Introduction
Clear cell renal cell carcinoma (CCRCC) represents
the most common subtype of renal cancer, which is frequently
observed to exhibit alterations in the von Hippel-Lindau gene
(1). Although nephrectomy,
radiotherapy and immunotherapy have been applied in the treatment
of CCRCC, the prognosis of advanced CCRCC remains poor, due to a
lack of early symptoms, signs and laboratory abnormalities
(2). Therefore, it is worth focusing
on novel biomarkers that are involved in the carcinogenic process
of CCRCC development.
Cytochrome C (Cyto C), located on the inner surface
of mitochondria, is a heme-containing metalloprotein and
multifunctional enzyme, that is involved in cell apoptosis
(3). As a mitochondrial biomarker,
Cyto C is released from mitochondria into the extracellular space
and the bloodstream via permeabilization of injured mitochondria
within 1 h of apoptosis induction (4). Therefore, Cyto C is considered to be a
critical mediator and biomarker in mitochondria-mediated apoptosis
(5). Furthermore, the translocation
of Cyto C is a rapid and apoptosis-specific process, which occurs
not only under normal physiological conditions but also exists in
patients with cancer (6). Previous
studies have demonstrated that serum Cyto C is an indicator of
outcome during therapy of various types of cancer, including
leukemia and lung cancer (6,7), indicating that Cyto C may be involved
in tumor initiation and progression. Nevertheless, the clinical
relevance and function of Cyto C in CCRCC remain to be
elucidated.
Materials and methods
Cell culture
The CCRCC cell line 786-O was purchased from the
American Type Culture Collection (cat. no. CRL-1932), and cultured
in DMEM (HyClone; GE Healthcare Life Sciences) supplemented with
10% FBS. The cells were incubated in a humidified incubator at 37°C
with 5% CO2.
Clinical samples
In the present study, 10 pairs of tumor and
corresponding normal tissues were obtained from patients with CCRCC
(4 women and 6 men; mean age, 51.2±3.1 years), who underwent
surgical resection at the Suqian First Hospital (Suqian, China)
between January 2016 and December 2017. Normal tissues were
isolated 5 cm away from the corresponding tumor margin. Tissue
microarrays, including 150 CCRCC and 30 corresponding normal
tissues, were obtained from Shanghai BioChip Co Ltd. Clinical
information for the specimens included in the tissue microarray is
presented in Table SI. The
Tumor-Node-Metastasis staging (TNM; American Joint Committee on
Cancer, 2017) and grading Fuhrman grading systems were used for the
classification of patients with CCRCC (8).
Gene Expression Omnibus (GEO)
database
The mRNA levels of Cyto C in CCRCC were publicly
available from the National Center for Biotechnology Information
GEO database (accession no. GDS505) (9). The mRNA levels of Cyto C were compared
using a paired Student's t-test.
IHC staining and scoring
Slides of human tissue microarrays were
deparaffinized at 60°C by a series of xylene and decreasing
gradient of ethanol (100, 95, 85 and 75%) and heated in 10 mM
citrate buffer, pH 6.0, for 15 min in a microwave oven. Endogenous
peroxidase activity was blocked with 0.3% hydrogen peroxide and
0.1% saponin dissolved in TBS for 30 min at 37°C. Sections were
incubated with mouse polyclonal anti-Cyto C (1:100; cat. no.
ab133504; Abcam) at 4°C, followed by incubation with
avidin-biotin-peroxidase at 37°C (1:1; cat. no. COD-Nr5007; Dako;
Agilent Technologies, Inc.). Sections were stained with
3-diaminobenzidine and scored for the level of Cyto C expression
using the following system: 0, <1% cells with positive staining;
1, 1–25% positive cells; 2, 26–50% positive cells; 3, 51–75%
positive cells; and 4, 76–100% positive cells. The staining
intensity of Cyto C was scored as follows: 0, negative; 1 weak; 2,
moderate; and 3, high intensity (Fig.
