Introduction
Non-small-cell lung cancer (NSCLC) accounts for ~85%
of all lung cancer cases, demonstrating a high degree of mortality
and poor survival worldwide (1). In
recent years, substantial advances have been achieved in NSCLC
diagnosis and treatment; however, the 5-year survival rate has
remained unchanged at 15% (2).
Cisplatin (DDP) is a first-line drug used in NSCLC chemotherapy;
however, resistance to chemotherapy drugs administered after
surgeries limits the prognosis of patients, as well as the use of
DDP in clinical applications (3).
Therefore, the molecular mechanisms underlying DDP resistance need
to be urgently elucidated in order to improve the survival rate of
patients with NSCLC.
Long non-coding RNAs (lncRNAs) are non-protein
coding RNAs that are greater than 200 nucleotides in length
(3). Numerous studies have indicated
that lncRNAs are capable of regulating gene expression at the
transcriptional, post-transcriptional and epigenetic levels, and
serve an important role in human cancer development, prognosis and
drug resistance (4,5). Recent studies have associated certain
lncRNAs, including CCAT1 (6) and
TRPM2-AS (7), with DDP resistance in
lung cancer. Long intergenic non-coding RNA 00707 (LINC00707),
located on chromosome 10p14, has also been associated with the
development and progression of cancers, including hepatocellular
carcinoma (8), gastric cancer
(9) and lung cancer (10). However, the function and underlying
mechanism of LINC00707 in the progression of DDP resistance in
NSCLC remains largely unclear.
Recently, emerging evidence has revealed that
lncRNAs are able to serve as competing endogenous RNAs or microRNA
(miRNA/miR) sponges, and induce regulatory effects on miRNAs and
miRNA-targeted mRNAs (11). miRNAs
are a group of evolutionarily conserved, single-stranded,
non-coding RNAs (21–25 nucleotides in length) that regulate
post-transcriptional gene expression by targeting the
3′-untranslated regions (3′-UTRs) of their target mRNAs (12). Aberrant expression of various miRNAs
has been suggested to contribute to DDP resistance in human cancers
(13). miR-145, a tumor suppressor
miRNA, has been reported to be downregulated in several types of
human cancers, including lung cancer (14). Furthermore, Zhan et al
(15) reported that miR-145 promoted
multidrug resistance protein 1 (MRP1) mRNA degradation and,
therefore, sensitized gallbladder cancer cells to DDP. However,
whether LINC00707 acts as an miR-145 sponge in order to regulate
DDP resistance in NSCLC cells remains to be investigated.
The aim of the present study was to investigate the
role and potential regulatory mechanism of LINC00707 in
DDP-resistance progression in NSCLC.
Materials and methods
Cell culture and transfection
DDP-resistant A549 cells (A549/DDP) and parental
A549 cells were obtained from The Cancer Institute of the Chinese
Academy of Sciences. The cells were maintained in RPMI-1640 medium
containing 10% FBS (both HyClone; GE Healthcare Life Sciences) and
1% penicillin-streptomycin (Gibco; Thermo Fisher Scientific, Inc.)
in a humidified incubator with 5% CO2 at 37°C. To
maintain the DDP-resistant phenotype, 2 µM DDP (Sigma-Aldrich;
Merck KGaA) was also added to the culture media of A549/DDP cells.
LINC00707 siRNA (si-LINC00707; 5′-GCAGGAACAUCACCAUCUUUU-3′), siRNA
negative control (si-NC; 5′-UUCUCCGAACGUGUCACGUTT-3′), miR-145
mimic (5′-GUCCAGUUUUCCCAGGAAUCCCU-3′), miRNA negative control (NC,
5′-UCACAACCUCCUAGAAAGAGUAGA-3′), miR-145 inhibitor
(5′-AGGGAUUCCUGGGAAAACUGGAC-3′) and negative control (inhibitor NC,
5′-UCUACUCUUUCUAGGAGGUUGUGA-3) were all purchased from Shanghai
GenePharma Co., Ltd. The transfection of above siRNAs or miRNA
mimics (final concentration: 50 nM) into A549/DDP cells
(4×105/per well of 6-well plate) was performed using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol. Cells
were collected for further experiments 48 h after transfection.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was isolated from the cells using TRIzol
reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to
the manufacturer's protocol. RNA (1 µg) was reversed to cDNA using
a High Capacity cDNA Reverse Transcription kit (cat. no. 4368814,
Thermo Fisher Scientific, Inc.) according to the manufacturer's
protocol. RT-qPCR was performed using the ABI 7500 RT-PCR system
(Applied Biosystems; Thermo Fisher Scientific, Inc.) with a
SYBR® Premix Ex Taq™ kit and TaqMan miRNA assay (both
Takara Biotechnology Co., Ltd.). The primers were synthesized by
Shanghai GenePharma Co., Ltd. Primer names and sequences are
provided in Table I. 18s rRNA was
used as internal references for lncRNA, mRNA and miRNA. U6 small
nuclear RNA was used as internal references for miRNA. The relative
expression levels were quantified using the 2−∆∆Cq
method (16). RT-qPCR reactions were
performed in triplicate with the following conditions: 95°C for 2
min; 40 cycles of 95°C for 15 sec and 60°C for 1 min.
 | Table I.Primers for reverse
transcription-quantitative PCR. |
Table I.
Primers for reverse
transcription-quantitative PCR.
