Introduction
Patients with hepatocellular carcinoma (HCC)
generally have a poor prognosis even after curative resection.
Their 5 year survival and tumor recurrence rates were 30–50% and
70–85%, respectively, after hepatic resection (1). One reason for these poor outcomes is
the tendency for vascular invasion that occurs with HCC (2). As the tumor grows, it invades
neighboring vessels via unknown mechanisms. Without vascular
invasion, the 5 year survival rates of patients with HCC who have
undergone radical hepatic resection are reportedly 50–70% (3). However, if the tumor invades the
adjacent vessels, the prognosis is poor despite curative treatments
such as hepatic resection (4).
Recently, it has been shown that outcomes after hepatic resection
for HCC can be negatively influenced not only by gross vascular
invasion, but also by microvascular invasion (McVI). McVI is
defined as a tumor within a vascular space lined by endothelium,
which is identified by microscopy (5). However, McVI is difficult to detect
with conventional diagnostic tools before surgery. This presents a
practical challenge, and many studies have focused on methods for
the preoperative prediction of McVI (6).
The mechanism by which HCC tumor cells invade
adjacent vessels has yet to be determined; several studies have
focused on issues associated with controlling this invasion
(7). Recent studies have shown that
the epidermal growth factor receptor (EGFR) signaling pathway, Ras,
and the Janus kinase/signal transducer and activator of
transcription (JAK/STAT) signaling pathway are dysregulated in HCC,
promoting cell motility, invasion, and tumor metastasis (8,9).
Mounting evidence demonstrates that epithelial-mesenchymal
transition (EMT) participates in the aggressive phenotypical
changes in cancer cells, and plays a major role in the invasion of
cancer cells into adjacent tissues (10,11).
Furthermore, several types of small non-coding RNA have been shown
to play critical roles in tumorigenesis and tumor progression, with
some regulating EMT directly or indirectly, thereby affecting tumor
invasion and metastasis (7,12).
MicroRNAs (miRNAs) are a class of
endogenously-expressed small non-coding RNAs. Recent studies have
identified clinically significant abnormal patterns of miRNA
expression in HCC, with some showing a marked association with
aggressive tumor phenotypes or poor survival (13). However, few studies have investigated
the relationship between McVI and aberrant miRNA expression.
Therefore, the present study aimed to identify miRNAs that act as
important contributors to the McVI of HCC.
Materials and methods
Study data
All molecular data from The Cancer Genome Atlas
Liver Hepatocellular Carcinoma database (TCGA LIHC) were downloaded
from the data portals (https://tcga-data.nci.nih.gov and http://gdac.broadinstitute.org/), where clinical
data are also publicly accessible. Integrated analysis with merged
molecular and clinical data is possible because unique TCGA barcode
structure is used to identify each molecular sample with the
corresponding patient. Information for VI of HCC is written to a
variable called ‘vascular_invasion’ in five different forms: None,
Micro, Macro, Not Available, and Unknown. The vascular invasion
(VI) status of each tumor sample is segregated into four groups:
Absence of VI (‘None’), McVI (‘Micro’), gross VI (‘Macro’), and no
available information about VI (‘Not Available’ or ‘Unknown’). The
patients, along with their corresponding clinical data and miRNA
expression profiles from the database, were categorized into two
groups according to the presence or absence of McVI. Patients
lacking values regarding vascular invasion were included for the
survival analysis of the candidate miRNAs. Descriptive statistics
were used to compare patient characteristics including sex, age,
etiology, laboratory data, types of surgery, and pathologic
data.
miRNA expression data were generated by the Illumina
HiSeq 2000 sequencing platform (Illumina, Inc.). Raw read counts
were used for the analysis of differentially expressed miRNAs and
mRNA. The processed RNA-Seq data, normalized according to the reads
per million (RPM) values of miRNAs and mRNAs, were also used in
survival and correlation analyses. The present study complied with
the publication guidelines provided by TCGA (http://cancergenome.nih.gov/publications/publicationguidelines).
All TCGA data are now available without restrictions on their use
in publications or presentations.
Screening for differentially expressed
miRNAs and prediction of their targets
The differentially expressed sequence (DESeq2)
package of the statistics software R version 3.3.1 was used to
assess the differentially expressed miRNAs based on the raw read
counts (14,15). Significant differential miRNA
expression was determined by false discovery rate (FDR) correction
for multiple hypothesis testing (using Benjamini-Hochberg-adjusted
P-values) with an FDR threshold <0.05. We used a validated
target module of the MiRWalk2.0 (zmf.umm.uni-heidelberg.de/mirwalk2) database to
identify the target genes of the differentially expressed miRNAs
(16).
Statistical analysis
Descriptive statistics of categorical variables
focused on frequencies and proportions. All continuous variables
were expressed as mean ± standard deviation. A chi-squared test and
t-test were used to compare, respectively, proportions and means.
Recurrence-free survival (RFS) and overall survival (OS) were
estimated for the two patient groups. The optimal cut-off values
for continuous variables of clinical parameters for use in
Kaplan-Meier survival analysis were estimated by receiver operating
characteristic (ROC) curve analysis. Survival rates and curves were
estimated by the Kaplan-Meier method and compared using the
log-rank test. The median disease free survival time, median
overall survival time, and the 95% confidence interval were also
measured. Cox regression analysis was used to define factors that
determined overall and disease-free survival rates by including
variables with statistical significance (P<0.05) for univariate
analysis. The statistical analysis was performed using the Epi,
GGally, MoonBook, and Survival packages of R version 3.3.1
(15).
Functional analysis
The Database for Annotation, Visualization, and
Integrated Discovery (DAVID) (https://david.ncifcrf.gov/) was used for Kyoto
Encyclopedia of Genes and Genome (KEGG) pathway analysis to extract
functional pathways involved in the mechanism of McVI. Cancer
Hallmarks Analytics Tool (CHAT) was used to determine the
association between selective miRNAs and documented evidence of
their roles in the hallmarks of cancer (17).
Results
Patient classification according to
vascular invasion status
Five of the 377 HCC patients had no miRNA expression
data in the TCGA LIHC database. The remaining 372 patients were
categorized into four groups according to their vascular invasion
(VI) status: Group A (n=206) had tumors without VI; Group B (n=94)
showed McVI; Group C (n=17) had gross VI; and Group D (n=55) were
patients with no available information about VI. The aim of this
study was to identify miRNAs selectively expressed in HCC
accompanied by McVI compared to HCC without VI and to investigate
their prognostic value. Seventeen patients in Group C were excluded
because they had HCC tumors with gross VI (Macro VI). For the tumor
nodes metastasis (TNM) stage based on the criteria established by
the Liver Cancer Study Group of Japan (LCSGJ), VI is one of three
factors for determining the T stage with tumor size and numbers.
