Introduction
Melanoma is a tumor that originates in melanocytes
of the skin or other parts of the body (1). The main function of melanocytes is to
produce melanin via melanogenesis, a multistep biochemical process
regulated by L-tyrosine, L-DOPA and other hormones (2,3).
Melanogenesis leads to the upregulation of hypoxia-inducible factor
1, which modulates the cellular metabolism of melanoma (4). A previous study has demonstrated that
pigmentation level is associated with the overall and disease-free
survival time of patients with stage III and IV melanoma (5). In the United States, >91,000
individuals were diagnosed with cutaneous melanoma in 2018, and
>9,000 patients succumbed to the disease in the same period
(6). Since melanoma tends to spread
lymphogenously and hematogenously, patients with inoperable
metastatic melanoma exhibit median survival times between 8 and 12
months (7). Therefore, melanoma
poses a serious threat to life.
Gene mutations in melanoma may activate multiple
signaling pathways that regulate proliferation,
epithelial-mesenchymal transition, invasion and metastasis in an
abnormal manner (8). For example,
BRAF mutations, predominantly V600E, occur in 40–50% of all
melanomas, whereas NRAS proto-oncogene, GTPase and neurofibromin 1
mutations occur in ~20 and 15% of melanomas, respectively (9). Targeted therapy and immunotherapy have
been demonstrated to be effective treatment methods (10,11).
BRAF/mitogen-activated protein kinase kinase inhibitors, as well as
antibodies against cytotoxic T-lymphocyte-associated protein 4 and
programmed cell death protein 1 have been used for treatment of
metastatic melanoma, with patient response rates ranging between 20
and 70% (12). Although these
breakthrough treatments have prolonged progression-free survival to
a certain extent, drug resistance still limits their effectiveness
(13). For example, immune-based
therapy is subject to limitations, such as the prevention of the
generation of an immunosuppressive environment (14). Therefore, there remains a need for
novel markers of prognosis and novel therapeutic drugs for
melanoma.
Weighted gene co-expression network analysis (WGCNA)
is widely used to analyze genetic expression data, locate modules
of highly correlated genes and identify potential biomarkers, as
well as therapeutic targets. Thus, the present study aimed to
utilize WGCNA to identify novel biomarkers associated with melanoma
prognosis. Additionally, the present study aimed to determine the
proximity between disease-associated proteins and drug targets in
the human protein-protein interactome in order to identify
potential drugs for the treatment of melanoma.
Materials and methods
Data processing
Melanoma transcriptome dataset GSE65904 (15) was downloaded from the Gene Expression
Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo). GSE65904 comprised
214 samples from patients with melanoma, no non-tumor tissue
samples or healthy subjects were included. Illumina HumanHT-12V4.0
expression beadchip was used as the sequencing platform. Clinical
information of patients, including sex, age, tumor stage, distant
metastasis and survival state, was collected. The GEO query package
in R v2.52.0 (https://git.bioconductor.org/packages/GEOquery) was
used to process the data. If the expression of a gene was not
significant compared with the background value (standard probe) in
>25% of all samples (P>0.05), the probe was removed from
further analysis. A total of 10,566 genes were obtained.
Weighted co-expression network
construction
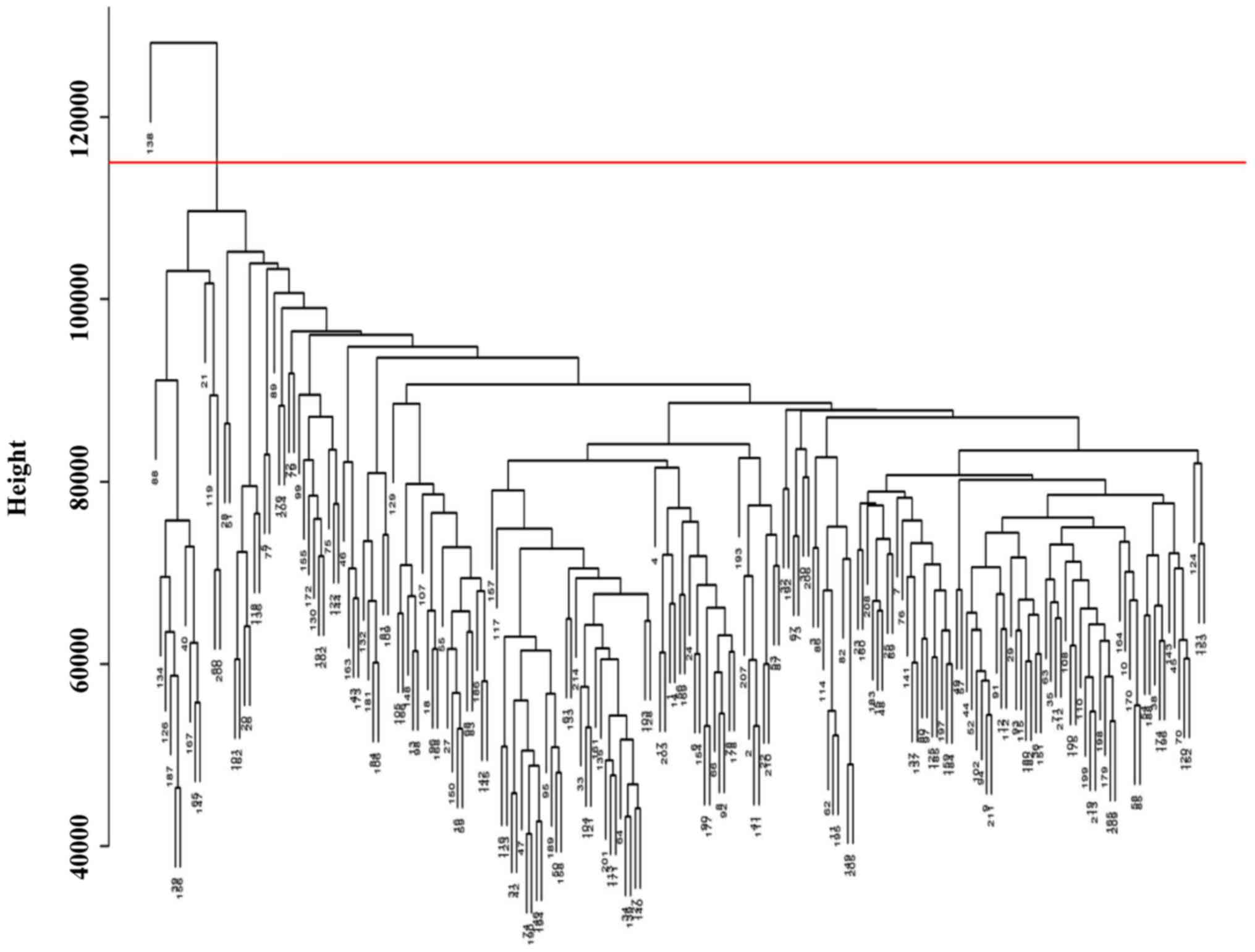
The top 50% most differentially expressed genes
(5,283 genes) were selected for WGCNA analysis following analysis
of variance using R 3.3.2 (https://www.r-project.org/) (16). These genes were used for screening
and cluster analysis of all samples, as well as to identify
outliers, following which one patient was removed from the study
(Fig. 1). The gene expression data
of the patients was used to construct the co-expression network,
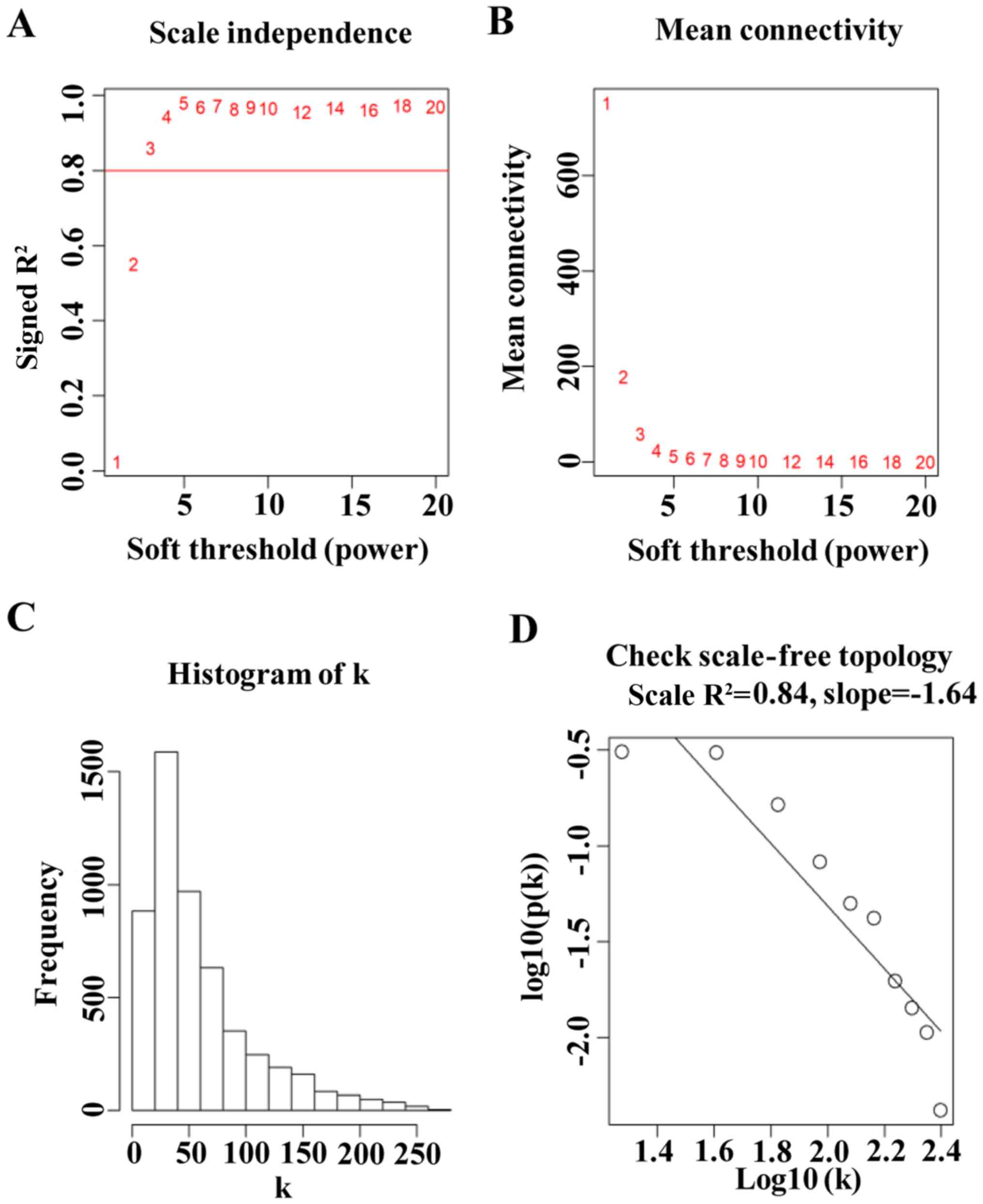
and the WGCNA algorithm was utilized for analysis (16). To ensure that the nodes of the
constructed co-expression network conformed to the power rate
distribution, appropriately soft threshold was selected (β=3),
which enabled the deletion of low mutual correlation relationships.