| Gene | Primer sequence (5′ →
3′) |
|---|
| LINC00707 | F:
GCTGCACATTGAACCAGATA |
|
| R:
ATGTTCCAGTCCAGTCTCAT |
| Bcl-2 | F:
TCATGTGTGTGGAGAGCGTC |
|
| R:
AGCCTCCGTTATCCTGGATC |
| Bax | F:
GATGCGTCCACCAAGAAGCT |
|
| R:
CGGCCCCAGTTGAAGTTG |
| MRP1 | F:
GGCTCAAGGAGTATTCAGAG |
|
| R:
CCATCGATGATGATCTCTCC-3 |
| P-gp | F:
TCATCGAGTCACTGCCTAAT |
|
| R:
CTATGGCAATGCGTTGTTTC |
| 18s rRNA | F:
CCTGGATACCGCAGCTAGGA |
|
| R:
GCGGCGCAATACGAATGCCCC |
| miR-145 | F:
GCGCTCCAGCTGGGGTCCAGTTTTCCCAGGAATC |
|
| R:
CTCAACTGGTGTCGTGGA |
| U6 | F:
CTCGCTTCGGCAGCACA |
|
| R:
AACGCTTCACGAATTTGCGT |
Cell proliferation assay
The sensitivity of cells to DDP treatment was
determined using a CellTiter 96® Non-Radioactive Cell
Proliferation Assay kit (Promega Corporation). In brief,
transfected cells were seeded in triplicate into 96-well plates at
a density of 4×104 cells/well in 100 µl RPMI-1640 medium
(HyClone; GE Healthcare Life Sciences). After 12 h, A549 and
A549/DDP cells were treated with different concentrations of DDP
(5, 10, 20, 40 and 80 µM) for 1, 2 and 3 days. Subsequently, MTT
(10 µl; 5 mg/ml) was added into each well and incubated for 4 h at
37°C. Then 100 µl of dimethyl sulfoxide was added to dissolve the
solution and solubilize the crystals. The optical density was
detected at 570 nm using a microplate reader (Bio-Rad Laboratories,
Inc.). The in vitro DDP activity was expressed in terms of
concentrations capable of suppressing cell proliferation by 50%
(IC50). This assay was performed in triplicate.
Flow cytometric analysis of
apoptosis
The Annexin V-FITC Apoptosis Detection Kit (Nanjing
KeyGen Biotech Co., Ltd.) was used to evaluate cell apoptosis.
Briefly, A549/DDP cells (106 cells/ml) were harvested 48
h after transfection and washed twice with ice-cold PBS. The cells
were then resuspended in 500 µl of binding buffer. Next, the cells
were stained with 5 µl of Annexin V-FITC and 5 µl of propidium
iodide, and incubated at 25°C for 15 min in the dark according to
the manufacturer's protocol. Cell apoptosis was measured via
FACSCalibur flow cytometry (BD Biosciences). Results were analyzed
using BD FACSDiva software (version 8.0; BD Biosciences). This
assay was performed in triplicate. Apoptotic rate was calculated
using the sum of early apoptotic and late apoptotic cells.
Western blotting
Total protein was extracted from the cells using
RIPA solution containing phenylmethylsulfonyl fluoride (Beyotime
Institute of Biotechnology). Protein concentration was measured
using BCA reagent (Beyotime Institute of Biotechnology). Total
protein (30 µg) was separated via SDS-PAGE (10% gel) and then
transferred to PVDF membranes (EMD Millipore). After blocking with
5% non-fat milk at 25°C for 2 h, the membranes were subsequently
incubated overnight at 4°C with the following primary antibodies:
Anti-Bcl-2 (1:1,000; cat. no. 4223); anti-Bax (1:1,000; cat. no.
5023); anti-MRP1 (1:1,000; cat. no. 14685); anti-P-glycoprotein
(P-gp; 1:1,000; cat. no. 12683); and anti-GAPDH (1:5,000; cat. no.
5174) (all purchased from Cell Signaling Technology, Inc.). Next,
membranes were washed with TBS containing 1% Tween 20 (TBST),
followed by incubation with horseradish peroxidase-linked secondary
antibodies (1:5,000; cat. no. 4050-05; Southern Biotech) at 25°C
for 2 h. The membranes were then washed with TBST. Protein bands
were visualized via chemiluminescence using an ECL kit (Thermo
Fisher Scientific, Inc.). Parallel blotting of GAPDH served as the
internal reference.
Luciferase reporter assay
The putative binding sites between LINC00707 and
miR-145 were predicted using StarBase v2.0 (http://starbase.sysu.edu.cn/agoClipRNA.php?source=lncRNA).
Wild-type (WT) and mutant (MUT) LINC00707 containing putative
miR-145 binding sequences were generated and cloned downstream of
the luciferase reporter vector, psi-CHECK-2 (Promega Corporation).
They were subsequently co-transfected with luciferase plasmids (0.5
µg/per well), and miR-145 mimics (final concentration, 50 nM) or
miRNA negative controls (final concentration, 50 nM) using the
Lipofectamine 2000 reagent. Luciferase activities were detected 48
h post-transfection using a Dual-Luciferase Reporter Assay System
(Promega Corporation). Renilla luciferase activity was
normalized to Firefly luciferase activity. This assay was performed
in triplicate.
Statistical analysis
Statistical analyses were performed using SPSS
software (version 19.0; IBM Corporation). All data are expressed as
the mean ± standard deviation from three independent experiments.
Differences between two groups were identified using Student's
t-test, and differences between more than two groups were
identified using ANOVA followed by a least significant difference
post hoc test. P<0.05 was considered to indicate a statistically
significant difference.