According to the Barcelona Clinic for Liver Cancer (BCLC) staging,
HCC with gross VI is classified as advanced stage. However, it
remains unclear how much microvascular invasion (McVI) provides
prognostic information for patients with HCC from the viewpoint of
the extent of tumor invasion or extension. Considered the first
step of metastatic dissemination via the vascular route, the
prognostic impact of McVI may be intuitively placed between non-VI
and gross invasion of vessels. However, there is no strong evidence
to support this speculation. Therefore, we decided to exclude
patients with HCC accompanied by gross VI (Group C) in our study.
Patients in Group D (no available information about VI) were
included when conducting univariate survival analysis for each
clinicopathological factor and selected miRNA. However, Group D
patients were automatically excluded in multivariate survival
analysis because they had missing values.
Table I summarizes
the clinical characteristics of the patients in Groups A and B. The
patients with McVI (Group B) had significantly higher preoperative
serum creatinine levels (P=0.003); however, the percentage of
patients who had undergone major operations was significantly lower
(P<0.001) than those in Group A. There were also notable
differences in tumor node metastasis (TNM) stage between the two
groups, with patients in Group B exhibiting more advanced stages
(P<0.04). Apart from these differences, the two groups were
comparable. The overall median follow-up period was 13 months
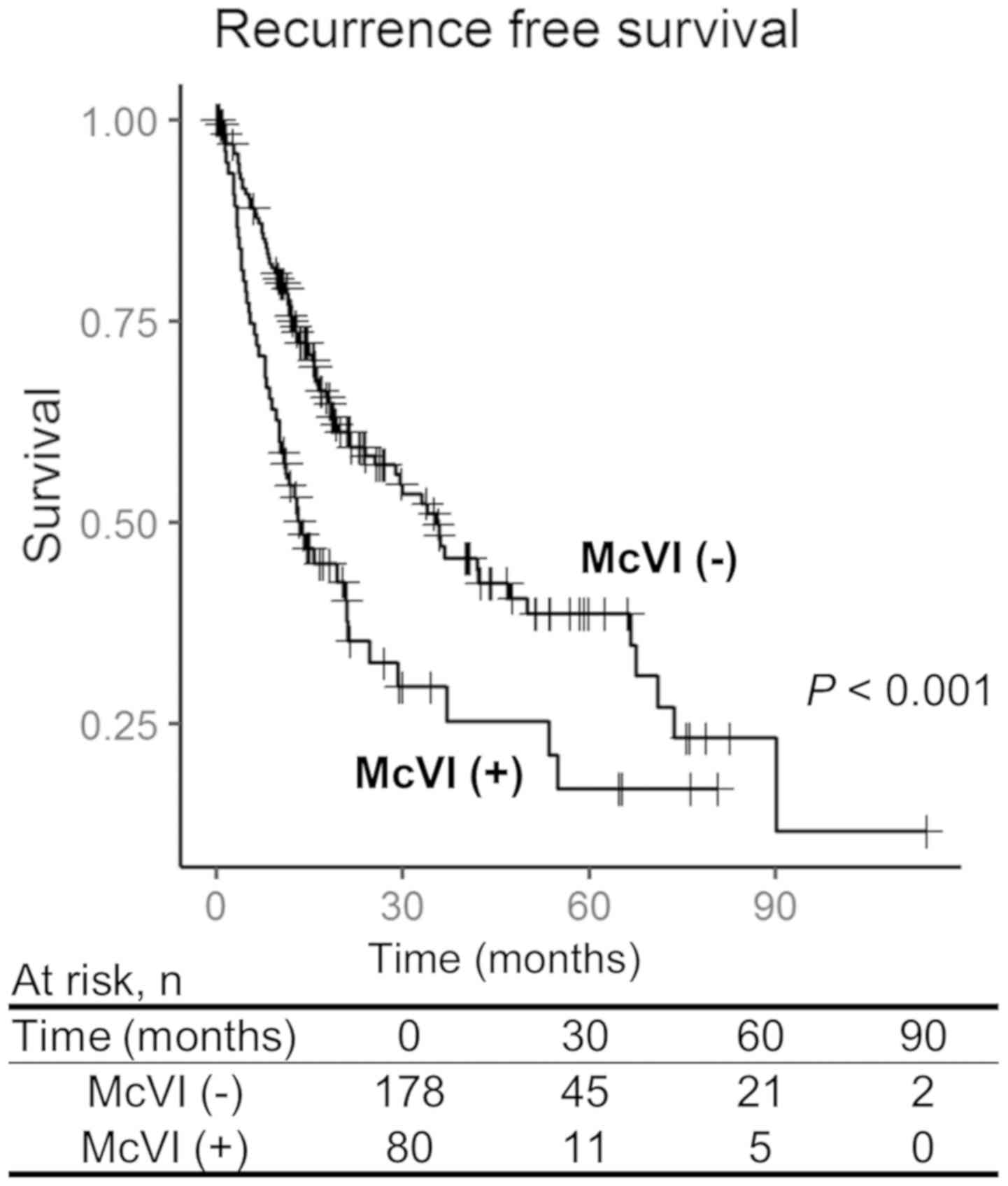
(range: 0–114 months). The survival analysis showed significantly
worse RFS for the patients with McVI (3 year RFS rates: Group A,
49.3%; Group B, 28.2%; P<0.001) (Fig.
1). There was no statistically significant difference in
overall survival rate between the groups.
 | Table I.Comparison of hepatocellular carcinoma
patient clinicopathological data with no vascular invasion (Group
A) or with microvascular invasion (Group B). |
Table I.
Comparison of hepatocellular carcinoma
patient clinicopathological data with no vascular invasion (Group
A) or with microvascular invasion (Group B).
| Factors | Group A (n=206) | Group B (n=94) | P-value (A vs.