The distribution of network nodes conformed to the power rate
distribution at β=3. Further investigation of the distribution of
node degrees in the co-expression network revealed that the degree
of nodes conformed to the power law distribution. This indicated
that the constructed co-expression network was a scale-free
network, conforming to the characteristics of common biological
networks. The average linkage hierarchical clustering method
(17) was used to cluster all
genes.
Identification of clinically
significant modules
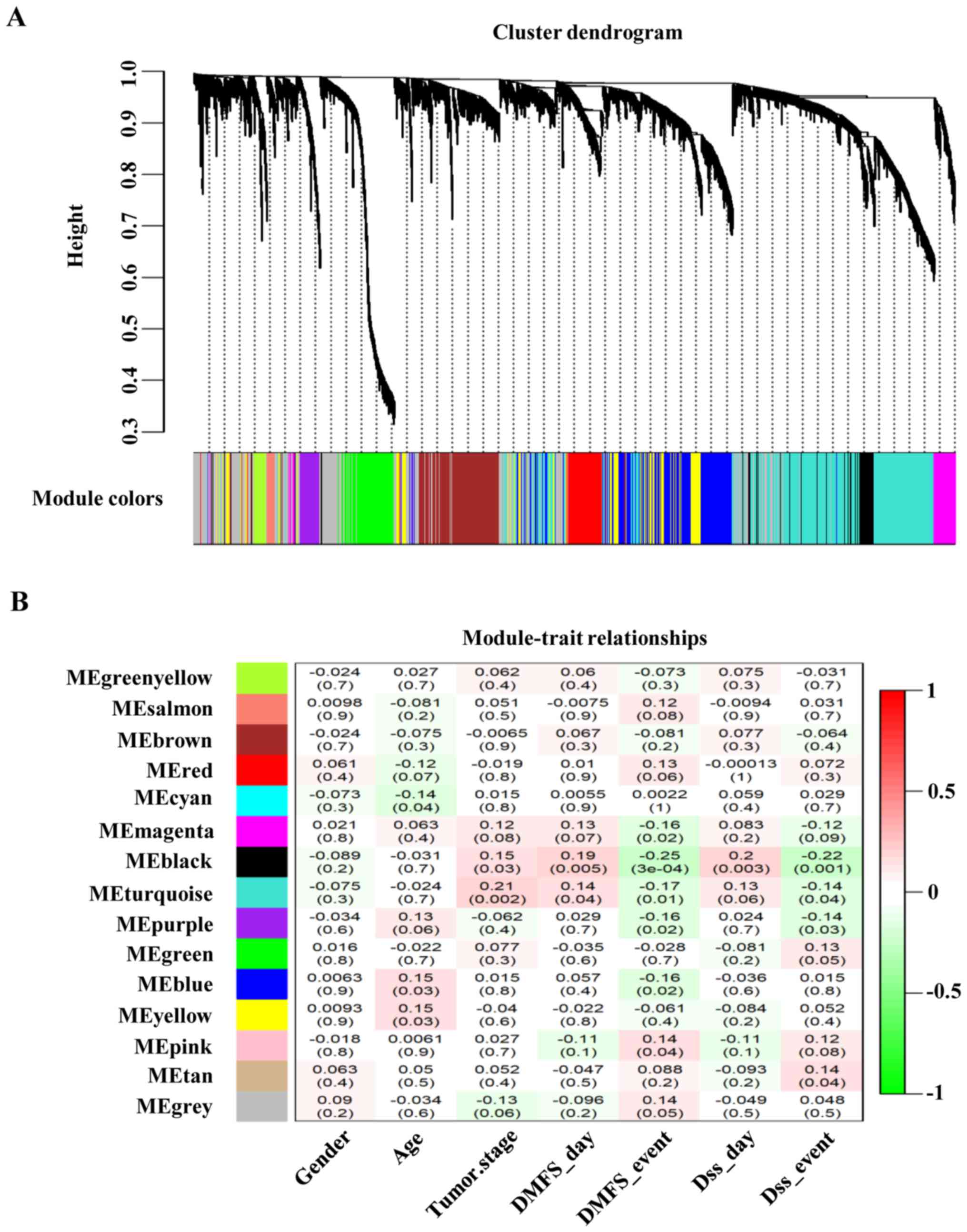
To obtain the gene modules that were associated with
clinical phenotypes, the correlation between modules and clinical
phenotypes was determined. Module eigengenes (MEs) were considered
as characteristics of all genes in a certain module. The
association between MEs and clinical characteristics was analyzed
to determine a clinically significant module for further use.
Gene Ontology (GO) and pathway
enrichment analysis
The ClusterProfiler package (https://github.com/GuangchuangYu/clusterProfiler)
in R v3.12.0 was used to determine the functions of the enriched
genes from the two modules in Fig. 3
(black and turquoise modules) in GO (18) and the Kyoto Encyclopedia of Genes and
Genomes (KEGG) (19) pathway
analysis, respectively. Genes in the clinically significant module
were categorized into three functional groups: Biological process
(BP), cellular component (CC) and molecular function (MF).
Identification and validation of hub
genes
To identify genes associated with melanoma
prognosis, the association between each gene and clinical
characteristics was evaluated, as well as the association between
each gene and core modules, such as module membership (MM) and gene
significance (GS). MM is defined by the correlation between the
gene expression profile and MEs, whereas GS is defined by the
association between a gene and external traits. Genes with
|MM+GS|=5% in the aforementioned modules (black and turquoise
modules) in Fig. 3 were selected as
potentially prognostic genes; all other genes were removed. To
further analyze the association between these genes, the remaining
candidate genes were input into STRING (https://string-db.org/) to construct a protein-protein
interaction (PPI) network using Cytoscape v3.2 (20).
To verify whether the identified genes were
associated with tumor progression and prognosis, the association
between each gene and survival was determined using the R survival
package v2.41-3 (https://cran.r-project.org/web/packages/survminer/index.html).
Clinical and RNA-sequencing data from 417 patients with melanoma
were downloaded from The Cancer Genome Atlas (TCGA) database
(https://cancergenome.nih.gov/) using the
TCGA biolinks package in R v2.12.3 (https://git.bioconductor.org/packages/TCGAbiolinks).
Overall survival was analyzed using the log-rank test. In addition,
the ggpubr package v0.2.1 (http://cran.r-project.org/web/packages/ggpubr/index.html)
was used to demonstrate the mRNA expression of hub genes in primary
and metastatic tumor, and the two groups were compared by Student's
t-test. Receiver operating characteristic (ROC) curve and area
under the curve (AUC) values were obtained using the pROC package
v1.15.0 (http://cran.r-project.org/web/packages/ROCR) to
evaluate the efficiency of the genes in distinguishing metastatic
and non-metastatic tumors.
Screening candidates for
treatment
Drug-target information of Food and Drug
Administration (FDA)-approved drugs was obtained from DrugBank
(https://www.drugbank.ca/). The exclusion of drugs
that had no known targets in the interactome resulted in a total of
1,269 unique drugs and 1,185 targets selected for further analysis.
Notably, only pharmacological targets (‘Targets’ section in
DrugBank), excluding enzymes, carriers and transporters typically
shared among different drugs, were considered. The protein
interaction information was obtained from a previously published
study, which contained data from 15 databases (21). Among these, 15,969 nodes and 217,160
mutual relationships were identified in the PPI networks. The
prognostic genes of melanoma were mapped to the PPI network. The
Igraph package v1.2.4.1 (https://igraph.org/) was used to estimate the shortest
distance between each target and a particular prognostic gene for
each FDA-approved drug (21).
