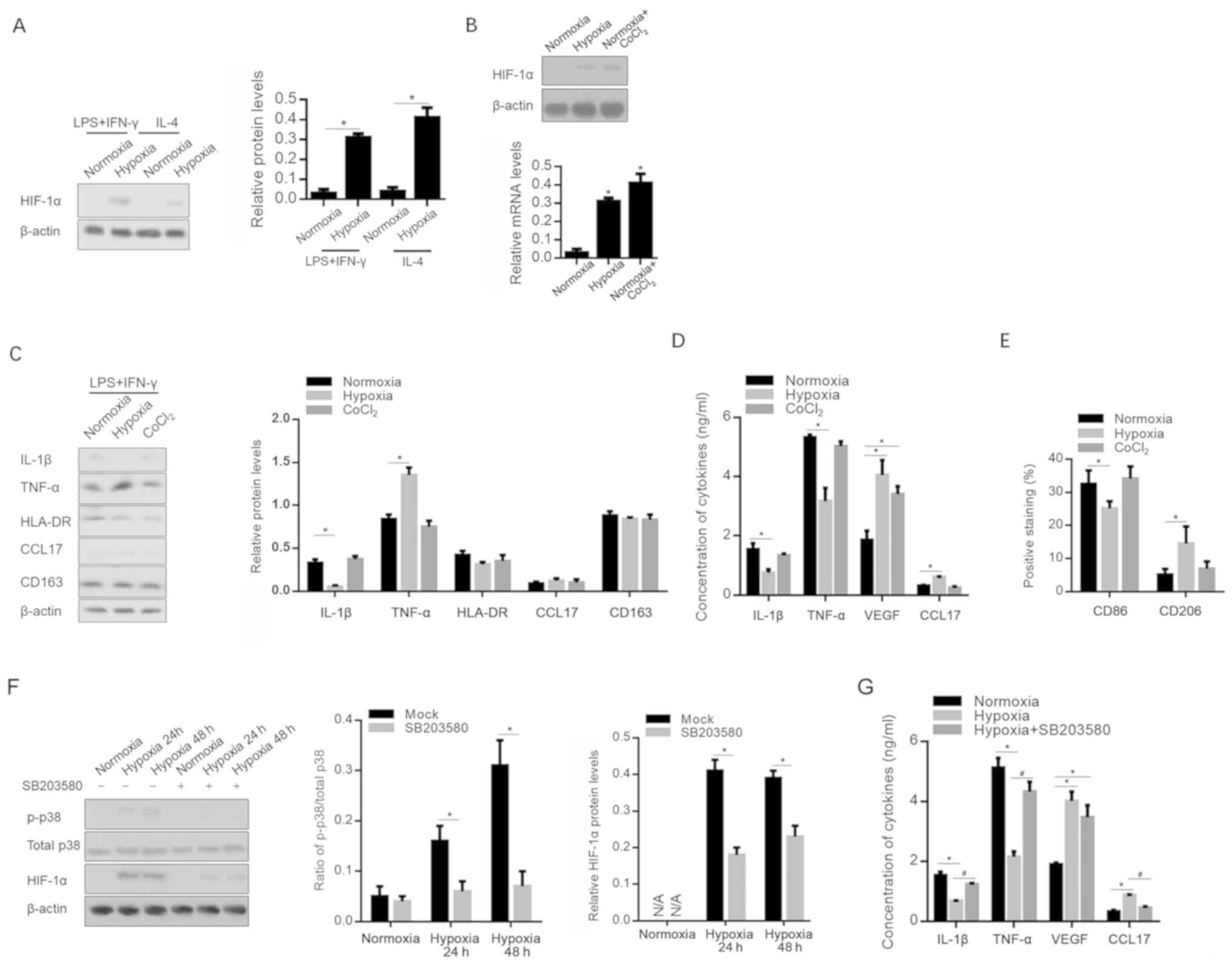 | Figure 4.Hypoxia affects the inflammatory
microenvironment in a p38-dependent and HIF-1α-independent manner.