Results
LINC00707 expression is upregulated in
A549/DDP cells
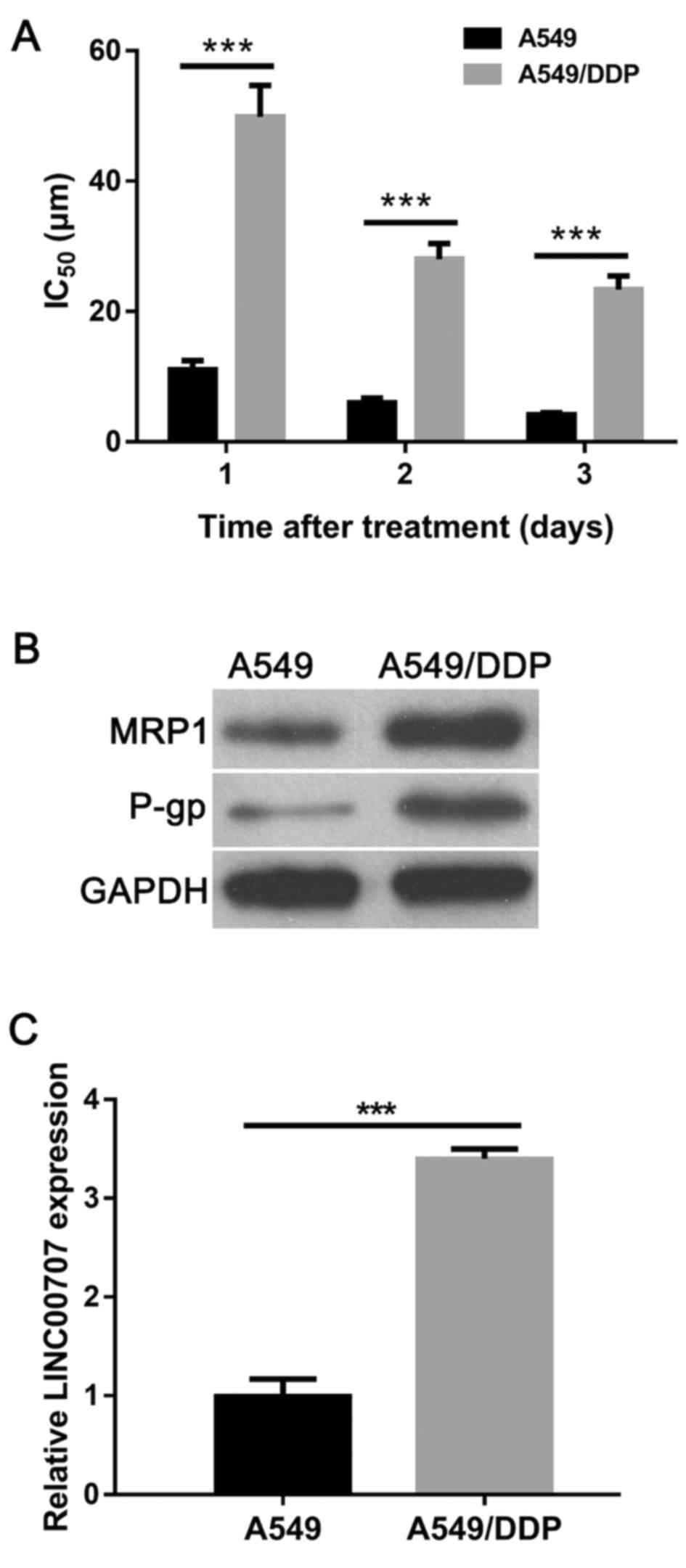
To confirm the DDP-resistant phenotype of purchased
A549/DDP cells, the half-maximal inhibitory concentration
(IC50) value of DDP, and the protein expression levels
of multidrug-resistance-related proteins MRP1 and P-gp were
examined in A549/DDP and parental A549 cells. The results showed
that the IC50 value of DDP in A549/DDP cells was
4.5-fold, 4.6-fold or 5.6-fold higher at 1, 2 or 3 days,
respectively, when compared with A549 cells (Fig. 1A). In addition, the protein
expression levels of MRP1 and P-gp were markedly increased in
A549/DDP cells when compared with those in A549 cells (Fig. 1B). These results verified the
DDP-resistant phenotype of A549/DDP cells. The RT-qPCR results
showed that LINC00707 expression was significantly upregulated in
A549/DDP cells when compared with in A549 cells (Fig. 1C). Therefore, this finding suggested
that LINC00707 may contribute to DDP resistance in A549/DDP
cells.
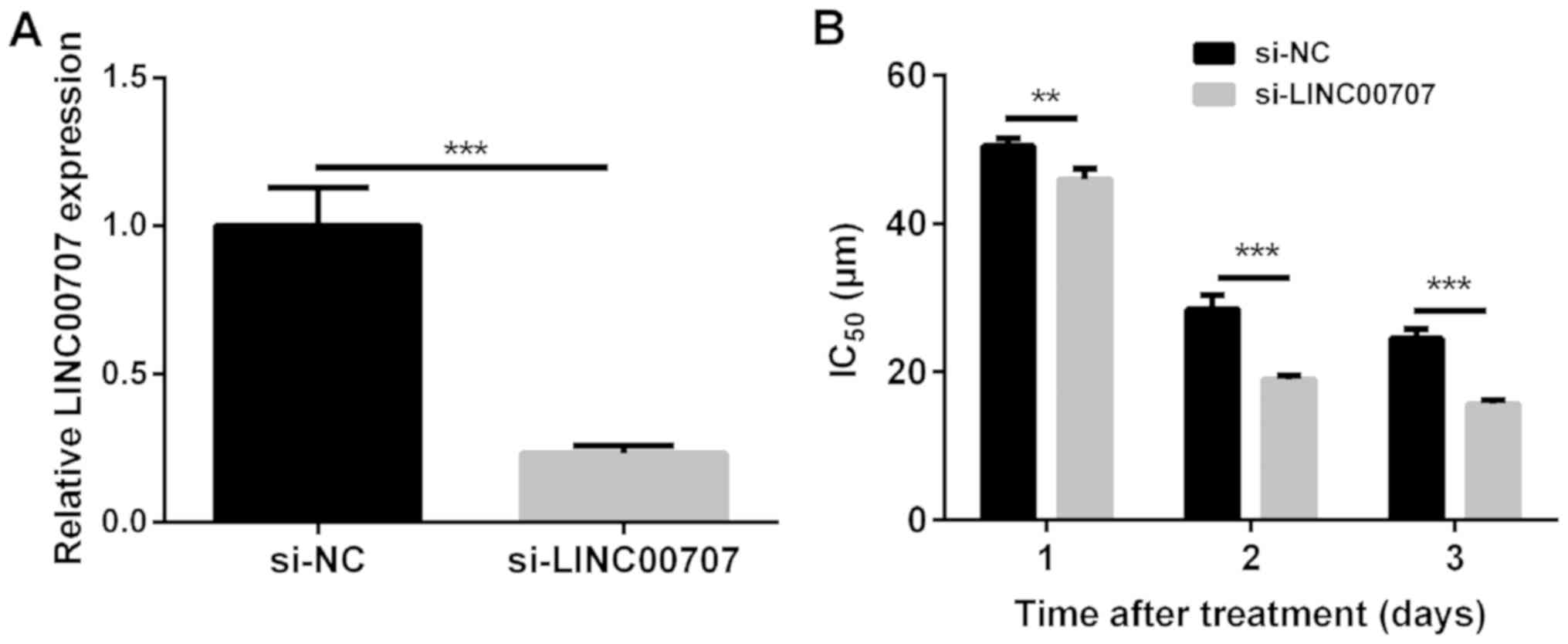
LINC00707 knockdown reduces the
IC50 value of DDP in A549/DDP cells
LINC00707 expression levels were observed to be
significantly decreased in A549/DDP cells transfected with
si-LINC00707 when compared with cells transfected with si-NC in the
control group (Fig. 2A). An MTT
assay revealed that si-LINC00707 significantly reduced the
IC50 value of DDP in A549/DDP cells compared with the
si-NC group (Fig. 2B). Thus, these
results indicated that LINC00707 knockdown induces DDP sensitivity
in A549/DDP cells.