B) | Group D (n=55) |
|---|
| Sex |
|
| 0.687 |
|
| Male | 136 (66.0%) | 65 (69.1%) |
| 42 (76.4%) |
|
Female | 70
(34.0%) | 29 (30.9%) |
| 13 (23.6%) |
| Age |
59.9±13.5 | 58.1±14.2 | 0.738 | 59.5±11.6 |
| Viral
hepatitis |
|
| 0.097 |
|
| No | 81
(39.9%) | 46 (51.1%) |
| 19 (34.5%) |
|
Yes | 122 (60.1%) | 44 (48.9%) |
| 36 (65.5%) |
| History of other
malignancy |
|
| 0.849 |
|
| No | 185 (89.8%) | 83 (88.3%) |
| 54 (98.2%) |
|
Yes | 21
(10.2%) | 11 (11.7%) |
| 1 (1.8%) |
| Platelet count
(×1,000/µl) | 207±91 | 229±118 | 0.257 | 227±93 |
| Serum creatinine
(mg/dl) | 0.9±0.4 | 1.3±1.1 | 0.003 | 0.8±0.2 |
| Serum albumin
(g/dl) | 3.7±1.0 | 4.0±1.1 | 0.062 | 3.8±0.8 |
| Serum total
bilirubin (mg/dl) | 0.8±0.6 | 0.9±0.5 | 0.450 | 0.6±0.3 |
| Child-Turcotte-Pugh
classification |
|
| 0.329 |
|
| A | 144 (90.0%) | 58 (92.1%) |
| 8 (100.0%) |
| B or
C | 16
(10.0%) | 5 (7.9%) |
| 0 (0.0%) |
| α-fetoprotein
(ng/ml) | 15,300±15,700 | 13,200±46,800 | 0.875 | 2,100±6,300 |
| Types of hepatic
resection |
|
| <0.001 |
|
|
Major | 133 (64.6%) | 36 (39.1%) |
| 21 (38.9%) |
|
Minor | 73 (35.4%) | 56 (60.9%) |
| 33 (61.1%) |
| Histologic grading
by Edmondson and Steiner's classification |
|
| 0.174 |
|
|
1–2 | 131 (63.9%) | 51 (54.8%) |
| 41 (77.4%) |
|
3–4 | 74
(36.1%) | 42 (45.2%) |
| 12 (22.6%) |
| Residual tumor |
|
| 0.433 |
|
|
Negative | 181 (96.3%) | 84 (93.3%) |
| 45 (93.8%) |
|
Positive | 7
(3.7%) | 6 (6.7%) |
| 3 (6.2%) |
| Ishak fibrosis
staging system |
|
| 0.267 |
|
| 0 | 52 (36.9%) | 16 (28.6%) |
| 1 (11.1%) |
|
1–4 | 36 (25.5%) | 18 (32.1%) |
| 5 (55.6%) |
|
5–6 | 53 (37.6%) | 22 (39.3%) |
| 3 (33.3%) |
| Grade of liver
inflammation |
|
| 0.282 |
|
| 0 | 64 (46.4%) | 32 (43.2%) |
| 12 (92.3%) |
|
1–2 | 65 (47.1%) | 33 (44.6%) |
| 1 (7.7%) |
|
3–4 | 9 (6.5%) | 9
(12.2%) |
| 0 (0.0%) |
| American Joint
Committee on Cancer TNM stage |
|
| 0.040 |
|
|
I–II | 164 (85.0%) | 65 (73.9%) |
| 21 (42.0%) |
|
III–IV | 29
(15.0%) | 23 (26.1%) |
| 29 (58.0%) |
miRNAs were differentially expressed
on the basis of the absence or presence of McVI
The expression levels of 540 miRNAs in the 300
samples were obtained from the TCGA expression data. Four miRNAs
were differentially expressed between the patients without VI
(Group A) and those with McVI (Group B): miRNA-141 [log2
fold change (FC) of 0.80 and FDR=0.005), miRNA-582 (log2
FC of 0.55 and FDR=0.045), and miRNA-9 (log2 FC of 1.22
and FDR <0.001) were upregulated to a greater extent in Group B,
while miRNA-675 was downregulated to a greater extent in Group B
(log2 FC of −0.99 and FDR=0.005) (Fig. 2A).
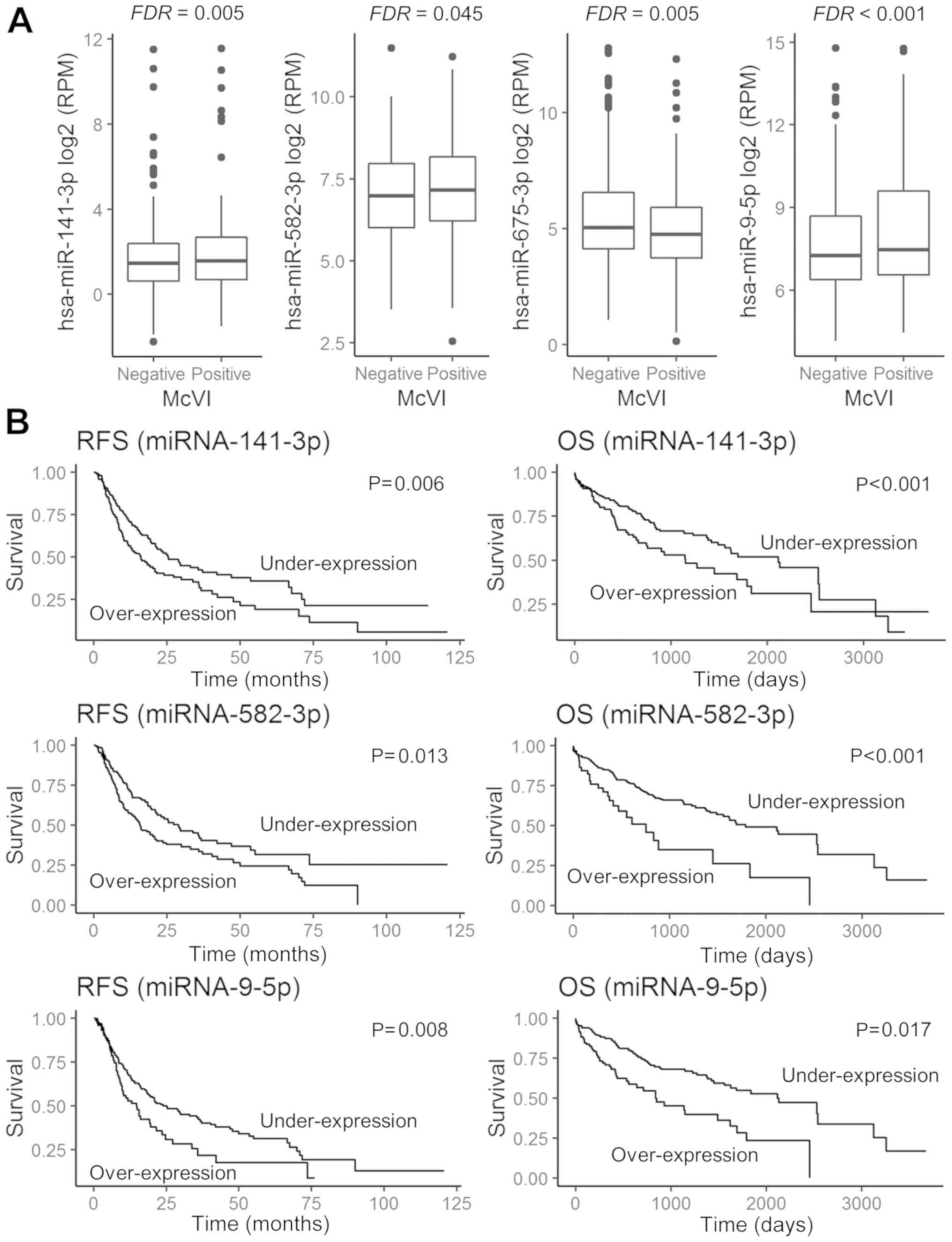 | Figure 2.Prognostic impact and expression of
selected miRNAs, according to the absence or presence of
microvascular invasion (McVI) in HCC tissue. (A) Comparison of
miRNA-141-3p, miRNA-582-3p, miRNA-675-3p, and miRNA-9-5p
expressions in HCC tissue with McVI or without. (B) RFS and OS
curves between upregulated and downregulated miRNA-141-3p,
miRNA-582-3p and miRNA-9-5p expression. Under-expression is
indicated by expression values less than the cut-off value, while
overexpression is indicated by expression values more than the
cut-off value. miRNA, microRNA; FDR, false discovery rate; RFS,
recurrence-free survival; OS, overall survival; RPM, reads per
million. |
Next, we investigated the discriminatory power of