HIF-1α protein expression level was measured following (A) hypoxia
and (B) CoCl2 treatment. (C) mRNA levels of IL-1β,
TNF-α, VEGF, HLA-DR, CCL17 and CD163 were measured after hypoxic
exposure or CoCl2 treatment. *P<0.05, vs.
Normoxia-exposed group. (D) Release of IL-1β, TNF-α, VEGF and CCL17
in supernatant was measured by ELISA. (E) CD86 and CD206 expression
was also assessed. *P<0.05, vs. Normoxia-exposed group. (F) p38
and phosphorylated p38 levels were measured under hypoxic
conditions with or without 1 µM SB203580. (G) IL-1β, TNF-α, VEGF
and CCL17 in the supernatant was measured by ELISA under hypoxic
conditions with or without 1 µM SB203580. *P<0.05, vs.
Normoxia-exposed group; #P<0.05, vs. Hypoxia-exposed
group. HIF, hypoxia inducible factor; 1β, interleukin-β; TNF-α,
tumor necrosis factor-α; VEGF, vascular endothelial growth factor;
HLA-DR, human leukocyte antigen-DR; CCL17, chemokine (C-C motif)
ligand 17; p-, phosphorylated; LPS, lipopolysaccharide; IFN-γ,
interferon-γ. |
Considering that p38 is a central mediator in the
production of pro-inflammatory cytokines (30), and that it is activated by hypoxia in
several cell types (31), p38 and
phosphorylated p38 were detected after hypoxia, with or without the
p38 signaling inhibitor SB203580. As shown in Fig. 4F, hypoxia significantly promoted the
phosphorylation of p38. This was eradicated by the addition of 1 µM
SB203580. Notably, it was observed that the addition of 1 µM
SB203580 significantly decreased the level of HIF-1α expression
after hypoxia. This indicates that SB203580 may decrease
HIF-1α-activated p38 via different pathways. Importantly,
pro-inflammatory cytokines decreased by hypoxia were recovered by
the addition of SB203580, indicating that hypoxia-activated p38
signaling is, at least in part, responsible for the production of
pro-inflammatory cytokines (Fig.
4G).
Discussion
The findings of the present study demonstrate that
hypoxia can modify the polarization of macrophages, regulate the
inflammatory microenvironment, and consequently regulate the
malignant behaviors of A549 lung cancer cells. It was also
indicated that the effects of hypoxia on the inflammatory
microenvironment are regulated via the p38-signaling pathway.
In hypoxic conditions, macrophages are able to
responsively accumulate HIF-1α and HIF-2α (32), and stimulation by different mediators
can induce different isotypes of HIF. IL-4-stimulated polarization
of macrophages towards a wound-healing phenotype is associated with
increased levels of HIF-2α, which is barely detected in classically
activated macrophages (33). HIF-1α
and −2α are detectable in hypoxic tumor microenvironments; this
indicates that in hypoxia-modified inflammatory microenvironments,
macrophages can alter their phenotype, resulting in a mixed
macrophage population and allowing simultaneous accumulation of
HIF-1α and −2α (34,35). Populations of macrophages with mixed
phenotypes tightly regulate the inflammatory microenvironment by
producing both pro- and anti-inflammatory cytokines and chemokines.
Thus, determining how hypoxia affects the polarization of
macrophages is important for further understanding the effects of
hypoxia on the inflammatory microenvironment (36). There has been much research into the
effect of hypoxia on macrophage polarization. Despite this, whether
hypoxia or HIF contribute to macrophage polarization remains
unknown (36). In the present study,
M1- and M2-polarized macrophages were stimulated under hypoxia, and
it was found that hypoxia affected the ratio of CD86 and CD206
expression in M1-polarized macrophages without affecting expression
in M2-polarized macrophages. This may indicate that hypoxia
modifies the polarization of macrophages towards the M2
phenotype.
To understand the effects of hypoxia on the
inflammatory microenvironment, macrophages were exposed to hypoxia
during stimulation. The data revealed that hypoxia significantly
decreased the release of pro-inflammatory cytokines, and increased
the expression of VEGF, which is reported to regulate macrophage
functions, including tumor promotion (37,38).