Standardization-based approximation indicated that lower values
were associated with an increased likelihood that the drug may act
on melanoma and prevent its progression.
Results
Weighted co-expression network
construction and key module identification
Following a cluster analysis of all samples, one
sample in GSE65904 was removed from subsequent analysis due to bias
(Fig. 1; Table I). To ensure a scale-free network, it
must satisfy R2> 0.8 (Fig.
2A), and the mean connectivity should be conserved as much as
possible (Fig. 2B). Furthermore, the
degree distribution of nodes in the co-expression network was
investigated further and the degree of nodes conforms to power law
distribution (Fig. 2C and D). The
WGCNA package in R was used to place genes with similar expression
patterns into modules through average linkage clustering; a total
of 15 modules were identified (Fig.
3A). The black module exhibited the strongest association with
tumor metastasis-free survival and disease-specific death survival
(Fig. 3), whereas the turquoise
module exhibited the strongest association with tumor stage.
Therefore, these two modules were considered to be clinically
significant and were selected for further analysis.
 | Table I.Summary of number and corresponding
GSM in GSE65904. |
Table I.
Summary of number and corresponding
GSM in GSE65904.
| Number | GSM |
|---|
| 1 | GSM1608593 |
| 2 | GSM1608594 |
| 3 | GSM1608595 |
| 4 | GSM1608596 |
| 5 | GSM1608597 |
| 6 | GSM1608598 |
| 7 | GSM1608599 |
| 8 | GSM1608600 |
| 9 | GSM1608601 |
| 10 | GSM1608602 |
| 11 | GSM1608603 |
| 12 | GSM1608604 |
| 13 | GSM1608605 |
| 14 | GSM1608606 |
| 15 | GSM1608607 |
| 16 | GSM1608608 |
| 17 | GSM1608609 |
| 18 | GSM1608610 |
| 19 | GSM1608611 |
| 20 | GSM1608612 |
| 21 | GSM1608613 |
| 22 | GSM1608614 |
| 23 | GSM1608615 |
| 24 | GSM1608616 |
| 25 | GSM1608617 |
| 26 | GSM1608618 |
| 27 | GSM1608619 |
| 28 | GSM1608620 |
| 29 | GSM1608621 |
| 30 | GSM1608622 |
| 31 | GSM1608623 |
| 32 | GSM1608624 |
| 33 | GSM1608625 |
| 34 | GSM1608626 |
| 35 | GSM1608627 |
| 36 | GSM1608628 |
| 37 | GSM1608629 |
| 38 | GSM1608630 |
| 39 | GSM1608631 |
| 40 | GSM1608632 |
| 41 | GSM1608633 |
| 42 | GSM1608634 |
| 43 | GSM1608635 |
| 44 | GSM1608636 |
| 45 | GSM1608637 |
| 46 | GSM1608638 |
| 47 | GSM1608639 |
| 48 | GSM1608640 |
| 49 | GSM1608641 |
| 50 | GSM1608642 |
| 51 | GSM1608643 |
| 52 | GSM1608644 |
| 53 | GSM1608645 |
| 54 | GSM1608646 |
| 55 | GSM1608647 |
| 56 | GSM1608648 |
| 57 | GSM1608649 |
| 58 | GSM1608650 |
| 59 | GSM1608651 |
| 60 | GSM1608652 |
| 61 | GSM1608653 |
| 62 | GSM1608654 |
| 63 | GSM1608655 |
| 64 | GSM1608656 |
| 65 | GSM1608657 |
| 66 | GSM1608658 |
| 67 | GSM1608659 |
| 68 | GSM1608660 |
| 69 | GSM1608661 |
| 70 | GSM1608662 |
| 71 | GSM1608663 |
| 72 | GSM1608664 |
| 73 | GSM1608665 |
| 74 | GSM1608666 |
| 75 | GSM1608667 |
| 76 | GSM1608668 |
| 77 | GSM1608669 |
| 78 | GSM1608670 |
| 79 | GSM1608671 |
| 80 | GSM1608672 |
| 81 | GSM1608673 |
| 82 | GSM1608674 |
| 83 | GSM1608675 |
| 84 | GSM1608676 |
| 85 | GSM1608677 |
| 86 | GSM1608678 |
| 87 | GSM1608679 |
| 88 | GSM1608680 |
| 89 | GSM1608681 |
| 90 | GSM1608682 |
| 91 | GSM1608683 |
| 92 | GSM1608684 |
| 93 | GSM1608685 |
| 94 | GSM1608686 |
| 95 | GSM1608687 |
| 96 | GSM1608688 |
| 97 | GSM1608689 |
| 98 | GSM1608690 |
| 99 | GSM1608691 |
| 100 | GSM1608692 |
| 101 | GSM1608693 |
| 102 | GSM1608694 |
| 103 | GSM1608695 |
| 104 | GSM1608696 |
| 105 | GSM1608697 |
| 106 | GSM1608698 |
| 107 | GSM1608699 |
| 108 | GSM1608700 |
| 109 | GSM1608701 |
| 110 | GSM1608702 |
| 111 | GSM1608703 |
| 112 | GSM1608704 |
| 113 | GSM1608705 |
| 114 | GSM1608706 |