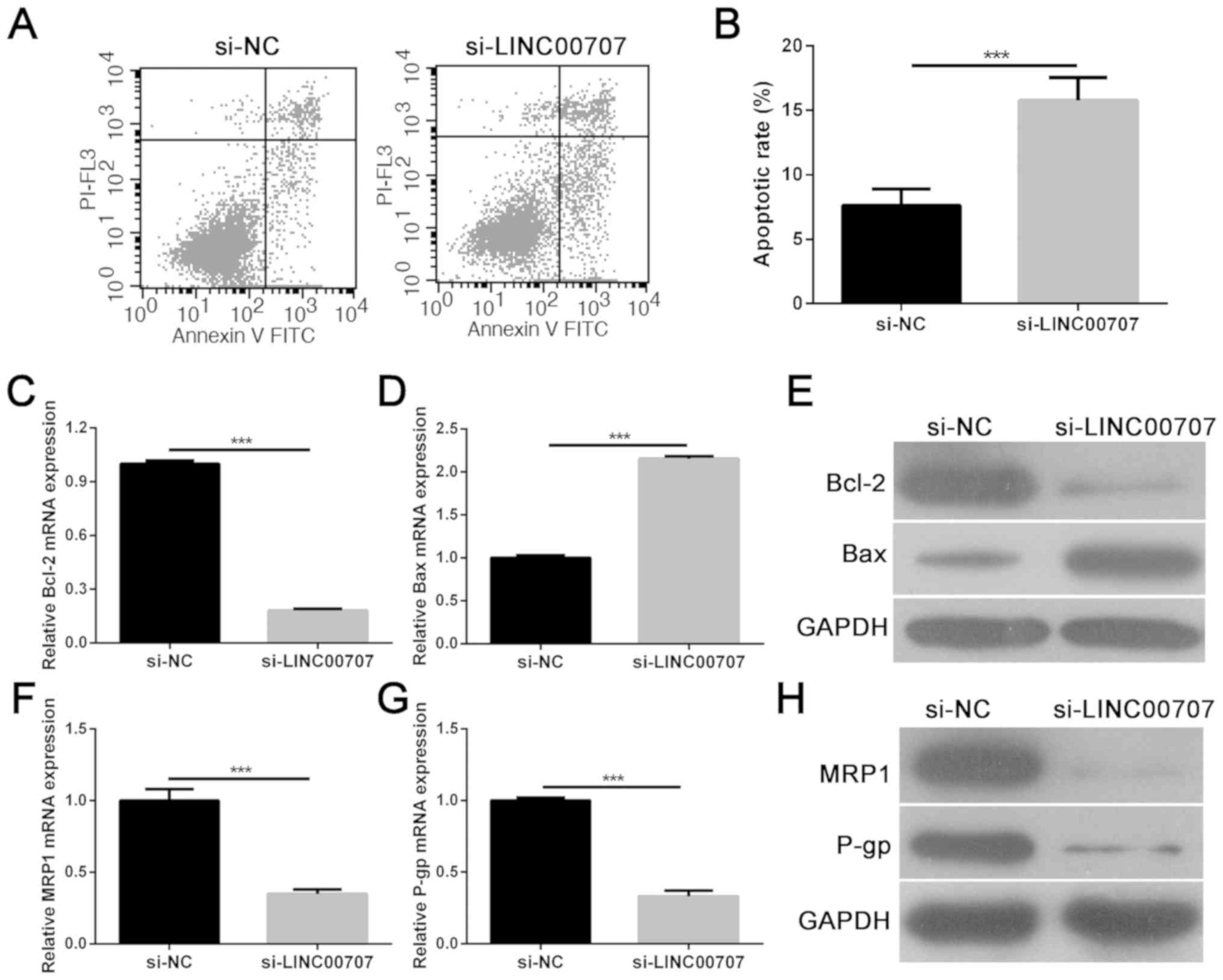
LINC00707 knockdown enhances apoptosis
and influences the expression of multidrug-resistance-related
proteins in A549/DDP cells
Flow cytometric analysis demonstrated that LINC00707
knockdown significantly accelerated A549/DDP cell apoptosis when
compared with the si-NC group (Fig. 3A
and B). Furthermore, the expression levels of the
apoptosis-associated proteins Bcl-2 and Bax were analyzed in
A549/DDP cells transfected with si-LINC00707 or si-NC using RT-qPCR
and western blotting. Results showed that LINC00707 knockdown
significantly inhibited Bcl-2 mRNA and protein expression, and
promoted Bax mRNA and protein expression in A549/DDP cells when
compared with the si-NC group (Fig.
3C-E). Moreover, multidrug-resistance-related proteins, MRP1
and P-gp, were selected as detection indexes to analyze the effect
of LINC00707 knockdown on DDP resistance in A549/DDP cells. The
results showed that LINC00707 knockdown significantly decreased
MRP1 and P-gp mRNA and protein levels in A549/DDP cells when
compared with the si-NC group (Fig.
3F-H). Thus, these results indicated that LINC00707 knockdown
enhances apoptosis and DDP sensitivity in A549/DDP cells.
LINC00707 functions as a molecular
miR-145 sponge in A549/DDP cells
Bioinformatics analysis was used to determine the
putative binding sites between LINC00707 and miR-145, and the
results revealed a miR-145 binding site in LINC00707 (Fig. 4A). RT-qPCR demonstrated that miR-145
expression was significantly decreased in A549/DDP cells when
compared with in parental A549 (Fig.