This indicates that hypoxia may promote an inflammatory
microenvironment. In M2-polarized macrophages, hypoxia increased
the VEGF mRNA level without altering that of VEGF protein,
indicating that hypoxia-induced VEGF is not secreted. Considering
that hypoxia-induced HIF-1α expression may contribute to the
promotion of an inflammatory microenvironment, CoCl2 was
employed to induce HIF-1α expression. However, only a minor effect
was observed following CoCl2 treatment, indicating that
hypoxia may modify the microenvironment independently of HIF-1α
signaling. As a limitation of the present study, the results were
not verified in vivo. In further investigations, it would be
worth evaluating the potential modification of hypoxia on the tumor
microenvironment in a mouse model (39).
The link between p38-mitogen-activated protein
kinase (MAPK) activation and hypoxia, and the resulting regulation
of physiological processes, is well established (40). It is reported that under hypoxic
conditions, the p38 MAPK signaling complex is also strongly
associated with inflammation-mediated apoptotic cell death in a
variety of cell types (41). Also
reported is that the inhibition of p38 reduces the release of
pro-inflammatory cytokines under hypoxic conditions (42). Thus, in the present study, the levels
of p38 and phosphorylated p38 (p-p38) were determined after hypoxic
exposure. SB203580, a p38 MAPK signaling inhibitor, was employed.
According to the results, p-p38 was significantly increased without
altering the p38 protein level, and was completely inhibited in the
presence of SB203580. Notably, the hypoxia-induced regulation of
cytokines was significantly reversed by treatment with SB203580,
demonstrating that p-p38 is critical for hypoxia-regulated cytokine
release. Furthermore, SB203580 treatment was found to significantly
decrease HIF-1α expression, which indicates an interesting point
for further investigation. No significant effect on malignant
behavior was detected from the conditioned medium of
SB203580-treated cells, which included proliferation, colony and
tumor formation. However, the effect of SB203580 on cytokine levels
indicates its potential effects on these malignant behaviors, which
should be explored in further investigations.
The data of the present study suggest that hypoxia
during macrophage polarization towards the M1 or M2 phenotype
significantly modifies the release of cytokines, and thus regulates
the inflammatory microenvironment. Moreover, it was revealed that
hypoxia-induced p38 phosphorylation, but not HIF-1α, is necessary
for regulating the inflammatory microenvironment, and that a
hypoxia-modified inflammatory microenvironment may contribute to
the promotion of malignant behaviors in A549 lung cancer cells.
These results indicate a novel approach to targeting cancer cells
by modifying the inflammatory microenvironment via the regulation
of macrophage polarization.
Acknowledgements
The authors would like to thank Tao Hong for
language editing.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
XK, CC, YS, GX and DL designed the experiments. XK
performed cell culture and data analysis. QC, JL and YT performed
gene expression analysis and protein analysis. XH, WQ, AC and HW
performed research on molecular mechanisms. GX and DL supervised
the experiments, writing and revisions.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Liu Y and Cao X: The origin and function
of tumor-associated macrophages. Cell Mol Immunol. 12:1–4. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Biswas SK, Sica A and Lewis CE: Plasticity
of macrophage function during tumor progression: Regulation by
distinct molecular mechanisms. J Immunol. 180:2011–2017. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Biswas SK and Mantovani A: Macrophage
plasticity and interaction with lymphocyte subsets: Cancer as a
paradigm. Nat Immunol. 11:889–896. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
van Ravenswaay Claasen HH, Kluin PM and
Fleuren GJ: Tumor infiltrating cells in human cancer. On the
possible role of CD16+ macrophages in antitumor cytotoxicity. Lab
Invest. 67:166–174. 1992.PubMed/NCBI
|
|
5
|
Coussens LM and Werb Z: Inflammation and
cancer. Nature. 420:860–867. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Franklin RA, Liao W, Sarkar A, Kim MV,
Bivona MR, Liu K, Pamer EG and Li MO: The cellular and molecular
origin of tumor-associated macrophages. Science. 344:921–925. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Schultze JL, Schmieder A and Goerdt S:
Macrophage activation in human diseases. Semin Immunol. 27:249–256.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sica A and Mantovani A: Macrophage
plasticity and polarization: In vivo veritas. J Clin Invest.