| 115 | GSM1608707 |
| 116 | GSM1608708 |
| 117 | GSM1608709 |
| 118 | GSM1608710 |
| 119 | GSM1608711 |
| 120 | GSM1608712 |
| 121 | GSM1608713 |
| 122 | GSM1608714 |
| 123 | GSM1608715 |
| 124 | GSM1608716 |
| 125 | GSM1608717 |
| 126 | GSM1608718 |
| 127 | GSM1608719 |
| 128 | GSM1608720 |
| 129 | GSM1608721 |
| 130 | GSM1608722 |
| 131 | GSM1608723 |
| 132 | GSM1608724 |
| 133 | GSM1608725 |
| 134 | GSM1608726 |
| 135 | GSM1608727 |
| 136 | GSM1608728 |
| 137 | GSM1608729 |
| 138 | GSM1608730 |
| 139 | GSM1608731 |
| 140 | GSM1608732 |
| 141 | GSM1608733 |
| 142 | GSM1608734 |
| 143 | GSM1608735 |
| 144 | GSM1608736 |
| 145 | GSM1608737 |
| 146 | GSM1608738 |
| 147 | GSM1608739 |
| 148 | GSM1608740 |
| 149 | GSM1608741 |
| 150 | GSM1608742 |
| 151 | GSM1608743 |
| 152 | GSM1608744 |
| 153 | GSM1608745 |
| 154 | GSM1608746 |
| 155 | GSM1608747 |
| 156 | GSM1608748 |
| 157 | GSM1608749 |
| 158 | GSM1608750 |
| 159 | GSM1608751 |
| 160 | GSM1608752 |
| 161 | GSM1608753 |
| 162 | GSM1608754 |
| 163 | GSM1608755 |
| 164 | GSM1608756 |
| 165 | GSM1608757 |
| 166 | GSM1608758 |
| 167 | GSM1608759 |
| 168 | GSM1608760 |
| 169 | GSM1608761 |
| 170 | GSM1608762 |
| 171 | GSM1608763 |
| 172 | GSM1608764 |
| 173 | GSM1608765 |
| 174 | GSM1608766 |
| 175 | GSM1608767 |
| 176 | GSM1608768 |
| 177 | GSM1608769 |
| 178 | GSM1608770 |
| 179 | GSM1608771 |
| 180 | GSM1608772 |
| 181 | GSM1608773 |
| 182 | GSM1608774 |
| 183 | GSM1608775 |
| 184 | GSM1608776 |
| 185 | GSM1608777 |
| 186 | GSM1608778 |
| 187 | GSM1608779 |
| 188 | GSM1608780 |
| 189 | GSM1608781 |
| 190 | GSM1608782 |
| 191 | GSM1608783 |
| 192 | GSM1608784 |
| 193 | GSM1608785 |
| 194 | GSM1608786 |
| 195 | GSM1608787 |
| 196 | GSM1608788 |
| 197 | GSM1608789 |
| 198 | GSM1608790 |
| 199 | GSM1608791 |
| 200 | GSM1608792 |
| 201 | GSM1608793 |
| 202 | GSM1608794 |
| 203 | GSM1608795 |
| 204 | GSM1608796 |
| 205 | GSM1608797 |
| 206 | GSM1608798 |
| 207 | GSM1608799 |
| 208 | GSM1608800 |
| 209 | GSM1608801 |
| 210 | GSM1608802 |
| 211 | GSM1608803 |
| 212 | GSM1608804 |
| 213 | GSM1608805 |
| 214 | GSM1608806 |
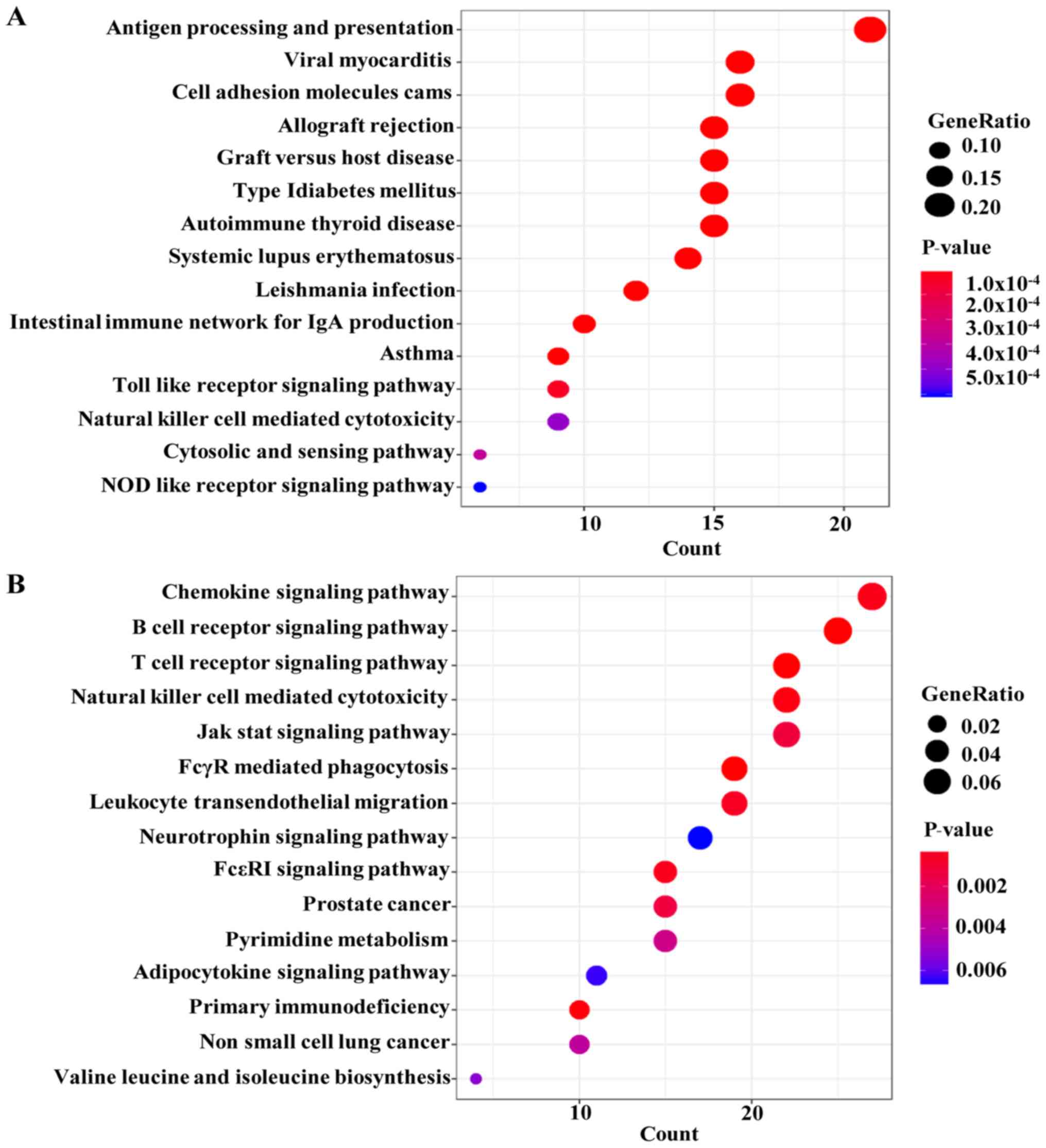
GO and KEGG pathway enrichment
analysis
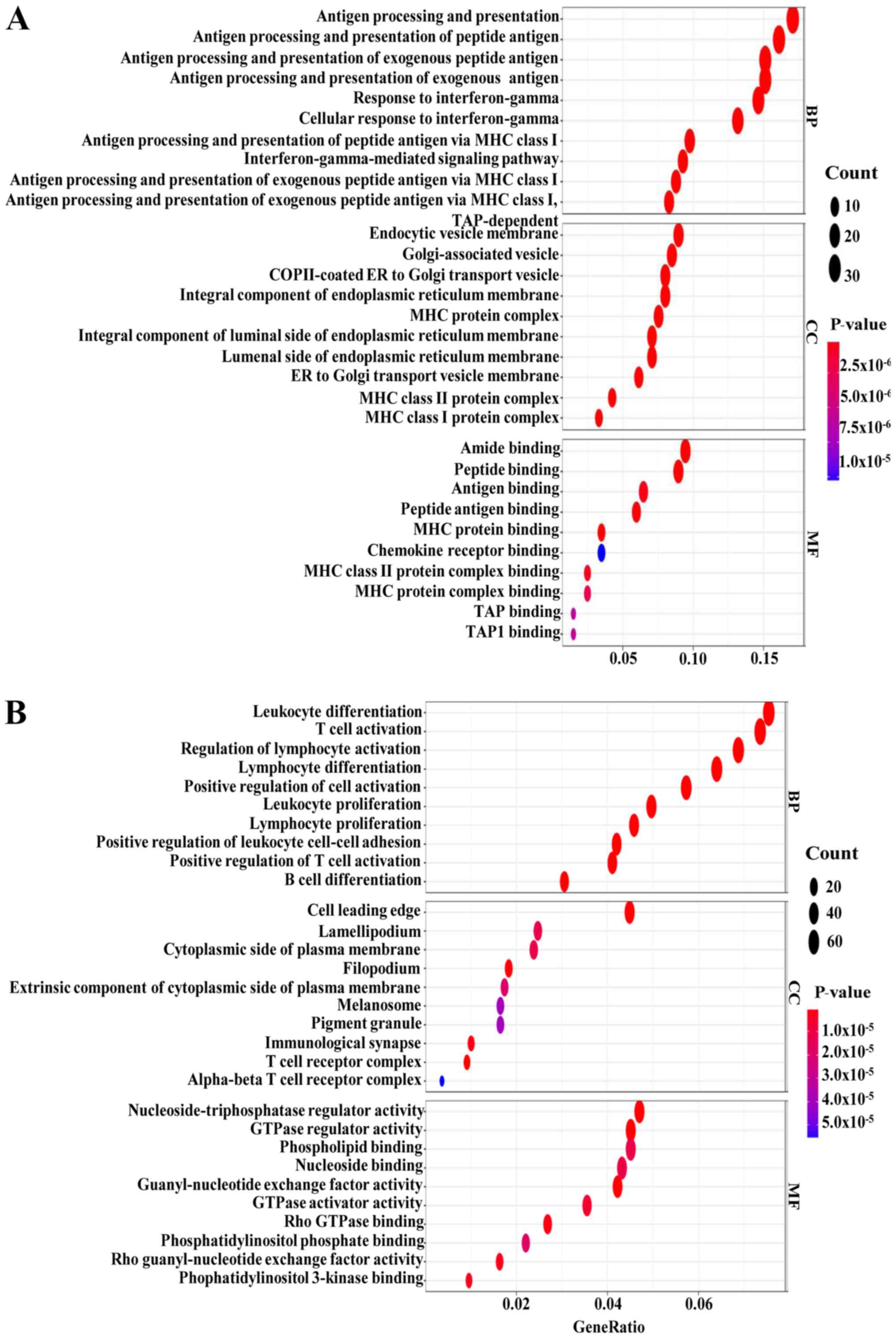
The genes in the clinically significant modules were
categorized into functional groups: BP, CC and MF. The genes in the
black module were mainly enriched in ‘antigen processing and
presentation’, ‘antigen processing and presentation of peptide
antigen’ and ‘antigen processing and presentation of exogenous
peptide antigen’ in the BP group, ‘endocytic vesicle membrane’,
‘Golgi-associated vesicle’ and ‘COPII-coated ER to Golgi transport
vesicle’ in the CC group, and ‘amide binding’, ‘peptide binding’
and ‘antigen binding’ in the MF group (Fig. 4A). The results of the KEGG pathway
analysis demonstrated that genes in the black module were mainly
involved in ‘antigen processing and presentation’, ‘viral
myocarditis’, ‘cell adhesion molecules cams’ and ‘allograft
rejection’, among others (Fig.
5A).
The genes in the turquoise module were mainly
enriched in ‘leukocyte differentiation’, ‘T cell activation’ and
‘regulation of lymphocyte activation’ in the BP group, ‘cell
leading edge’, ‘lamellipodium’ and ‘cytoplasmic side of plasma
membrane’ in the CC group and ‘nucleoside-triphosphatase regulator
activity’, ‘GTPase regulator activity’ and ‘phospholipid binding’
in the MF group (Fig. 4B). The
results of the KEGG pathway analysis demonstrated that the genes in
the turquoise module were mainly involved in ‘chemokine signaling
pathway’, ‘B cell receptor signaling pathway’ and ‘T cell receptor
signaling pathway’, among others (Fig.
5B).