the four miRNAs to assess the presence and the absence of McVI. To
evaluate the predictive value, we used the ROC curve to analyze
sensitivity and specificity. Each cut-off value was as follows:
2.03 RPM for miRNA-141-3p, 211.98 RPM for miRNA-582-3p, 23.93 RPM
for miRNA-675-3p, and 65.37 RPM for miRNA-9-5p. Table II demonstrates the association
between the number of aberrantly expressed miRNAs and McVI of
resected HCC specimens in 300 patients. Most patients (90%) without
McVI showed no aberrant expression patterns in these four miRNAs.
There were increases in the prevalence of McVI as the number of
aberrantly expressed miRNAs increased.
 | Table II.Univariate analysis of factors
predictive of recurrence-free survival. |
Table II.
Univariate analysis of factors
predictive of recurrence-free survival.
| Factors | No. of
patients | MDFSTa (95% CI) | P-value |
|---|
| Sex |
|
| 0.368 |
|
Male | 216 | 21.6
(17.6–33.0) |
|
|
Female | 92 | 19.4
(14.8–36.7) |
|
| Age (years) |
|
| 0.167 |
|
<51 | 255 | 21.0
(17.6–35.8) |
|
|
≥51 | 53 | 21.2
(14.1–33.9) |
|
| Hepatitis B or C
infection status |
|
| 0.385 |
|
Negative | 119 | 19.2
(14.9–33.0) |
|
|
Positive | 182 | 23.0
(17.6–36.0) |
|
| Platelet count
(×1,000/µl) |
|
| 0.173 |
|
<282 | 207 | 28.9
(21.0–37.1) |
|
|
≥282 | 48 | 19.6
(14.2–55.1) |
|
| Serum creatinine
(mg/dl) |
|
| 0.077 |
|
<1.1 | 172 | 21.0
(18.3–33.0) |
|
|
≥1.1 | 78 | 36.0
(23.9–73.6) |
|
| Serum albumin
(g/dl) |
|
| 0.363 |
|
≥4.1 | 119 | 29.7
(21.0–55.1) |
|
|
<4.1 | 132 | 20.2
(15.7–36.7) |
|
| Serum total
bilirubin (mg/dl) |
|
| 0.680 |
|
<0.8 | 142 | 23.0
(19.2–40.4) |
|
|
≥0.8 | 111 | 29.3
(21.0–50.0) |
|
| Child-Turcotte-Pugh
classification |
|
| 0.604 |
| A | 187 | 23.6
(19.2–42.0) |
|
| B or
C | 19 | 23.9 (8.6-NA) |
|
| α-fetoprotein
(ng/ml) |
|
| 0.129 |
|
<85,150 | 229 | 25.5
(19.4–36.0) |
|
|
≥85,150 | 8 | 30.2 (13.1-NA) |
|
| Extent of
resection |
|
| <0.001 |
|
Major | 160 | 35.6
(23.0–53.5) |
|
|
Minor | 147 | 14.2
(10.2–20.9) |
|
| Residual tumor |
|
| 0.031 |
| No | 276 | 23.0
(18.6–33.9) |
|
|
Yes | 13 | 12.6 (10.8-NA) |
|
| Histologic grading
by Edmondson and Steiner's classification |
|
| 0.760 |
|
I–II | 195 | 21.6
(18.4–29.7) |
|
|
III–IV | 109 | 20.9
(12.6–47.0) |
|
| Ishak fibrosis
staging system |
|
| 0.101 |
| 0 | 57 | 23.9 (15.4-NA) |
|
|
1–4 | 55 | 24.8
(18.2–36.7) |
|
|
5–6 | 72 | 18.3
(12.9–47.0) |
|
| Grade of liver
inflammation |
|
| 0.950 |
| 0 | 88 | 23.9
(20.9–40.4) |
|
|
1–2 | 90 | 24.8
(18.2–47.0) |
|
|
3–4 | 15 | 18.3 (9.7-NA) |
|
| American Joint
Committee on Cancer TNM stage |
|
| 0.005 |
|
I–II | 180 | 33.9
(24.8–55.1) |
|
|
III–IV | 40 | 13.1
(8.6–21.0) |
|
|
MicroRNA-141-3p |
|
| 0.006 |
|
Underexpression | 164 | 25.3
(21.0–47.0) |
|
|
Overexpression | 158 | 16.1
(11.7–21.6) |
|
|
MicroRNA-582-3p |
|
| 0.013 |
|
Underexpression | 143 | 28.9
(21.2–47.0) |
|
|
Overexpression | 179 | 15.7
(12.6–21.6) |
|
|
MicroRNA-675-3p |
|
| 0.878 |
|
Underexpression | 95 | 21.0
(16.1–35.6) |
|
|
Overexpression | 227 | 20.9
(15.7–33.0) |
|
| MicroRNA-9-5p |
|
| 0.008 |
|
Underexpression | 247 | 23.9
(19.2–36.7) |
|
|
Overexpression | 75 | 14.8
(9.76–23.6) |
|
Survival analysis
The ROC-curve-determined optimal cutoff value of
miRNA expression in the study participants (355 patients) was used
to classify the patients into under- and over-expression groups.