122:787–795. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Martinez FO, Sica A, Mantovani A and
Locati M: Macrophage activation and polarization. Front Biosci.
13:453–461. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Jeannin P, Paolini L, Adam C and Delneste
Y: The roles of CSFs on the functional polarization of
tumor-associated macrophages. FEBS J. 285:680–699. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Murdoch C, Muthana M and Lewis CE: Hypoxia
regulates macrophage functions in inflammation. J Immunol.
175:6257–6263. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lewis C and Murdoch C: Macrophage
responses to hypoxia: Implications for tumor progression and
anti-cancer therapies. Am J Pathol. 167:627–635. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Roda JM, Wang Y, Sumner LA, Phillips GS,
Marsh CB and Eubank TD: Stabilization of HIF-2α induces sVEGFR-1
production from tumor-associated macrophages and decreases tumor
growth in a murine melanoma model. J Immunol. 189:3168–3177. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Burke B, Giannoudis A, Corke KP, Gill D,
Wells M, Ziegler-Heitbrock L and Lewis CE: Hypoxia-induced gene
expression in human macrophages: Implications for ischemic tissues
and hypoxia-regulated gene therapy. Am J Pathol. 163:1233–1243.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lewis JS, Lee JA, Underwood JC, Harris AL
and Lewis CE: Macrophage responses to hypoxia: Relevance to disease
mechanisms. J Leukoc Biol. 66:889–900. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Huber R, Meier B, Otsuka A, Fenini G,
Satoh T, Gehrke S, Widmer D, Levesque MP, Mangana J, Kerl K, et al:
Tumour hypoxia promotes melanoma growth and metastasis via High
Mobility Group Box-1 and M2-like macrophages. Sci Rep. 6:299142016.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Didkowska J, Wojciechowska U, Manczuk M
and Łobaszewski J: Lung cancer epidemiology: Contemporary and
future challenges worldwide. Ann Transl Med. 4:1502016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Dizon DS, Krilov L, Cohen E, Gangadhar T,
Ganz PA, Hensing TA, Hunger S, Krishnamurthi SS, Lassman AB,
Markham MJ, et al: Clinical cancer advances 2016: Annual report on
progress against cancer from the American Society of Clinical
Oncology. J Clin Oncol. 34:987–1011. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ugel S, De Sanctis F, Mandruzzato S and
Bronte V: Tumor-induced myeloid deviation: When myeloid-derived
suppressor cells meet tumor-associated macrophages. J Clin Invest.
125:3365–3376. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
Quatromoni JG and Eruslanov E:
Tumor-associated macrophages: Function, phenotype, and link to
prognosis in human lung cancer. Am J Transl Res. 4:376–389.
2012.PubMed/NCBI
|
|
21
|
Meng X, Kong FM and Yu J: Implementation
of hypoxia measurement into lung cancer therapy. Lung Cancer.
75:146–150. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Karetsi E, Ioannou MG, Kerenidi T, Minas
M, Molyvdas PA, Gourgoulianis KI and Paraskeva E: Differential
expression of hypoxia-inducible factor 1α in non-small cell lung
cancer and small cell lung cancer. Clinics (Sao Paulo).
67:1373–1378. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Swinson DE, Jones JL, Cox G, Richardson D,
Harris AL and O'Byrne KJ: Hypoxia-inducible factor-1 alpha in non
small cell lung cancer: Relation to growth factor, protease and
apoptosis pathways. Int J Cancer. 111:43–50. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chavez-Galan L, Olleros ML, Vesin D and
Garcia I: Much more than M1 and M2 macrophages, there are also
CD169(+) and TCR(+) macrophages. Front Immunol.
6:2632015.PubMed/NCBI
|
|
26
|
Roszer T: Understanding the mysterious M2
macrophage through activation markers and effector mechanisms.
Mediators Inflamm. 2015:8164602015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wan J, Ma J, Mei J and Shan G: The effects
of HIF-1alpha on gene expression profiles of NCI-H446 human small
cell lung cancer cells. J Exp Clin Cancer Res. 28:1502009.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wan J, Chai H, Yu Z, Ge W, Kang N, Xia W
and Che Y: HIF-1α effects on angiogenic potential in human small
cell lung carcinoma. J Exp Clin Cancer Res. 30:772011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Goldberg MA and Schneider TJ: Similarities
between the oxygen-sensing mechanisms regulating the expression of
vascular endothelial growth factor and erythropoietin. J Biol Chem.