4B). As shown in Fig. 1C,
LINC00707 expression was significantly upregulated in A549/DDP
cells when compared with that in parental A549 cells. Therefore, it
was hypothesized that LINC00707 may lead to this result by sponging
miR-145. miR-145 levels were significantly upregulated in A549/DDP
cells following transfection with miR-156 mimics (Fig. 4C). Dual-luciferase reporter assay
results indicated that cells co-transfected with WT-LINC00707 and
miR-145 mimics exhibited significantly weakened luciferase
activity, whereas no significant difference was identified in
luciferase activity in cells co-transfected with MUT-LINC00707 and
miR-145 mimics compared with the NC group (Fig. 4D). LINC00707 knockdown markedly
promoted miR-145 expression in A549/DDP cells (Fig. 4E). Overall, these results suggest
that LINC00707 may function as a molecular sponge by competitively
binding to miR-145.
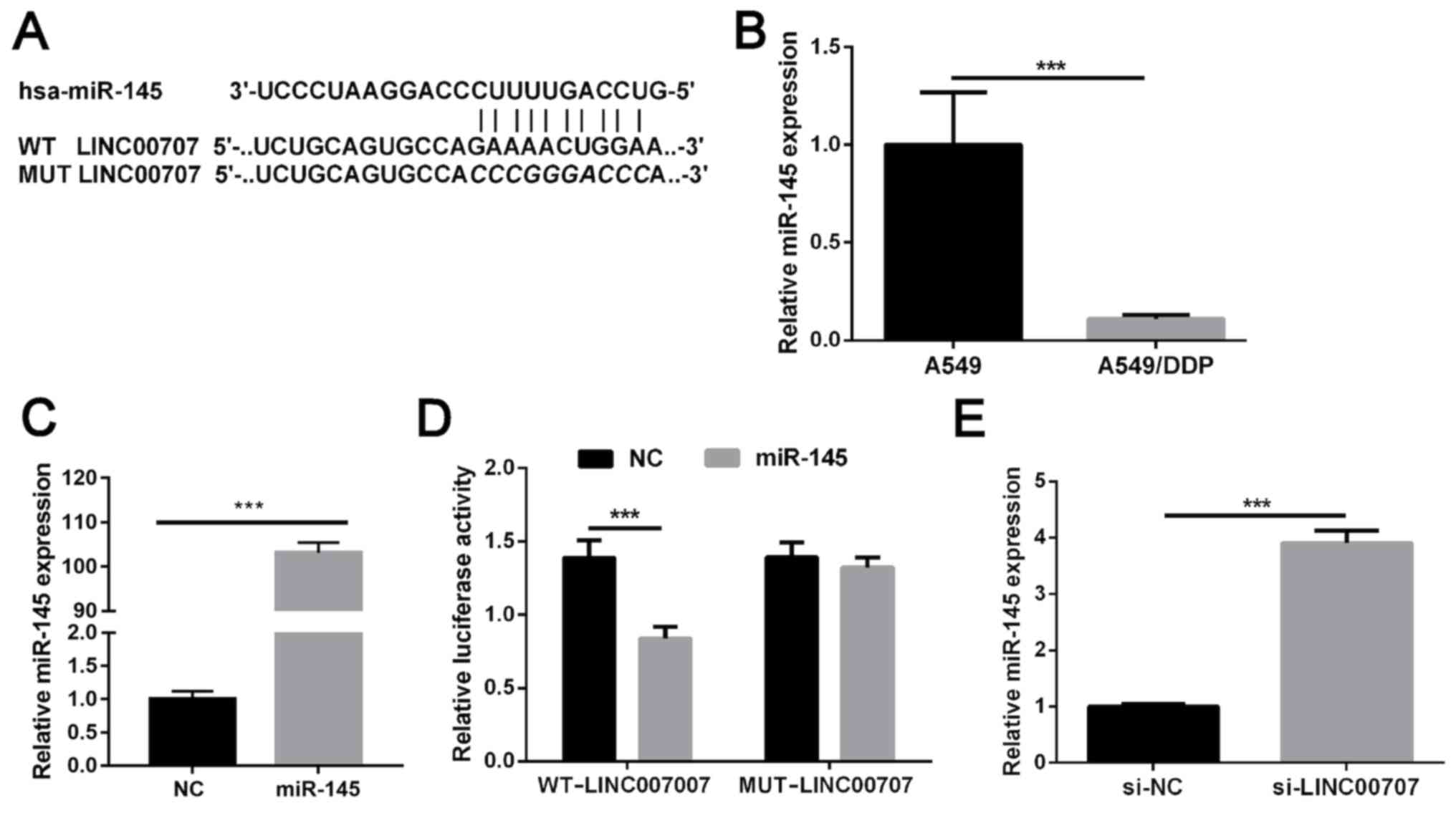 | Figure 4.LINC00707 functions as a molecular
miR-145 sponge in A549/DDP cells. (A) Predicted binding sites
between LINC00707 and miR-145. (B) Relative expression of miR-145
in A549/DDP cells and parental A549 cells was measured using
RT-qPCR. ***P<0.001 vs. A549. (C) miR-145 was upregulated in
cells transfected with miR-145 mimics. ***P<0.001 vs. NC. (D)
Luciferase activity was measured in cells co-transfected with
either WT-LINC00707 or MUT-LINC00707, and miRNA negative control or
miR-145 mimics. ***P<0.001 vs. WT-LINC00707. (E) Relative
expression levels of miR-145 in A549/DDP cells transfected with
either si-LINC00707 or si-NC were measured using RT-qPCR.
***P<0.001 vs. si-NC. LINC00707, long intergenic non-coding RNA
00707; miR, microRNA; RT-qPCR, reverse transcription-quantitative
PCR; A549/DDP cells, DDP-resistant A549 cells; si-LINC00707,
LINC00707 siRNA; si-NC, siRNA negative control; WT, wild-type; MUT,
mutant; NC, negative control. |
miR-145 downregulation weakens the
effect of LINC00707 knockdown in A549/DDP cells
To gain insight into the mechanism via which
LINC00707 knockdown enhanced DDP sensitivity in A549/DDP cells, an
miR-145 inhibitor or inhibitor NC was further transfected into
A549-DDP cells transfected with si-LINC00707. RT-qPCR analysis
revealed that miR-145 inhibitor significantly downregulated miR-145
expression in A549/DDP cells compared with inhibitor NC (Fig. 5A). Additionally, it was demonstrated
that the miR-145 inhibitor significantly downregulated miR-145
expression in A549-DDP cells transfected with si-LINC00707 when
compared with the control group (Fig.