Identification and validation of hub
genes
Genes with |MM+GS|=5% in the black and turquoise
modules were selected as candidate prognostic genes, and all other
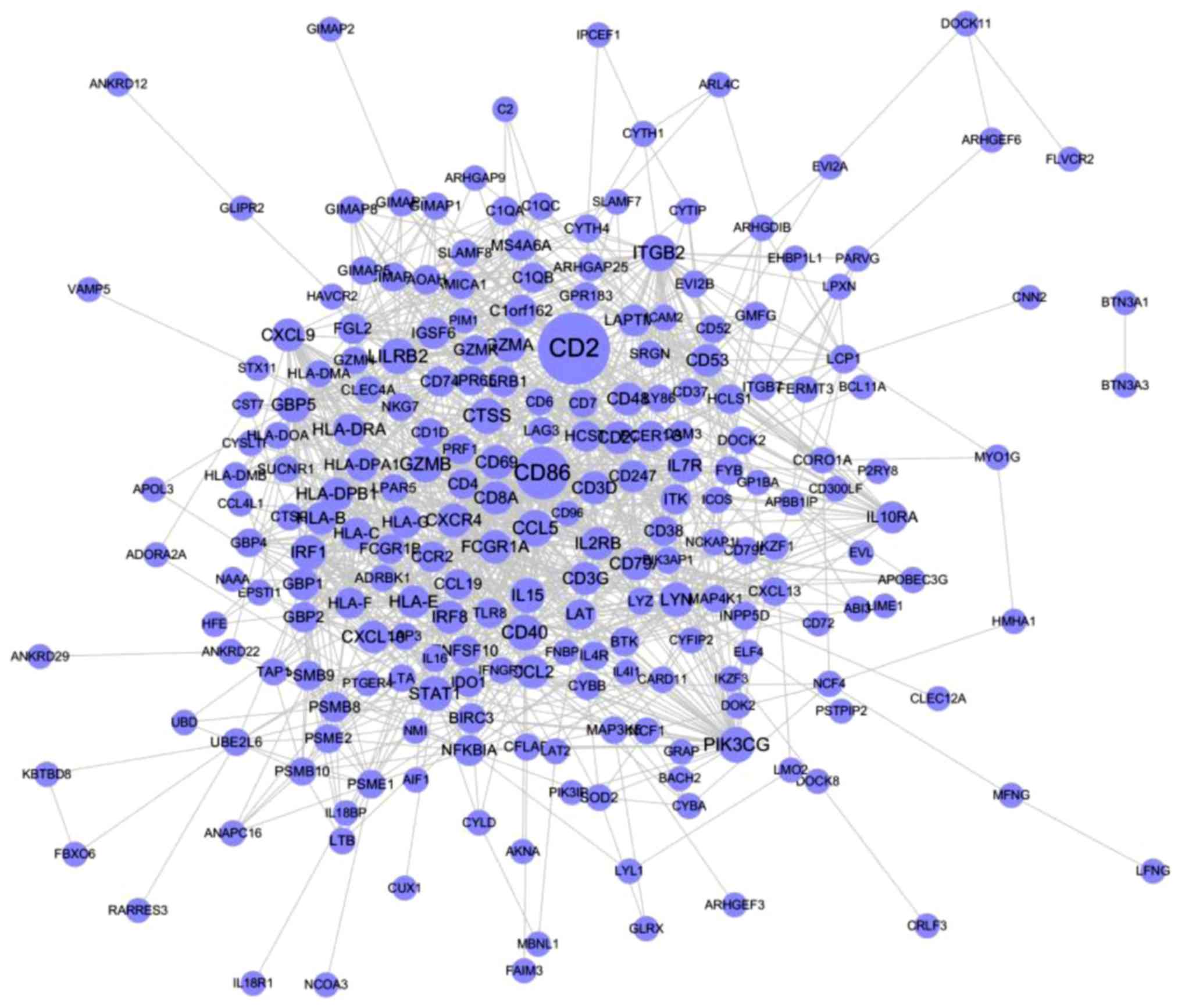
genes were removed. A PPI network of all genes in the black and
turquoise modules was constructed using Cytoscape. The network
comprised 222 nodes and 1,416 edges according to the STRING
database (Fig. 6). Among those,
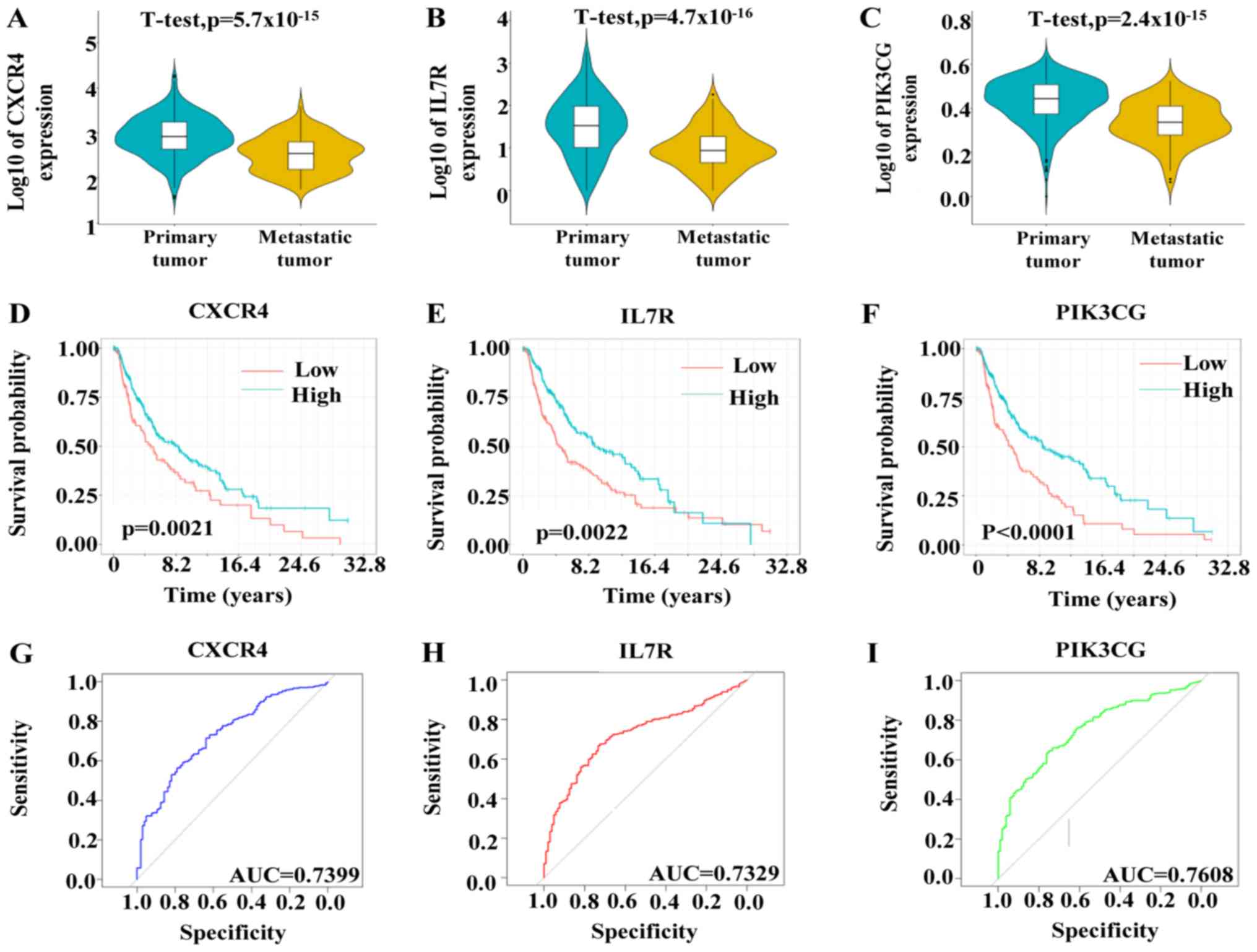
C-X-C motif chemokine receptor 4 (CXCR4), interleukin 7 receptor
(IL7R) and phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic
subunit γ (PIK3CG) were positively associated with overall survival
(Fig. 7D-F). Based on TCGA data, the
expression levels of CXCR4, IL7R and PI3KG were upregulated in
primary tumors compared with metastatic tumors (Fig. 7A-C). In addition, the ROC curves
indicated that CXCR4, IL7R and PI3KG exhibited excellent efficacy
for diagnosing primary and metastatic tumor tissues (Fig. 7G-I).