The RFS cut-off values for miRNA-141-3p, miRNA-582-3p,
miRNA-675-3p, and miRNA-9-5p were 3.34 RPM, 125.79 RPM, 21.15 RPM,
and 552.91 RPM, respectively. The OS cut-off value was 4.75 RPM for
miRNA-141-3p, 441.09 RPM for miRNA-582-3p, 20.96 RPM for
miRNA-675-3p, and 546.69 RPM for miRNA-9-5p. Kaplan-Meier analysis
indicated that miRNA-141-3p, miRNA-582-3p, and miRNA-9-5p
overexpression was significantly associated with both poor RFS
(P=0.008, P=0.006, and P=0.013, respectively) and worse OS
(P=0.008, P<0.001, and P<0.001, respectively) (Fig. 2B). However, HCC patients exhibiting
under-expression of miRNA-675 showed no significant difference in
survival outcome compared to those with over-expression of
miRNA-675 (Tables II and III).
 | Table III.Univariate analysis of factors
predictive of overall survival. |
Table III.
Univariate analysis of factors
predictive of overall survival.
| Factors | No. of
patients | MOSTa (95% CI) | P-value |
|---|
| Sex |
|
| 0.265 |
|
Male | 241 | 1836 (1423-NA) |
|
|
Female | 112 | 1450
(887–2532) |
|
| Ages (years) |
|
| 0.004 |
|
<70 | 274 | 2456 (1624-NA) |
|
|
≥70 | 79 | 1147
(785–1791) |
|
| Hepatitis B or C
infection status |
|
| 0.013 |
|
Negative | 144 | 1135
(837–1791) |
|
|
Positive | 202 | 2456 (1685-NA) |
|
| Platelet count
(×1,000/µl) |
|
| 0.141 |
|
<282 | 233 | 2456 (1685-NA) |
|
|
≥282 | 55 | 1135
(848–2542) |
|
| Serum creatinine
(mg/dl) |
|
| 0.746 |
|
<0.78 | 80 | 1372 (887-NA) |
|
|
≥0.78 | 203 | 2456 (1694-NA) |
|
| Serum albumin
(g/dl) |
|
| 0.419 |
|
<4.3 | 85 | 3258 (1560-NA) |
|
|
≥4.3 | 198 | 1694 (1423-NA) |
|
| Serum total
bilirubin (mg/dl) |
|
| 0.144 |
|
<1 | 201 | 2131
(1624–3258) |
|
| ≥1 | 84 | NA (1622-NA) |
|
| Child-Turcotte-Pugh
classification |
|
| 0.911 |
| A | 209 | 2542 (2131-NA) |
|
| B or
C | 20 | 987 (601-NA) |
|
| α-fetoprotein
(ng/ml) |
|
| 0.086 |
|
<85,150 | 259 | 2456 (1685-NA) |
|
|
≥85,150 | 8 | NA (633-NA) |
|
| Extent of
resection |
|
| 0.061 |
|
Major | 189 | 2116 (1624-NA) |
|
|
Minor | 161 | 1149 (887-NA) |
|
| Residual tumor |
|
| 0.113 |
| No | 309 | 1791
(1450–3125) |
|
|
Yes | 16 | 837 (385-NA) |
|
| Histologic grading
by Edmondson and Steiner's classification |
|
| 0.224 |
|
I–II | 222 | 1791
(1423–2542) |
|
|
III–IV | 127 | NA (1149-NA) |
|
| Ishak fibrosis
staging system |
|
| 0.913 |
| 0 | 69 | 2456 (931-NA) |
|
|
1–4 | 59 | 1791 (1372-NA) |
|
|
5–6 | 77 | NA (1685-NA) |
|
| Grade of liver
inflammation |
|
| 0.133 |
| 0 | 108 | 2456 (1624-NA) |
|
|
1–2 | 99 | NA (1368-NA) |
|
|
3–4 | 17 | NA (359–27.9) |
|
| American Joint
Committee on Cancer TNM stage |
|
| 0.001 |
|
I–II | 180 | NA (2456-NA) |
|
|
III–IV | 40 | 1791 (660-NA) |
|
|
MicroRNA-141-3p |
|
| <0.001 |
|
Underexpression | 240 | 2116 (1560-NA) |
|
|
Overexpression | 130 | 1149
(743–1836) |
|
|
MicroRNA-582-3p |
|
| <0.001 |
|
Underexpression | 312 | 1791
(1490–3258) |
|
|
Overexpression | 58 | 757 (415-NA) |
|
|
MicroRNA-675-3p |
|
| 0.172 |
|
Underexpression | 113 | 1271 (837-NA) |
|
|
Overexpression | 257 | 2116 (1423-NA) |
|
| MicroRNA-9-5p |
|
| 0.017 |
|
Underexpression | 274 | 2116 (1560-NA) |
|
|
Overexpression | 96 | 837 (558–1694) |
|
Prediction of associated pathway and
functional analysis of selected miRNAs
We identified experimentally validated target genes
of miRNA-141-3p, miRNA-582-3p, and miRNA-9-5p using miRNA-mRNA
pairs from the MiRWalk2.0 databases (15) There were a total of 459 validated
target genes identified for these three selected miRNAs. In order
to estimate the regulatory effects, 459 genes were selected for
KEGG pathway analysis using the DAVID (https://david.ncifcrf.gov/) functional annotation
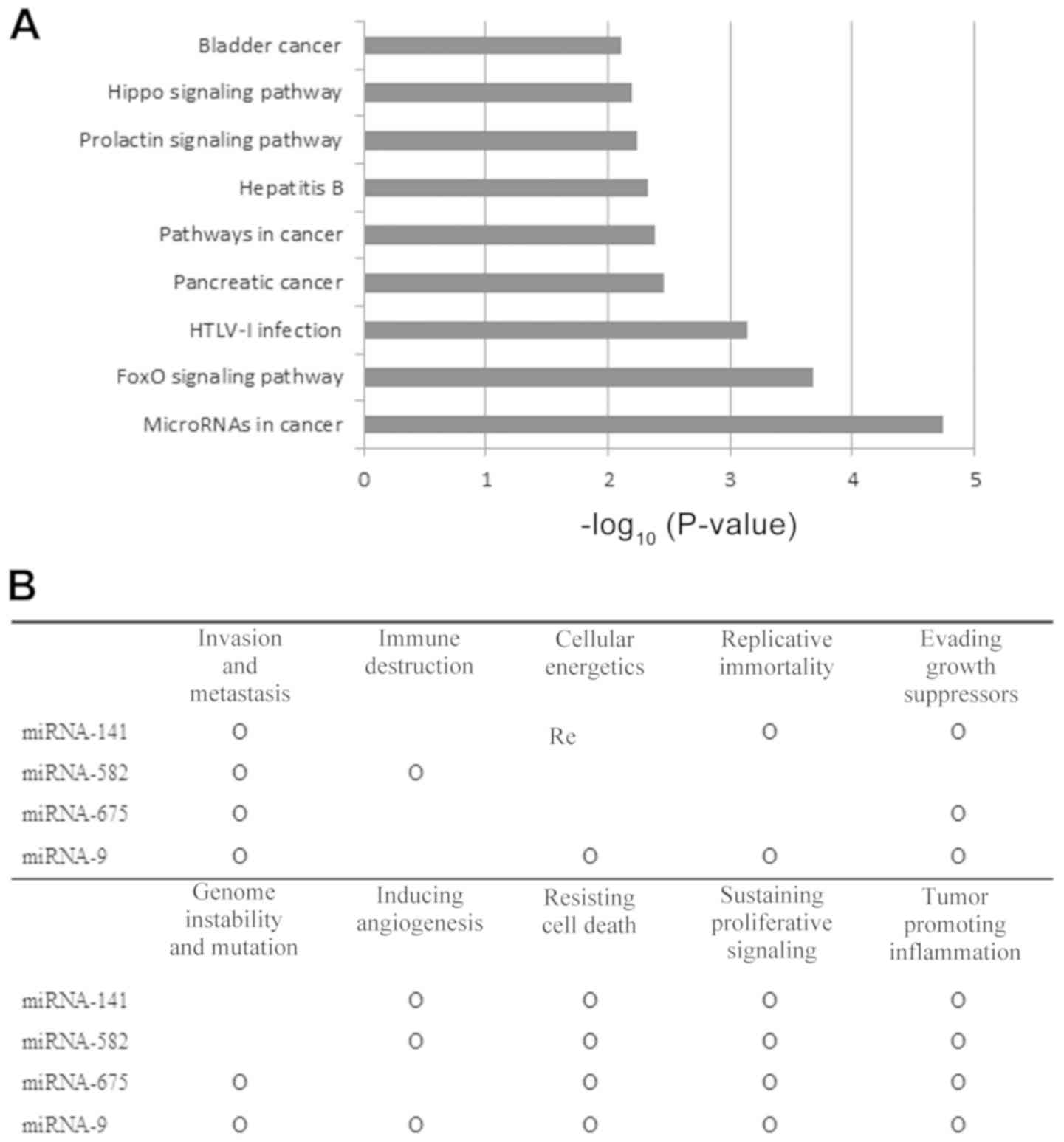
tool. Twenty-four genes were enriched in microRNAs associated with
cancer, while 14 genes were enriched in the FoxO signal pathway
(Fig. 3A). A text mining analysis
was conducted to determine which ‘hallmarks of cancer’ were
affected when these miRNAs were dysregulated. All of the selected
miRNAs were associated with at least six hallmarks of cancer.