5B). It was further explored whether miR-145 downregulation
reversed the effect of LINC00707 knockdown in A549/DDP cells. An
MTT assay showed that miR-145 downregulation significantly
increased the IC50 value of DDP in A549/DDP cells
transfected with si-LINC00707 (Fig.
5C). Flow cytometric analysis also demonstrated that miR-145
downregulation inhibited apoptosis in A549/DDP cells transfected
with si-LINC00707 (Fig. 5D and E).
Furthermore, western blotting results indicated that miR-145
downregulation promoted Bcl-2, MRP1 and P-gp expression, while
reducing Bax expression in A549/DDP cells transfected with
si-LINC00707 and miR-145 inhibitor (Fig.
5F and G). Thus, these results indicated that miR-145
downregulation may reverse the effect of LINC00707 knockdown in
A549/DDP cells.
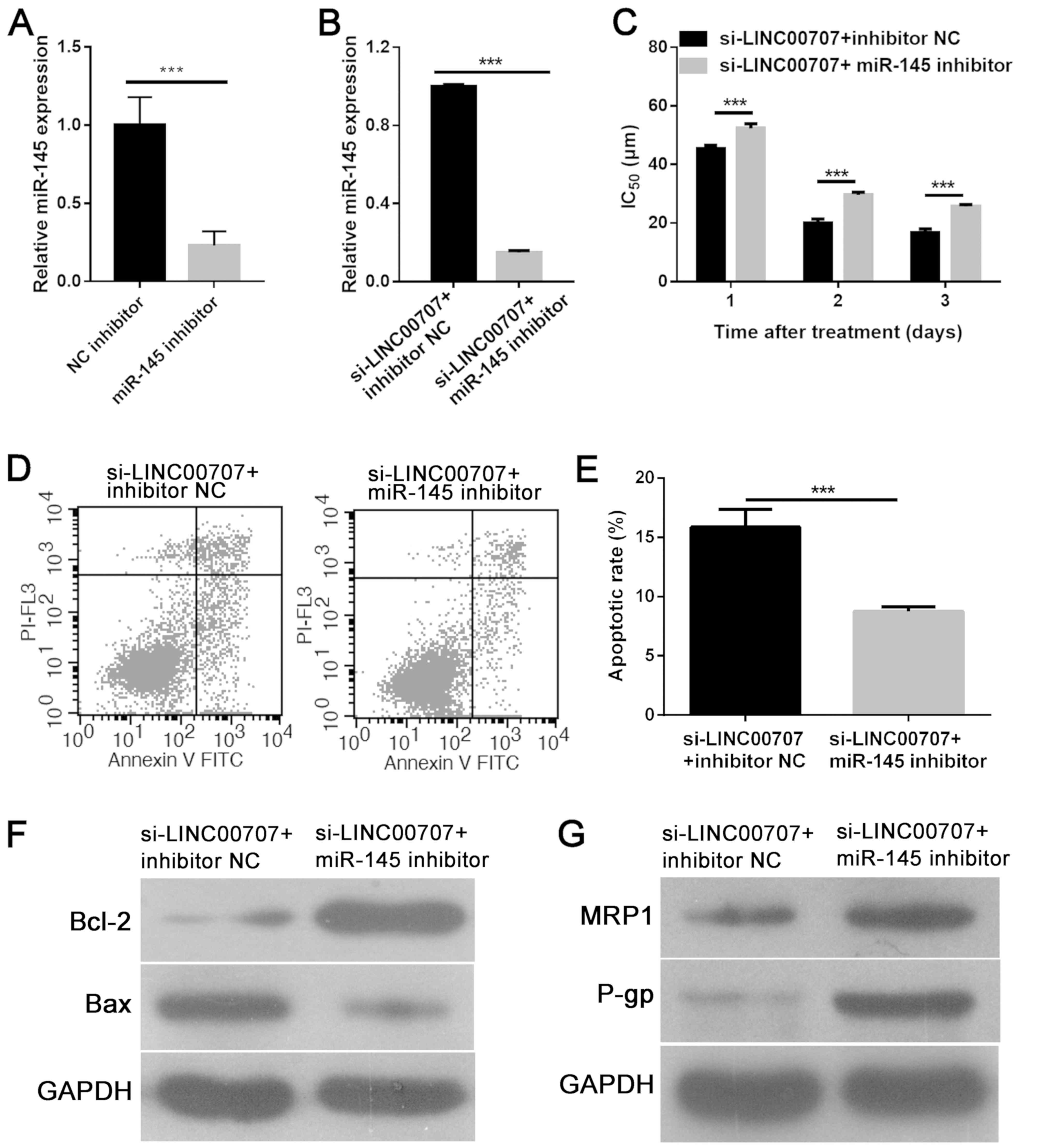 | Figure 5.miR-145 downregulation reverses the
effect of LINC00707 knockdown in A549/DDP cells. (A) miR-145 was
downregulated in A549/DDP cells transfected with miR-145 inhibitor.
(B) Relative expression levels of miR-145 in A549/DDP cells
transfected with si-LINC00707 + inhibitor NC or si-LINC00707 +
miR-145 inhibitor were measured using RT-qPCR. (C) IC50
values of DDP in A549/DDP cells transfected with either
si-LINC00707 + inhibitor NC or si-LINC00707 + miR-145 inhibitor
were measured using an MTT assay. (D-E) Apoptotic cell rates of
A549/DDP cells transfected with either si-LINC00707 + inhibitor NC
or si-LINC00707 + miR-145 inhibitor were measured using flow
cytometry. Protein expression levels of (F) Bcl-2 and Bax, and (G)
MRP1 and P-gp in A549/DDP cells transfected with either
si-LINC00707 + inhibitor NC or si-LINC00707 + miR-145 inhibitor
were measured using western blotting. ***P<0.001 vs.
si-LINC00707 + inhibitor NC. MRP1, multidrug resistance protein 1;
P-gp, P-glycoprotein; IC50, half-maximal inhibitory
concentration; LINC00707, long intergenic non-coding RNA 00707;
miR, microRNA; si-LINC00707, LINC00707 siRNA; NC, negative control;
A549/DDP cells, DDP-resistant A549 cells; RT-qPCR, reverse
transcription-quantitative PCR. |
Discussion
DDP is considered to be a classical chemotherapeutic
drug used for treating patients with NSCLC. However, DDP resistance
among patients with NSCLC presents a significant barrier towards
successful chemotherapy (17).