Screening candidates for
treatment
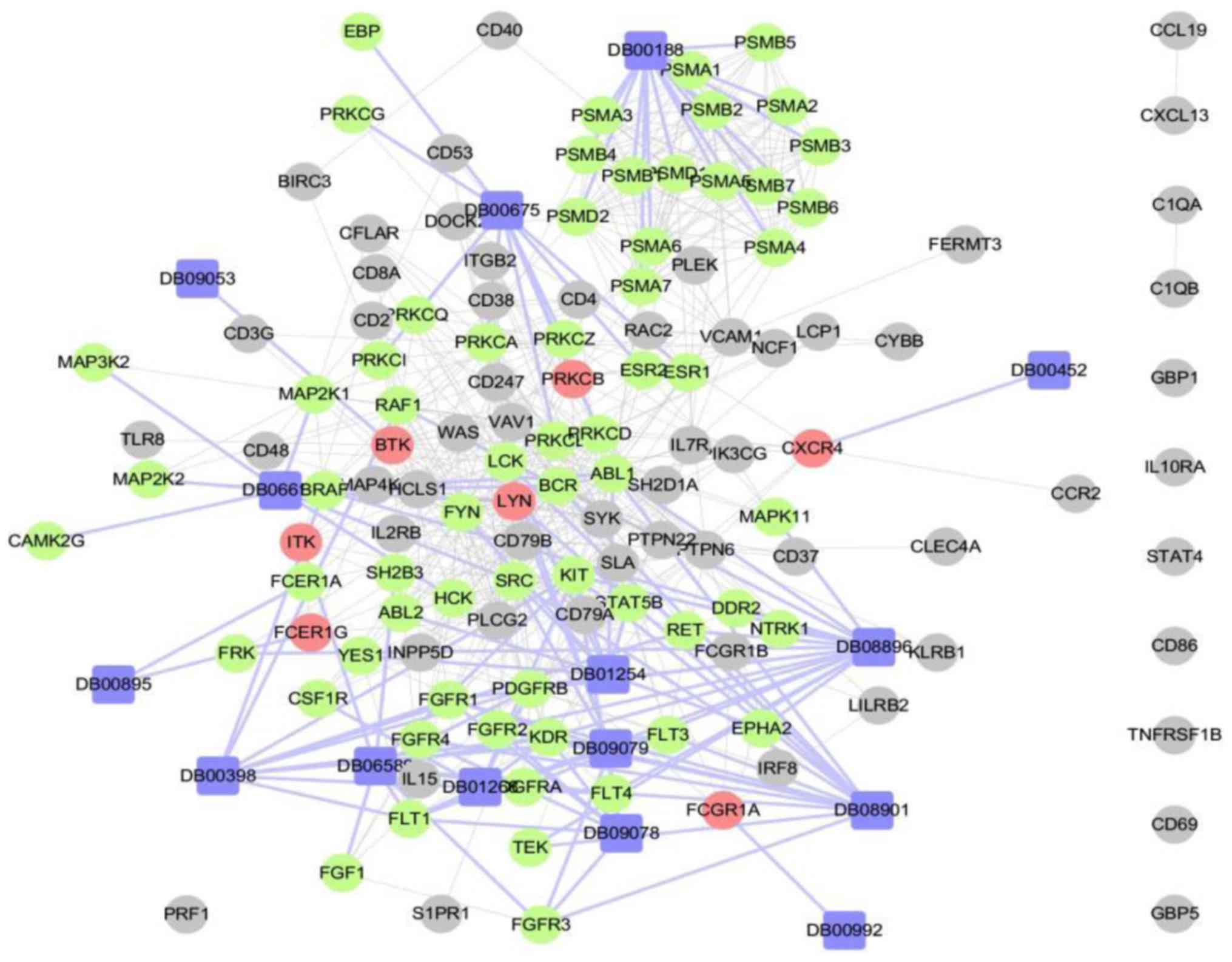
Using genes which were identified as hub nodes in
the PPI network (degree >30) associated with prognosis
(P<0.05) and metastasis (AUC >0.7) as potential targets in
the drug-gene interaction analysis (Fig.
8), the top 15 drugs ranked by the proximity of genes and drugs
were screened as possible treatments for melanoma. The screened
drugs could be divided into several major categories, including
tyrosine kinase inhibitors (TKIs), vascular endothelial growth
factor receptor (VEGFR) inhibitors, estrogen receptor modulators,
proteasome inhibitors, Burton's tyrosine kinase (BTK) inhibitors
and Raf kinase inhibitors. The top 15 drugs are: Ponatinib,
nintedanib, tamoxifen, framycetin, regorafenib, dasatinib,
sunitinib, bosutinib, benzylpenicilloyl polylysine, ibrutinib,
pazopanib, methyl aminolevulinate, bortezomib, sorafenib,
lenvatinib.
Discussion
Among skin tumors, melanoma is the most malignant
(22). High recurrence and
metastasis rates affect the efficacy of melanoma treatment
(23). The effects of conventional
chemotherapy, immunotherapy and targeted therapy remain limited.
Thus, identifying novel molecular targets and exploring therapeutic
drugs for melanoma is important. In the present study, the GEO
database was used to obtain genetic and clinical information from
patients with melanoma, construct a co-expression network, select
the most significant module and identify three hub genes: CXCR4,
IL7R and PIK3CG. TCGA, which was used for further verification,
revealed that three aforementioned specific molecules: CXCR4, IL7R
and PIK3CG identified in melanoma tissues were associated with
prognosis and metastasis. In addition, the top 15 drugs ranked by
the proximity of genes and drugs were screened using a network
screening method, and a drug-gene network was constructed.
CXCR4, which is a receptor of C-X-C motif chemokine
12 (CXCL12), is located on the surface of >23 human tumors, for
example breast cancer, ovarian cancer, glioma, pancreatic cancer
and prostate cancer (24). CXCL12
binds to CXCR4, which activates several extra- and intracellular
signaling pathways, including the nuclear factor κB,
Ca2+-dependent protein tyrosine kinase 2β, PI3K-Akt and
mitogen-activated protein kinase signaling pathways (25). In various types of cancer, such as
oral (26), esophageal (27), gastric, colon, liver, pancreatic,
thyroid and ovarian cancer (28),
and leukemia (29), CXCR4 expression
is strongly associated with chemotaxis, invasion, angiogenesis and
cell proliferation, all of which are involved in tumorigenesis and
cancer. However, the results of the present study indicated that,
compared with primary tumors, CXCR4 is downregulated in metastatic
tumors, and is therefore associated with good prognosis in patients
with melanoma. Mitchell et al (30) demonstrated that most of melanoma
cases with mitosis, ulceration and regression were CXCR4-negative.
Patients with American Joint Committee on Cancer (AJCC) stage
(31) I and II melanoma exhibit
higher expression of CXCR4 compared with those with AJCC stages III
and IV, and a proportion of patients with AJCC stage III–IV
melanoma are CXCR4-negative (30).
Therefore, the role of CXCR4 as a biomarker warrants further
investigation.
IL7R, which is expressed in immune cells, is crucial
for the survival, development and homeostasis of the immune system
(32). IL-7Rα activates Janus
kinases 1 and 3, promoting the function of signal transducer and
activator of transcription 5, which leads to the modulation of gene
expression, as well as the activation of anti-apoptotic and
pro-survival signaling pathways (33). Thus, IL7R is classified as an
oncogene associated with several tumors, including esophageal and
prostate cancer (34). However, a
bioinformatics study has demonstrated that patients with colon
cancer lacking IL7R (two cases of mortality out of three cases) had
a median survival time of 34 months compared with patients with
normal IL7R status, whose survival time was 45 months (35). Studies on the association between
IL7R and melanoma, as well as the association between IL7R and
metastasis, are lacking.
The PI3K signaling pathway modulates various
biological processes, including cell proliferation, survival,
motility, death and metabolism (36). Aberrations in these processes are
pivotal for the pathogenesis of cancer. Based on structural
differences, PI3K can be divided into several subunits, including
PIK3CA, PIK3CB, PIK3CD and PIK3CG (37). A previous study has revealed that
PIK3CG is expressed at undetectable levels in glioblastoma cells,
and that blocking this specific subunit does not cause cytotoxicity
(38). Another study has
demonstrated that PIK3CG is downregulated in colorectal cancer,
whereas 12 other genes in the PI3K-AKT signaling pathway are
upregulated (39). However, a
bioinformatics-based study reported that PIK3CG is significantly
associated with melanoma metastasis to regional lymph nodes, which
contradicted the results of the present study, suggesting that
further investigation may be required to clarify the role of PIK3CG
in the metastasis of melanoma (40).
In the present study, the GEO database, which
comprised 214 melanoma samples, and TCGA database, which included
417 patients, were selected to verify the roles of the identified
genes. Double validation and a large number of samples contributed
to the reliability of the candidate genes. However, a limitation of
the present study was a lack of clinical or experimental
validation. Further study is required to verify the role of CXCR4,
IL7R and PI3K3CG in melanoma.
The analysis of the association between genes and
FDA-approved drugs demonstrated that the top 15 drugs were TKIs,
VEGFR inhibitors, estrogen receptor modulators, proteasome
inhibitors, Bcr-Abl kinase inhibitors, BTK inhibitors, Raf kinase
inhibitors, framycetin, benzylpenicilloyl polylysine and methyl
aminolevulinate. TKIs that function by blocking the Bcr-Abl
tyrosine-kinase included dasatinib, ponatinib and bosutinib, which
are used to treat chronic myelogenous leukemia and acute
lymphocytic leukemia (41). Other
drugs, including nintedanib, regorafen, sunitinib, pazopanib,
sorafenib and lenvatinib inhibit several receptor tyrosine kinases,
including platelet-derived growth factors, VEGFR, fibroblast growth
factor receptors and Raf family kinases, which inhibit tumor
angiogenesis and tumor cell proliferation (42). Ibrutinib, a BTK inhibitor, is used to
treat chronic lymphocytic leukemia (43). Tamoxifen, a selective estrogen
receptor modulator, is used for the treatment and prevention of
estrogen receptor-positive breast cancer (44). Bortezomib was the first therapeutic
proteasome inhibitor to be tested in humans; it serves a role in
cell cycle arrest and apoptosis, and is approved in the United
States for the treatment of relapsed multiple myeloma and mantle
cell lymphoma (45). Framycetin,
which is an antibiotic, is used to treat leg ulcers and other
conditions associated with wound healing (46). Benzylpenicilloyl polylysine is used
as a skin-testing reagent for individuals with a history of
penicillin allergy (47). Methyl
aminolevulinate, which is metabolized into phototoxic compounds,
such as protopophyrin IX, may represent a candidate for
photodynamic therapy, as it can induce oxidative damage to the cell
(48).