especially, ‘invasion and metastasis,’ ‘resisting cell death,’
‘sustaining proliferative signaling,’ and ‘tumor promoting
inflammation’ (Fig. 3B).
Discussion
In this study, we used publicly available data from
the TCGA repositories to identify miRNAs associated with McVI and
poor prognosis of HCC. Our results showed that the expression
levels of miRNA-141, −582, and −9 were significantly higher in HCC
with McVI, while the expression level of miRNA-675 was lower
compared to HCC without VI. Furthermore, the overexpression of
miRNA-141-3p, −582-3p, and −9-5p was significantly associated with
both poor RFS and OS after hepatic resection for HCC. Several
recent studies have focused on the relationship between cancer
metastasis and the aberrant expression of miRNAs (18). One study showed an association
between the decreased expression of miRNA-199a and McVI (19). However, to the best of our knowledge,
no previous study has investigated the association between
miRNA-141/-582/-675/-9 and McVI in HCC tissue samples.
McVI is a well-known factor that contributes to
tumor recurrence and poor prognosis; its incidence in surgically
resected HCC has been reported to be 15–52% (20). Notably, McVI was found to be a
predictor of early tumor recurrence following hepatic resection for
early-stage HCC (21). A recent
study showed that its prognostic significance was comparable to
that of gross VI limited to segmental or sectional vascular
branches of HCC (5). An
international consensus conference recommended that patients with
HCC accompanied by McVI should not be included as candidates for
liver transplantation because of this severely negative oncologic
feature (22). For this reason, many
studies have focused on how to predict McVI of HCC preoperatively
due to the poor sensitivity and specificity of conventional
modalities such as positron emission tomography and computed
tomography scan for detecting McVI (6).
VI is considered to reflect the aggressiveness of
the tumor and is a well-established negative prognostic factor
after hepatic resection for HCC (23). However, information is limited
regarding the underlying mechanism for VI. One possibility is that
VI may be purely coincidental, with high arterial pressure in the
tumor resulting in the spread of cancer cells to neighboring
vessels. However, recent studies of the mechanism of tumor
metastasis have shown the importance of phenotype changes in
individual tumor cells (10,24). Genomic data analyses have identified
unique genes and non-coding RNAs that play important roles in
metastasis (25,26).
miRNAs represent a class of endogenously-expressed
small non-coding RNAs, approximately 22 nucleotides in length, that
have the capacity to epigenetically regulate gene expression
(27). miRNAs primarily function as
post-transcriptional regulators through the degradation of their
target mRNA or by inhibiting translation (28). They are emerging as powerful
regulators of critical biological processes, including the cell
cycle, metabolism, development, and cell differentiation (29). There is also growing evidence that
miRNAs are associated with tumor progression, being involved in
cell migration, angiogenesis, and invasion (7,8,19,26).
miR-141 has been shown to be a prognostic factor for different
types of cancer. Recently, miRNA-141 was shown to be involved in
controlling cancer cell proliferation of colorectal cancer by
regulating the tumor-suppressor gene MAP2K4 (30).
In this study, the upregulation of miRNA-9 and
miR-582 was also associated with McVI, as well as a statistically
significant difference in survival. miRNA-9 has been shown to be a
predictor for different types of cancer. In various studies, high
miRNA-9 expression levels were related to worse survival rates and
a high risk of cancer metastasis in various carcinomas (31). Another study reported that miR-582
promotes cancer stem cell traits of non-small-cell lung cancer, and
that miR-582 inhibition potently inhibits tumor progression
(32).