Therefore, there is an urgent need to elucidate the molecular and
biological mechanisms underlying the development of DDP resistance.
In the present study, LINC00707 expression was found to be
significantly upregulated in A549/DDP cells. Correspondingly,
LINC00707 knockdown enhanced A549/DDP cell sensitivity towards DDP.
Moreover, it was demonstrated that miR-145 was a target of
LINC00707 and that miR-145 downregulation was capable of reversing
the effect of LINC00707 knockdown in A549/DDP cells.
A number of studies have shown that lncRNA
dysregulation fuels drug resistance in human cancers, including
NSCLC (18,19). For example, lncRNA maternally
expressed 3 was shown to be downregulated in DDP-resistant NSCLC
cells, and its overexpression enhanced DDP sensitivity in NSCLC
cells in vitro (18).
Therefore, comprehensive elucidation of lncRNA regulatory
mechanisms in drug resistance may provide a promising therapeutic
strategy for the treatment of NSCLC. LINC00707 was previously
identified to be an oncogene in various cancers. It was shown to be
upregulated in hepatocellular carcinoma cells, thereby promoting
hepatocellular carcinoma progression (8). Its expression was also shown to be
highly upregulated in gastric cancer tissues and cells, thus
promoting their proliferation and metastasis by interacting with
human antigen R (9). In lung cancer,
Ma et al (10) found that
LINC00707 expression was clearly upregulated in lung adenocarcinoma
tissues and cells; notably, LINC00707 promoted lung adenocarcinoma
progression by regulating cell division control protein 42.
However, the role of LINC00707 in the progression of DDP resistance
in NSCLC still remains unclear. Herein, it was revealed that
LINC00707 expression was highly upregulated in A549/DDP cells.
LINC00707 knockdown reduced the IC50 value of DDP in
A549/DDP cells. In addition, LINC00707 knockdown enhanced the
percentage of apoptotic A549/DDP cells, inhibited the expression of
anti-apoptotic protein Bcl-2 and promoted the expression of
pro-apoptotic protein Bax in A549/DDP cells. These results
indicated that LINC00707 knockdown enhances the DDP sensitivity of
A549/DDP cells. By investigating the underlying mechanism, it was
demonstrated that LINC00707 knockdown inhibited the expression of
MRP1 and P-gp. The official full name of MRP1 is ATP binding
cassette subfamily C member 1. The official full name of P-gp is
ATP binding cassette subfamily B member 1. Both MRP1 and P-gp are
members of the superfamily of ATP-binding cassette transporters,
which is involved in multidrug resistance. The increased expression
of MRP1 and P-gp usually represents the enhancement of multidrug
resistance (20). Therefore, the
results of the present study suggest that LINC00707 knockdown
enhances DDP sensitivity by weakening multidrug resistance.
Recent studies have proposed that lncRNAs, including
LINC00707, may serve as molecular miRNA sponges, thus affecting
their target gene expression indirectly. For example, Jia et
al (21) reported that LINC00707
sponged miR-370-3p to promote the osteogenesis of human bone
marrow-derived mesenchymal stem cells by increasing WNT2B. In
addition, Tu et al (22)
found that LINC00707 contributed to hepatocellular carcinoma
progression by sponging miR-206, which led to the upregulation of
CDK14. In the present study, miR-145 was identified as a target of
LINC00707. Changes in LINC00707 expression resulted in
corresponding changes in miR-145 expression, thereby indicating
that miR-145 expression was negatively regulated by LINC00707.
miR-145, a known tumor suppressor, has been commonly reported to be
downregulated in various types of human cancers, including
colorectal cancer (23), breast
cancer (24) and NSCLC (25), and has been shown to suppress tumor
cell proliferation, apoptosis, migration and invasion (26). In the present study, it was revealed
that miR-145 downregulation markedly reversed LINC00707
knockdown-induced cell dysfunction in A549-DDP cells. This was
determined by the increased IC50 value of DDP, reduced
apoptosis, increased Bcl-2, MRP1 and P-gp protein expression
levels, and attenuated Bax protein expression levels in A549/DDP
cells. Interestingly, Zhan et al (15) reported that miR-145 promoted MRP1
mRNA degradation by directly targeting its 3′-UTR, thereby
sensitizing gallbladder cancer cells to DDP. Similarly, in the
present study, it was demonstrated that miR-145 downregulation led
to upregulated MRP1 protein expression, thus suggesting that
miR-145 downregulation may enhance DDP activity in A549/DDP cells
by regulating the expression of MRP1. These results indicated that
silencing of LINC00707 enhances DDP sensitivity in NSCLC cells by
sponging miR-145.
There are certain limitations in this study.
Firstly, the effect of LINC00707 knockdown-induced DDP resistance
was analyzed only in A549/DDP cells; additional NSCLC cells were
not included. Furthermore, although miR-145 was identified as a
target of LINC00707, its target genes were not identified. Finally,
the effect of LINC00707 knockdown on A549/DDP cells was not
verified in vivo. Despite these limitations, the present
study indicated that LINC00707 contributed to the progression of
DDP resistance in NSCLC cells.
In conclusion, LINC00707 was identified to be highly
expressed in A549/DDP cells, and its knockdown was in turn found to
significantly enhance DDP sensitivity in A549/DDP cells by sponging
miR-145. Therefore, these findings suggest that LINC00707 may be a
potential target in the treatment of DDP-resistant NSCLC
patients.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author.