269:4355–4359. 1994.PubMed/NCBI
|
|
30
|
Bachstetter AD and Van Eldik LJ: The p38
MAP kinase family as regulators of proinflammatory cytokine
production in degenerative diseases of the CNS. Aging Dis.
1:199–211. 2010.PubMed/NCBI
|
|
31
|
Xu L, Pathak PS and Fukumura D:
Hypoxia-induced activation of p38 mitogen-activated protein kinase
and phosphatidylinositol 3′-kinase signaling pathways contributes
to expression of interleukin 8 in human ovarian carcinoma cells.
Clin Cancer Res. 10:701–707. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Takeda N, O'Dea EL, Doedens A, Kim JW,
Weidemann A, Stockmann C, Asagiri M, Simon MC, Hoffmann A and
Johnson RS: Differential activation and antagonistic function of
HIF-{alpha} isoforms in macrophages are essential for NO
homeostasis. Genes Dev. 24:491–501. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Imtiyaz HZ, Williams EP, Hickey MM, Patel
SA, Durham AC, Yuan LJ, Hammond R, Gimotty PA, Keith B and Simon
MC: Hypoxia-inducible factor 2alpha regulates macrophage function
in mouse models of acute and tumor inflammation. J Clin Invest.
120:2699–2714. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Mosser DM and Edwards JP: Exploring the
full spectrum of macrophage activation. Nat Rev Immunol. 8:958–969.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Talks KL, Turley H, Gatter KC, Maxwell PH,
Pugh CW, Ratcliffe PJ and Harris AL: The expression and
distribution of the hypoxia-inducible factors HIF-1alpha and
HIF-2alpha in normal human tissues, cancers, and tumor-associated
macrophages. Am J Pathol. 157:411–421. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Brune B, Dehne N, Grossmann N, Jung M,
Namgaladze D, Schmid T, von Knethen A and Weigert A: Redox control
of inflammation in macrophages. Antioxid Redox Signal. 19:595–637.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Douglas NC, Zimmermann RC, Tan QK,
Sullivan-Pyke CS, Sauer MV, Kitajewski JK and Shawber CJ: VEGFR-1
blockade disrupts peri-implantation decidual angiogenesis and
macrophage recruitment. Vasc Cell. 6:162014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Li N, Qin J, Lan L, Zhang H, Liu F, Wu Z,
Ni H and Wang Y: PTEN inhibits macrophage polarization from M1 to
M2 through CCL2 and VEGF-A reduction and NHERF-1 synergism. Cancer
Biol Ther. 16:297–306. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Almendros I, Gileles-Hillel A, Khalyfa A,
Wang Y, Zhang SX, Carreras A, Farré R and Gozal D: Adipose tissue
macrophage polarization by intermittent hypoxia in a mouse model of
OSA: Effect of tumor microenvironment. Cancer Lett. 361:233–239.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Fu S, Bai R, Zhao Z, Zhang Z, Zhang G,
Wang Y, Wang Y, Jiang D and Zhu D: Overexpression of
hypoxia-inducible factor-1α and vascular endothelial growth factor
in sacral giant cell tumors and the correlation with tumor
microvessel density. Exp Ther Med. 8:1453–1458. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Shologu N, Scully M, Laffey JG and O'Toole
D: Human mesenchymal stem cell secretome from bone marrow or
adipose-derived tissue sources for treatment of hypoxia-induced
pulmonary epithelial injury. Int J Mol Sci. 19(pii): E29962018.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Sakiyama S, dePerrot M, Han B, Waddell TK,
Keshavjee S and Liu M: Ischemia-reperfusion decreases protein
tyrosine phosphorylation and p38 mitogen-activated protein kinase
phosphorylation in rat lung transplants. J Heart Lung Transplant.
22:338–346. 2003. View Article : Google Scholar : PubMed/NCBI
|