Angiogenesis is a hallmark of several types of
tumor, including melanoma. The process of angiogenesis is crucial
for tumor development and metastasis (49). VEGF is one of the most important
cytokines responsible for tumor-mediated angiogenesis (50). VEGF is strongly expressed in melanoma
and serves a critical role in the progression of the disease
(51). In a phase II study of
sunitinib in patients with advanced melanoma, 4/31 (13%) patients
exhibited a partial response and 8 (26%) had stable disease
(52). Pazopanib, a VEGF and
platelet-derived growth factor inhibitor, has been used in
combination with paclitaxel in a phase II study of patients with
metastatic melanoma; the 6-month progression-free survival rate was
68%, and the 1-year overall survival rate was 48% (53). The objective response rate was 37%,
comprising one complete and 20 partial responses (54). A phase Ib study using lenvatinib
(E7080) in combination with temozolomide for the treatment of
advanced melanoma indicated an overall objective response rate of
18.8% (six patients), comprising all partial responses (55). SRC proto-oncogene, non-receptor
tyrosine kinase (SRC) is a promising target in the treatment of
solid types of cancer, including human melanoma; bosutinib, a SRC
inhibitor, which induces cell death via lysosomal membrane
permeabilization in melanoma cells, is a promising therapeutic
agent for melanoma treatment (56).
SRC inhibitor Dasatinib specifically inhibits p53 phosphorylation
in melanoma; however, a comprehensive validation is required
(57). Ibrutinib, a BTK inhibitor,
has been used to treat chronic lymphocytic leukemia/small
lymphocytic lymphoma and subsequent melanoma that occurs following
leukemia (58). Sorafenib, a Raf
inhibitor, is a first-line therapeutic agent used in advanced
melanoma (phase I and open-label phase II) trials with an overall
response rate of 12% with one complete response and nine partial
responses (59). Bortezomib
administration reduces the levels of proangiogenic cytokines in
plasma (60). A clinical trial has
indicated that tamoxifen therapy is not effective for treating
melanoma, and that the mode of action of antiestrogens in melanoma
is unclear (61). To the best of our
knowledge, ponatinib, nintedanib, regorafen, framycetin,
benzylpenicilloyl polylysine and methyl aminolevulinate have not
been used to treat melanoma.
The present study used WGCNA to construct a gene
co-expression network in order to determine the associations
between genes and modules and to explore the association between
the gene modules and clinical characteristics. Two significant
modules (black and turquoise modules) shown in Fig. 3, were identified to be associated
with the progression of melanoma. GO and KEGG pathway analyses
demonstrated that this module was mostly involved in functions
associated with antigen presentation. In addition, three hub genes,
CXCR4, IL7R and PI3K3CG, were identified and demonstrated to be
associated with the progression and prognosis of melanoma. Analysis
of the interaction between genes and drug targets of the top 15
drugs for melanoma enabled the construction of a network of
drug-gene interactions. Ponatinib, regorafen, nintedanib,
framycetin, benzyl penicilloyl polylysine and methyl
aminolevulinate, which were among the 15 drugs not currently used
to treat melanoma, may be potential novel therapeutic drugs for
this disease.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
JYG conceived and designed the study. XYD, QW, SMZ
and JHY acquired and interpreted the data. LW, CYW, YYX, XY, TFX
and YYP conducted the data analyses. LW, CYW and YYX wrote the
manuscript. JYG revised the manuscript. All authors read and
approved the final manuscript and agreed to be accountable for all
aspects of the research in ensuring that the accuracy or integrity
of any part of the work are appropriately investigated and
resolved.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
WGCNA
|
weighted gene co-expression network
analysis
|
|
GEO
|
Gene Expression Omnibus
|
|
ROC
|
receiver operating characteristic
|
|
GO
|
Gene Ontology
|
|
KEGG
|
Kyoto Encyclopedia of Genes and
Genomes
|
|
BP
|
biological process
|
|
CC
|
cellular component
|
|
MF
|
molecular function
|
|
MEs
|
module eigengenes
|
|
GS
|
gene significance
|
|
MM
|
module membership
|
|
PPI
|
protein-protein interaction
|
|
TKIs
|
tyrosine kinase inhibitors
|
|
BTK
|
Burton's tyrosine kinase
|
|
VEGFR
|
vascular endothelial growth factor
receptor
|
References
|
1
|
Colebatch AJ and Scolyer RA: Trajectories
of premalignancy during the journey from melanocyte to melanoma.
Pathology. 50:16–23. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Slominski A, Tobin DJ, Shibahara S and
Wortsman J: Melanin pigmentation in mammalian skin and its hormonal
regulation. Physiol Rev. 84:1155–1228. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Slominski A, Zmijewski MA and Pawelek J:
L-tyrosine and L-dihydroxyphenylalanine as hormone-like regulators
of melanocyte functions. Pigment Cell Melanoma Res. 25:14–27. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Slominski A, Kim TK, Brozyna AA,
Janjetovic Z, Brooks DL, Schwab LP, Skobowiat C, Jóźwicki W and
Seagroves TN: The role of melanogenesis in regulation of melanoma
behavior: Melanogenesis leads to stimulation of HIF-1α expression
and HIF-dependent attendant pathways. Arch Biochem Biophys.
563:79–93. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Slominski RM, Zmijewski MA and Slominski
AT: The role of melanin pigment in melanoma. Exp Dermatol.
24:258–259. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hyams DM, Cook RW and Buzaid AC:
Identification of risk in cutaneous melanoma patients: Prognostic
and predictive markers. J Surg Oncol. 119:175–186. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ugurel S, Rohmel J, Ascierto PA, Flaherty
KT, Grob JJ, Hauschild A, Larkin J, Long GV, Lorigan P, McArthur
GA, et al: Survival of patients with advanced metastatic melanoma:
The impact of novel therapies-update 2017. Eur J Cancer.
83:247–257. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Read J, Wadt KA and Hayward NK: Melanoma
genetics. J Med Genet. 53:1–14. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Koelblinger P, Dornbierer J and Dummer R:
A review of binimetinib for the treatment of mutant cutaneous
melanoma. Future Oncol. 13:1755–1766. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Silva IP and Long GV: Systemic therapy in
advanced melanoma: Integrating targeted therapy and immunotherapy
into clinical practice. Curr Opin Oncol. 29:484–492. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
da Silveira Nogueira Lima JP, Georgieva M,
Haaland B and de Lima Lopes G: A systematic review and network
meta-analysis of immunotherapy and targeted therapy for advanced
melanoma. Cancer Med. 6:1143–1153. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Appenzeller S, Gesierich A, Thiem A,
Hufnagel A, Jessen C, Kneitz H, Regensburger M, Schmidt C,
Zirkenbach V, Bischler T, et al: The identification of
patient-specific mutations reveals dual pathway activation in most
patients with melanoma and activated receptor tyrosine kinases in
BRAF/NRAS wild-type melanomas. Cancer. 125:586–600. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Winder M and Virós A: Mechanisms of drug
resistance in melanoma. Handb Exp Pharmacol. 249:91–108. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Slominski AT and Carlson JA: Melanoma
resistance: A bright future for academicians and a challenge for
patient advocates. Mayo Clin Proc. 89:429–433. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Cirenajwis H, Ekedahl H, Lauss M, Harbst
K, Carneiro A, Enoksson J, Rosengren F, Werner-Hartman L, Törngren
T, Kvist A, et al: Molecular stratification of metastatic melanoma
using gene expression profiling: Prediction of survival outcome and
benefit from molecular targeted therapy. Oncotarget. 6:12297–12309.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Tang J, Kong D, Cui Q, Wang K, Zhang D,
Gong Y and Wu G: Prognostic genes of breast cancer identified by
gene Co-expression network analysis. Front Oncol. 8:3742018.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Rahmani B, Zimmermann MT, Grill DE,
Kennedy RB, Oberg AL, White BC, Poland GA and McKinney BA:
Recursive indirect-paths modularity (RIP-M) for detecting community
structure in RNA-Seq Co-expression networks. Front Genet. 7:802016.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Gene Ontology Consortium: Gene ontology
consortium: Going forward. Nucleic Acids Res 43 (Database Issue).
D1049–D1056. 2015. View Article : Google Scholar
|
|
19
|
Kanehisa M, Sato Y, Kawashima M, Furumichi
M and Tanabe M: KEGG as a reference resource for gene and protein
annotation. Nucleic Acids Res. 44:D457–D462. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Su G, Morris JH, Demchak B and Bader GD:
Biological network exploration with Cytoscape 3. Curr Protoc
Bioinformatics. 47:8.13.1–24. 2014. View Article : Google Scholar
|
|
21
|
Cheng F, Desai RJ, Handy DE, Wang R,
Schneeweiss S, Barabási AL and Loscalzo J: Network-based approach
to prediction and population-based validation of in silico drug
repurposing. Nat Commun. 9:26912018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Rajabi P, Bagheri M and Hani M: Expression
of estrogen receptor alpha in malignant melanoma. Adv Biomed Res.
6:142017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Testori A, Ribero S and Bataille V:
Diagnosis and treatment of in-transit melanoma metastases. Eur J
Surg Oncol. 43:544–560. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Balkwill F: Cancer and the chemokine
network. Nat Rev Cancer. 4:540–550. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Nazari A, Khorramdelazad H and Hassanshahi
G: Biological/pathological functions of the CXCL12/CXCR4/CXCR7 axes
in the pathogenesis of bladder cancer. Int J Clin Oncol.