Authors' contributions
HZ was involved in study design, conducting all
experiments and preparing the manuscript. YL was involved in data
collection and literature analysis. WX was responsible for
performing the cell culture. KL and CL were responsible for
performing the western blot analysis. All of the authors read and
approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zhou K, Wang L, Cheng R, Liu X, Mao S and
Yan Y: Elemene increases autophagic apoptosis and drug sensitivity
in human cisplatin (DDP)-resistant lung cancer cell line
SPC-A-1/DDP By inducing beclin-1 expression. Oncol Res. 2017.(Epub
ahead of print). View Article : Google Scholar
|
|
3
|
Hu Y, Zhu QN, Deng JL, Li ZX, Wang G and
Zhu YS: Emerging role of long non-coding RNAs in cisplatin
resistance. OncoTargets Ther. 11:3185–3194. 2018. View Article : Google Scholar
|
|
4
|
Zhang S, Ma H, Zhang D, Xie S, Wang W, Li
Q, Lin Z and Wang Y: LncRNA KCNQ1OT1 regulates proliferation and
cisplatin resistance in tongue cancer via miR-211-5p mediated
Ezrin/Fak/Src signaling. Cell Death Dis. 9:7422018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wu F, Li J, Du X, Zhang W, Lei P and Zhang
Q: Long noncoding RNA AB019562 promotes cell proliferation and
metastasis in human hepatocellular carcinoma. Mol Med Rep.
16:69–74. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hu B, Zhang H, Wang Z, Zhang F, Wei H and
Li L: LncRNA CCAT1/miR-130a-3p axis increases cisplatin resistance
in non-small-cell lung cancer cell line by targeting SOX4. Cancer
Biol Ther. 18:974–983. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ma LY, Xie XW, Ma L, Pang JL, Xiong XM,
Zheng HD, Shen XL, Wen ZG and Wang HY: Downregulated long
non-coding RNA TRPM2-AS inhibits cisplatin resistance of non-small
cell lung cancer cells via activation of p53- p66shc pathway. Eur
Rev Med Pharmacol Sci. 21:2626–2634. 2017.PubMed/NCBI
|
|
8
|
Wang J, Luo Z, Yao T, Li W and Pu J:
LINC00707 promotes hepatocellular carcinoma progression through
activating ERK/JNK/AKT pathway signaling pathway. J Cell Physiol.
234:6908–6916. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Xie M, Ma T, Xue J, Ma H, Sun M, Zhang Z,
Liu M, Liu Y, Ju S, Wang Z and De W: The long intergenic
non-protein coding RNA 707 promotes proliferation and metastasis of
gastric cancer by interacting with mRNA stabilizing protein HuR.
Cancer Lett. 443:67–79. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ma T, Ma H, Zou Z, He X, Liu Y, Shuai Y,
Xie M and Zhang Z: The long intergenic noncoding RNA 00707 promotes
lung adenocarcinoma cell proliferation and migration by regulating
Cdc42. Cell Physiol Biochem. 45:1566–1580. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yuan Y, Haiying G, Zhuo L, Ying L and Xin
H: Long non-coding RNA LINC00339 facilitates the tumorigenesis of
non-small cell lung cancer by sponging miR-145 through targeting
FOXM1. Biomed Pharmacother. 105:707–713. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Liu J, Xing Y and Rong L: miR-181
regulates cisplatin-resistant non-small cell lung cancer via
downregulation of autophagy through the PTEN/PI3K/AKT pathway.
Oncol Rep. 39:1631–1639. 2018.PubMed/NCBI
|
|
14
|
Liu K, Chen H, You Q, Ye Q, Wang F, Wang
S, Zhang S, Yu K, Li W and Gu M: miR145 inhibits human
nonsmall-cell lung cancer growth by dual-targeting RIOK2 and NOB1.
Int J Oncol. 53:257–265. 2018.PubMed/NCBI
|
|
15
|
Zhan M, Zhao X, Wang H, Chen W, Xu S, Wang
W, Shen H, Huang S and Wang J: miR-145 sensitizes gallbladder
cancer to cisplatin by regulating multidrug resistance associated
protein 1. Tumour Biol. 37:10553–10562. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Thatcher N, Hirsch FR, Luft AV, Szczesna
A, Ciuleanu TE, Dediu M, Ramlau R, Galiulin RK, Bálint B, Losonczy
G, et al: Necitumumab plus gemcitabine and cisplatin versus
gemcitabine and cisplatin alone as first-line therapy in patients
with stage IV squamous non-small-cell lung cancer (SQUIRE): An
open-label, randomised, controlled phase 3 trial. Lancet Oncol.
16:763–774. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang P, Chen D, Ma H and Li Y: LncRNA MEG3
enhances cisplatin sensitivity in non-small cell lung cancer by
regulating miR-21-5p/SOX7 axis. Onco Targets Ther. 10:5137–5149.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chen QN, Wei CC, Wang ZX and Sun M: Long
non-coding RNAs in anti-cancer drug resistance. Oncotarget.
8:1925–1936. 2017.PubMed/NCBI
|
|
20
|
Tekchandani P, Kurmi BD and Paliwal SR:
Nanomedicine to deal with cancer cell biology in multi-drug
resistance. Mini Rev Med Chem. 17:1793–1810. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Jia B, Wang Z, Sun X, Chen J, Zhao J and
Qiu X: Long noncoding RNA LINC00707 sponges miR-370-3p to promote
osteogenesis of human bone marrow-derived mesenchymal stem cells
through upregulating WNT2B. Stem Cell Res Ther. 10:672019.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Tu J, Zhao Z, Xu M, Chen M, Weng Q, Wang J
and Ji J: LINC00707 contributes to hepatocellular carcinoma
progression via sponging miR-206 to increase CDK14. J Cell Physiol.
234:10615–10624. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Sheng N, Tan G, You W, Chen H, Gong J,
Chen D, Zhang H and Wang Z: MiR-145 inhibits human colorectal
cancer cell migration and invasion via PAK4-dependent pathway.
Cancer Med. 6:1331–1340. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ding Y, Zhang C, Zhang J, Zhang N, Li T,
Fang J, Zhang Y, Zuo F, Tao Z, Tang S, et al: miR-145 inhibits
proliferation and migration of breast cancer cells by directly or
indirectly regulating TGF-β1 expression. Int J Oncol. 50:1701–1710.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li JC and Zheng JQ: Effect of microRNA-145
on proliferation and apoptosis of human non-small cell lung cancer
A549 cells by regulating mTOR signaling pathway. J Cell Biochem.
2017.(Epub ahead of print). View Article : Google Scholar
|
|
26
|
Zhou X, Yue Y, Wang R, Gong B and Duan Z:
MicroRNA-145 inhibits tumorigenesis and invasion of cervical cancer
stem cells. Int J Oncol. 50:853–862. 2017. View Article : Google Scholar : PubMed/NCBI
|