Although this study demonstrated that the
downregulation of miRNA-675 was associated with McVI, miRNA-675
expression level was not associated with any statistically
significant difference in survival. Previous studies have found
that decreased miRNA-675 expression was involved in enhanced cell
proliferation and invasion, as well as patient survival, in
non-small cell lung cancer and pancreatic cancer (33,34). Its
role as a tumor suppressor with anti-oncogenic activity is believed
to have emanated from its inhibitory effect on GPR55 or ZEB1.
However, other studies have reported differing results. It has been
suggested that miRNA-675 and its primary precursor long non-coding
RNA H19 play an oncogenic role in gliomas by inhibiting cadherin 13
or retinoblastoma 1 (35,36). Further studies are necessary to
determine the exact roles of miRNA-675 in tumorigenesis and cancer
progression.
Lately, there has been a dramatic increase in the
number of genomic databases for diseases, including those for
cancer. TCGA provides a large volume of publicly available cancer
genomic data. Researchers interested in the molecular biology of
cancer can access this valuable source of data. In the present
study, we used the epigenetic profiles of HCC tumor samples to
acquire novel information regarding the role of miRNA-141/-582/-9
in the McVI of HCC. It is likely that, in the near future, newly
discovered molecular targets based on the TCGA database will
provide clinical applications, including those for the early
detection, treatment, and even prevention of cancers. However, the
effective translation of cancer genomics or proteomics into
clinical practice necessitates progress in analytics; this requires
close cooperation between bioinformaticians, mathematicians, and
oncologists.
One limitation of the present study is that it was
conducted using only one public database. External validation of
the findings in this study is currently impossible, as no other
genomic database (Gene Expression Omnibus and ArrayExpress) of HCC
exists that includes both miRNA profiles and McVI parameters in the
clinical data profiles. Furthermore, most clinical and
corresponding tissue sample data in the TCGA database come from
Western countries. Consequently, further studies with HCC samples
from Eastern countries are needed to validate the results of this
study. Another drawback of the present study is that it was based
on the analysis of secondary data. Consequently, there was a lack
of information on important perioperative data, such as liver
enzyme profiles, antiviral drug use, and postoperative progression
of underlying liver disease, all of which are believed to influence
tumor recurrence and de novo malignancy. It is known that miRNAs
inhibit target genes through the degradation of their target mRNAs
or by inhibiting translation. If a specific miRNA functions by
suppressing translation and not through mRNA degradation, it is
difficult to identify a target from miRNA and mRNA expression
profiles and their correlation analysis alone. In the TCGA
database, the protein expression data is limited compared with the
miRNA and mRNA data; experimental verification is therefore
required.
This study showed that miRNA-141/-582/-9 were
overexpressed to a greater extent in HCC tissues with McVI than in
those without VI, and that their high expression levels were
significantly associated with poor survival of patients after
hepatic resection for HCC. Therefore, these three miRNAs warrant
consideration as potential therapeutic targets for cancer
treatment.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National
Research Foundation of Korea (NRF) grant funded by the Korea
government (MSIP; Ministry of Science, ICT & Future Planning;
grant nos. 2017R1C1B1008436 and 2017R1C1B5076680).
Availability of data and materials
The datasets generated and analyzed during the
current study are available in The Cancer Genome Atlas Liver
Hepatocellular Carcinoma database (TCGA LIHC) repository,
https://tcga-data.nci.nih.gov and
http://gdac.broadinstitute.org/.
Authors' contributions
YP, SKS and MGP designed the present study. YP, SKS,
MGP, SKP and CWC analyzed and interpreted the data. YP and SKS
wrote and revised the manuscript. MGP, SKP and CWC reviewed the
manuscript. All authors discussed the study and reviewed the
manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Llovet JM, Schwartz M and Mazzaferro V:
Resection and liver transplantation for hepatocellular carcinoma.
Semin Liver Dis. 25:181–200. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Thuluvath PJ: Vascular invasion is the
most important predictor of survival in HCC, but how do we find it?
J Clin Gastroenterol. 43:101–102. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Llovet JM, Bru C and Bruix J: Prognosis of
hepatocellular carcinoma: The BCLC staging classification. Semin
Liver Dis. 19:329–338. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Peng ZW, Guo RP, Zhang YJ, Lin XJ, Chen MS
and Lau WY: Hepatic resection versus transcatheter arterial
chemoembolization for the treatment of hepatocellular carcinoma
with portal vein tumor thrombus. Cancer. 118:4725–4736. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Park YK, Song SK, Kim BW, Park SK, Chung
CW and Wang HJ: Prognostic significance of microvascular invasion
in tumor stage for hepatocellular carcinoma. World J Surg Oncol.
15:2252017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Shirabe K, Toshima T, Kimura K, Yamashita
Y, Ikeda T, Ikegami T, Yoshizumi T, Abe K, Aishima S and Maehara Y:
New scoring system for prediction of microvascular invasion in
patients with hepatocellular carcinoma. Liver Int. 34:937–941.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wang W, Lin H, Zhou L, Zhu Q, Gao S, Xie
H, Liu Z, Xu Z, Wei J, Huang X and Zheng S: MicroRNA-30a-3p
inhibits tumor proliferation, invasiveness and metastasis and is
downregulated in hepatocellular carcinoma. Eur J Surg Oncol.
40:1586–1594. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Dang Z, Shangguan J, Zhang C, Hu P, Ren Y,
Lv Z, Xiang H and Wang X: Loss of protocadherin-17 (PCDH-17)
promotes metastasis and invasion through hyperactivation of
EGFR/MEK/ERK signaling pathway in hepatocellular carcinoma. Tumour
Biol. 37:2527–2535. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Calvisi DF, Ladu S, Gorden A, Farina M,
Conner EA, Lee JS, Factor VM and Thorgeirsson SS: Ubiquitous
activation of Ras and Jak/Stat pathways in human HCC.