22:991–1000. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ishikawa T, Nakashiro K, Hara S, Klosek
SK, Li C, Shintani S and Hamakawa H: CXCR4 expression is associated
with lymph-node metastasis of oral squamous cell carcinoma. Int J
Oncol. 28:61–66. 2006.PubMed/NCBI
|
|
27
|
Wu J, Wu X, Liang W, Chen C, Zheng L and
An H: Clinicopathological and prognostic significance of chemokine
receptor CXCR4 overexpression in patients with esophageal cancer: A
meta-analysis. Tumour Biol. 35:3709–3715. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kodama J, Hasengaowa, Kusumoto T, Seki N,
Matsuo T, Ojima Y, Nakamura K, Hongo A and Hiramatsu Y: Association
of CXCR4 and CCR7 chemokine receptor expression and lymph node
metastasis in human cervical cancer. Ann Oncol. 18:70–76. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Konoplev S, Rassidakis GZ, Estey E,
Kantarjian H, Liakou CI, Huang X, Xiao L, Andreeff M, Konopleva M
and Medeiros LJ: Overexpression of CXCR4 predicts adverse overall
and event-free survival in patients with unmutated FLT3 acute
myeloid leukemia with normal karyotype. Cancer. 109:1152–1156.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Mitchell B, Leone D, Feller K, Menon S,
Bondzie P, Yang S, Park HY and Mahalingam M: Protein expression of
the chemokine receptor CXCR4 and its ligand CXCL12 in primary
cutaneous melanoma-biomarkers of potential utility? Hum Pathol.
45:2094–2100. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Balch CM, Gershenwald JE, Soong SJ,
Thompson JF, Atkins MB, Byrd DR, Buzaid AC, Cochran AJ, Coit DG,
Ding S, et al: Final version of 2009 AJCC melanoma staging and
classification. J Clin Oncol. 27:6199–6206. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Leung GA, Cool T, Valencia CH, Worthington
A, Beaudin AE and Forsberg EC: The lymphoid-associated interleukin
7 receptor (IL7R) regulates tissue-resident macrophage development.
Development. 146(pii): dev1761802019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Vitiello G, Losi GR, Amarante MK,
Ceribelli JR, Carmelo E and Watanabe M: Interleukin 7 receptor
alpha Thr244Ile genetic polymorphism is associated with
susceptibility and prognostic markers in breast cancer subgroups.
Cytokine. 103:121–126. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kim MJ, Choi SK, Hong SH, Eun JW, Nam SW,
Han JW and You JS: Oncogenic IL7R is downregulated by histone
deacetylase inhibitor in esophageal squamous cell carcinoma via
modulation of acetylated FOXO1. Int J Oncol. 53:395–403.
2018.PubMed/NCBI
|
|
35
|
Oliveira DM, Santamaria G, Laudanna C,
Migliozzi S, Zoppoli P, Quist M, Grasso C, Mignogna C, Elia L,
Faniello MC, et al: Identification of copy number alterations in
colon cancer from analysis of amplicon-based next generation
sequencing data. Oncotarget. 9:20409–20425. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Vivanco I and Sawyers CL: The
phosphatidylinositol 3-Kinase AKT pathway in human cancer. Nat Rev
Cancer. 2:489–501. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
37
|
Thorpe LM, Yuzugullu H and Zhao JJ: PI3K
in cancer: Divergent roles of isoforms, modes of activation and
therapeutic targeting. Nat Rev Cancer. 15:7–24. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Pridham KJ, Varghese RT and Sheng Z: The
role of Class IA phosphatidylinositol-4,5-bisphosphate 3-kinase
catalytic subunits in glioblastoma. Front Oncol. 7:3122017.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Semba S, Itoh N, Ito M, Youssef EM, Harada
M, Moriya T, Kimura W and Yamakawa M: Down-regulation of PIK3CG, a
catalytic subunit of phosphatidylinositol 3-OH kinase, by CpG
hypermethylation in human colorectal carcinoma. Clin Cancer Res.
8:3824–3831. 2002.PubMed/NCBI
|
|
40
|
Gorlov I, Orlow I, Ringelberg C, Hernando
E, Ernstoff MS, Cheng C, Her S, Parker JS, Thompson CL,
Gerstenblith MR, et al: Identification of gene expression levels in
primary melanoma associated with clinically meaningful
characteristics. Melanoma Res. 28:380–389. 2018.PubMed/NCBI
|
|
41
|
Abou DI, Jabbour E, Short NJ and Ravandi
F: Treatment of philadelphia chromosome-positive acute
lymphoblastic leukemia. Curr Treat Options Oncol. 20:42019.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zhao Y and Adjei AA: Targeting
angiogenesis in cancer therapy: Moving beyond vascular endothelial
growth factor. Oncologist. 20:660–673. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Deeks ED: Ibrutinib: A review in chronic
lymphocytic leukaemia. Drugs. 77:225–236. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Shaguft a and Ahmad I: Tamoxifen a
pioneering drug: An update on the therapeutic potential of
tamoxifen derivatives. Eur J Med Chem. 143:515–531. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Guerrero-Garcia TA, Mogollon RJ and
Castillo JJ: Bortezomib in plasmablastic lymphoma: A glimpse of
hope for a hard-to-treat disease. Leuk Res. 62:12–16. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Rai R, Shenoy MM, Viswanath V, Sarma N,
Majid I and Dogra S: Contact sensitivity in patients with venous
leg ulcer: A multi-centric Indian study. Int Wound J. 15:618–622.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Vemuri P, Harris KE, Suh LA and Grammer
LC: Preparation of benzylpenicilloyl-polylysine: A preliminary
study. Allergy Asthma Proc. 25:165–168. 2004.PubMed/NCBI
|
|
48
|
Yazdanyar S, Zarchi K and Jemec GBE: Pain
during topical photodynamic therapy-comparing methyl
aminolevulinate (Metvix®) to aminolaevulinic acid
(Ameluz®); an intra-individual clinical study.
Photodiagnosis Photodyn Ther. 20:6–9. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Jour G, Ivan D and Aung PP: Angiogenesis
in melanoma: An update with a focus on current targeted therapies.
J Clin Pathol. 69:472–483. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Ribatti D: Tumor refractoriness to
anti-VEGF therapy. Oncotarget. 7:46668–46677. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Ott PA, Hodi FS and Buchbinder EI:
Inhibition of immune checkpoints and vascular endothelial growth
factor as combination therapy for metastatic melanoma: An overview
of rationale, preclinical evidence, and initial clinical data.
Front Oncol. 5:2022015. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Decoster L, Vande BI, Neyns B, Majois F,
Baurain JF, Rottey S, Rorive A, Anckaert E, De Mey J, De Brakeleer
S and De Grève J: Biomarker analysis in a Phase II study of
sunitinib in patients with advanced melanoma. Anticancer Res.
35:6893–6899. 2015.PubMed/NCBI
|
|
53
|
Urbonas V, Schadendorf D, Zimmer L, Danson
S, Marshall E, Corrie P, Wheater M, Plummer E, Mauch C, Scudder C,
et al: Paclitaxel with or without trametinib or pazopanib in
advanced wild-type BRAF melanoma (PACMEL): A multicentre,
open-label, randomised, controlled phase II trial. Ann Oncol.
30:317–324. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Fruehauf JP, El-Masry M, Osann K,
Parmakhtiar B, Yamamoto M and Jakowatz JG: Phase II study of
pazopanib in combination with paclitaxel in patients with
metastatic melanoma. Cancer Chemother Pharmacol. 82:353–360. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Hong DS, Kurzrock R, Falchook GS, Andresen
C, Kwak J, Ren M, Xu L, George GC, Kim KB, Nguyen LM, et al: Phase
1b study of lenvatinib (E7080) in combination with temozolomide for
treatment of advanced melanoma. Oncotarget. 6:43127–43134. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Noguchi S, Shibutani S, Fukushima K, Mori
T, Igase M and Mizuno T: Bosutinib, an SRC inhibitor, induces
caspase-independent cell death associated with permeabilization of
lysosomal membranes in melanoma cells. Vet Comp Oncol. 16:69–76.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Skoko J, Rožanc J, Charles EM, Alexopoulos
LG and Rehm M: Post-treatment de-phosphorylation of p53 correlates
with dasatinib responsiveness in malignant melanoma. BMC Cell Biol.
19:282018. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Archibald WJ, Meacham PJ, Williams AM,
Baran AM, Victor AI, Barr PM, Sahasrahbudhe DM and Zent CS:
Management of melanoma in patients with chronic lymphocytic
leukemia. Leuk Res. 71:43–46. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Eisen T, Marais R, Affolter A, Lorigan P,
Robert C, Corrie P, Ottensmeier C, Chevreau C, Chao D, Nathan PD,
et al: Sorafenib and dacarbazine as first-line therapy for advanced
melanoma: Phase I and open-label phase II studies. Br J Cancer.
105:353–359. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Rossi UA, Finocchiaro L and Glikin GC:
Bortezomib enhances the antitumor effects of interferon-β gene
transfer on melanoma cells. Anticancer Agents Med Chem. 17:754–761.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Ribeiro MP, Santos AE and Custódio JB:
Rethinking tamoxifen in the management of melanoma: New answers for
an old question. Eur J Pharmacol. 764:372–378. 2015. View Article : Google Scholar : PubMed/NCBI
|