Gastroenterol. 130:1117–1128. 2006. View Article : Google Scholar
|
|
10
|
Bacigalupo ML, Manzi M, Espelt MV,
Gentilini LD, Compagno D, Laderach DJ, Wolfenstein-Todel C,
Rabinovich GA and Troncoso MF: Galectin-1 triggers
epithelial-mesenchymal transition in human hepatocellular carcinoma
cells. J Cell Physiol. 230:1298–1309. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang CH, Guo ZY, Chen ZT, Zhi XT, Li DK,
Dong ZR, Chen ZQ, Hu SY and Li T: TMPRSS4 facilitates
epithelial-mesenchymal transition of hepatocellular carcinoma and
is a predictive marker for poor prognosis of patients after
curative resection. Sci Rep. 5:123662015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Li PF, Chen SC, Xia T, Jiang XM, Shao YF,
Xiao BX and Guo JM: Non-coding RNAs and gastric cancer. World J
Gastroenterol. 20:5411–5419. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li L, Liu Y, Guo Y, Liu B, Zhao Y, Li P,
Song F, Zheng H, Yu J, Song T, et al: Regulatory MiR-148a-ACVR1/BMP
circuit defines a cancer stem cell-like aggressive subtype of
hepatocellular carcinoma. Hepatology. 61:574–584. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Love MI, Huber W and Anders S: Moderated
estimation of fold change and dispersion for RNA-seq data with
DESeq2. Genome Biol. 15:5502014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
R Core Team, . R: A language and
environment for statistical computingR Foundation for Statistical
Computing; Vienna: 2016, https://www.R-project.org/April 1–2018
|
|
16
|
Dweep H and Gretz N: MiRWalk2.0: A
comprehensive atlas of microRNA-target interactions. Nat Methods.
12:6972015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Baker S, Ali I, Silins I, Pyysalo S, Guo
Y, Hogberg J, Stenius U and Korhonen A: Cancer hallmarks analytics
tool (CHAT): A text mining approach to organize and evaluate
scientific literature on cancer. Bioinformatics. 33:3973–3981.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Fang F, Chang RM, Yu L, Lei X, Xiao S,
Yang H and Yang LY: MicroRNA-188-5p suppresses tumor cell
proliferation and metastasis by directly targeting FGF5 in
hepatocellular carcinoma. J Hepatol. 63:874–885. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Shi Y, Song Q, Yu S, Hu D and Zhuang X:
Microvascular invasion in hepatocellular carcinoma overexpression
promotes cell proliferation and inhibits cell apoptosis of
hepatocellular carcinoma via inhibiting miR-199a expression. Onco
Targets Ther. 8:2303–2310. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Rodríguez-Perálvarez M, Luong TV, Andreana
L, Meyer T, Dhillon AP and Burroughs AK: A systematic review of
microvascular invasion in hepatocellular carcinoma: Diagnostic and
prognostic variability. Ann Surg Oncol. 20:325–339. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Park YK, Song SK, Kim BW, Park SK, Lee JI,
Lim SS and Wang HJ: Conditional survival analysis demonstrates that
recurrence risk of surgically treated hepatocellular carcinoma
evolves with time. J Gastrointest Surg. 21:1237–1244. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Clavien PA, Lesurtel M, Bossuyt PM, Gores
GJ, Langer B and Perrier A; OLT for HCCConsensus Group, : OLT for
HCC consensus group. Recommendations for liver transplantation for
hepatocellular carcinoma: An international consensus conference
report. Lancet Oncol. 13:e11–e22. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Bai T, Chen J, Xie ZB, Wu FX, Wang SD, Liu
JJ and Li LQ: The efficacy and safety of postoperative adjuvant
transarterial embolization and radiotherapy in hepatocellular
carcinoma patients with portal vein tumor thrombus. Onco Targets
Ther. 9:3841–3848. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Guo HB, Zhang Y and Chen HL: Relationship
between metastasis-associated phenotypes and N-glycan structure of
surface glycoproteins in human hepatocarcinoma cells. J Cancer Res
Clin Oncol. 127:231–236. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hu X, Zhao Y, Wei L, Zhu B, Song D, Wang
J, Yu L and Wu J: CCDC178 promotes hepatocellular carcinoma
metastasis through modulation of anoikis. Oncogene. 36:4047–4059.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Budhu A, Jia HL, Forgues M, Liu CG,
Goldstein D, Lam A, Zanetti KA, Ye QH, Qin LX, Croce CM, et al:
Identification of metastasis-related microRNAs in hepatocellular
carcinoma. Hepatology. 47:897–907. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Iyengar BR, Choudhary A, Sarangdhar MA,
Venkatesh KV, Gadgil CJ and Pillai B: Non-coding RNA interact to
regulate neuronal development and function. Front cell Neurosci.
8:472014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Filipowicz W, Jaskiewicz L, Kolb FA and
Pillai RS: Post-transcriptional gene silencing by siRNAs and
miRNAs. Curr Opin Struct Biol. 15:331–341. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Cora D, Re A, Caselle M and Bussolino F:
MicroRNA-mediated regulatory circuits: Outlok and perspectives.
Phys Biol. 14:0450012017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ding L, Yu LL, Han N and Zhang BT: MiR-141
promotes colon cancer cell proliferation by inhibiting MAP2K4.
Oncol Lett. 13:1665–1671. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhang Y, Zhou J, Sun M, Sun G, Cao Y,
Zhang H, Tian R, Zhou L, Duan L, Chen X and Lun L: Prognostic value
of microRNA-9 in various cancers: A meta-analysis. Pathol Oncol
Res. 23:573–582. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Fang L, Cai J, Chen B, Wu S, Li R, Xu X,
Yang Y, Guan H, Zhu X, Zhang L, et al: Aberrantly expressed
miR-582-3p maintains lung cancer stem cell-like traits by
activating Wnt/beta-catenin signalling. Nat Commun. 6:86402015.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
He D, Wang J, Zhang C, Shan B, Deng X, Li
B, Zhou Y, Chen W, Hong J, Gao Y, et al: Down-regulation of
miR-675-5p contributes to tumor progression and development by
targeting pro-tumorigenic GPR55 in non-small cell lung cancer. Mol
Cancer. 14:732015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ma L, Tian X, Guo H, Zhang Z, Du C, Wang
F, Xie X, Gao H, Zhuang Y, Kornmann M, et al: Long noncoding RNA
H19 derived miR-675 regulates cell proliferation by down-regulating
E2F-1 in human pancreatic ductal adenocarcinoma. J Cancer.
9:389–399. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Shi Y, Wang Y, Luan W, Wang P, Tao T,
Zhang J, Qian J, Liu N and You Y: Long non-coding RNA H19 promotes
glioma cell invasion by deriving miR-675. PLoS One. 9:e862952014.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zheng Y, Lu X, Xu L, Chen Z, Li Q and Yuan
J: MicroRNA-675 promotes glioma cell proliferation and motility by
negatively regulating retinoblastoma 1. Hum Pathol. 69:63–71. 2017.
View Article : Google Scholar : PubMed/NCBI
|