S1). The final score of Cyto C expression was defined as the
staining intensity score multiplied by the score for the percentage
of positive cells. The cut-off value of the IHC score was
determined using X-tile statistical software version 3.6.1
(10). All slides were evaluated
using a light microscope independently by two pathologists who were
blinded to the clinical outcome of the patients. The present study
was performed according to the principles of the Declaration of
Helsinki and approved by the Ethics Committee of Suqian First
Hospital.
Validation analysis using the TCGA
database
The clinicopathological information and expression
data of the patients with CCRCC from The Cancer Genome Atlas
(TCGA)-database (http://www.tcga.org/) were downloaded
from The Human Protein Atlas database (https://www.proteinatlas.org).
Lentiviral infection and small
interfering (si)RNA tranfection procedures
For overexpression of Cyto C in CCRCC cells, the
Cyto C cDNA was cloned into a pLOV-CMV-eGFP-EF1a-PuroR lentiviral
vector (GeneChem, Inc.). Lentiviral particles were packaged using
Helper 1.0. CCRCC cells were infected with concentrated Cyto C-flag
lentivirus (MOI=2), and then selected and enriched with 4 µg/ml
puromycin for a week, and 2 µg/ml puromycin was sustained for
continuous selection pressure (termed the OECyto C line). The mock
group was infected with empty lentivirus.
For the knockdown experiments, 786-O cells were
transfected with siRNAs targeting Cyto C (15 nM; cat. no. sc-29292;
Santa Cruz Biotechnology, Inc.) and control siRNAs (siNC; cat. no.
sc-37007; Santa Cruz Biotechnology, Inc.) using
Lipofectamine® 3000 (Invitrogen; Thermo Fisher
Scientific, Inc.). After 6 h transfection, the culture was replaced
with fresh complete medium.
Western blotting
Cells were lysed with RIPA Lysis Buffer (Sangon
Biotech, Co., Ltd.) supplemented with Halt Protease Inhibitor
Cocktail (cat. no. 87786, Thermo Fisher Scientific, Inc.). The
lysate was centrifuged for 5 min at 12,000 × g to remove insoluble
substances. Protein concentration was detected by using
bicinchoninic acid assay Kit (Sangon Biotech, Co., Ltd.). Proteins
(30 µg) were separated by 10% SDS-PAGE and transferred onto
polyvinylidene fluoride membranes (EMD Millipore). Membranes were
blocked with 5% skimmed milk for 1 h, incubated with primary
antibodies at 4°C overnight and with secondary antibodies at for 1
h at room temperature. The antibodies used were as follows: Anti-
Cyto C (1:1,000; cat. no. 11940; Cell Signaling Technology, Inc.);
anti-GAPDH (1:1,000; cat. no. 5174; Cell Signaling Technology,
Inc.); and anti-cytochrome c oxidase subunit 4I1 (COXIV) antibody
(1:1,000; cat. no. ab33985; Abcam). SuperSignal West Femto Maximum
Sensitivity Substrate (Thermo Fisher Scientific, Inc.) was used to
detect the signal on the membranes, and signal was visualized using
a Chemiluminescence Imaging System (Fusion Solo S).
Cell apoptosis assay
To assess the extent of apoptosis, the FITC-Annexin
V/Dead Cell Apoptosis kit (Thermo Fisher Scientific, Inc.) with
FITC-Annexin V and propidium iodide (PI) double staining was used.
A total of 10 µl Annexin V and 1 µl PI (100 µg/ml) were added to
100 µl of cell suspension containing 1×105 cells.
Following incubation for 30 min at 37°C, 400 µl binding buffer was
added to each tube, and cells were analyzed using a FACSArray flow
cytometer (BD Biosciences) Data were analyzed using FlowJo 7.6
software (FlowJo LLC).
Cell Counting Kit-8 (CCK-8) assay
The mock and OECyto C cells were seeded into 96-well
plates at a concentration of 500 cells/well, and cultured for 0,
24, 48, 72 and 96 h. A solution of 10 µl CCK-8 (Beyotime Institute
of Biotechnology) and 90 µl DMEM was added to each well and
incubated for 1 h at 37°C. Measurements of optical density were
collected at 480 nm using a Multiskan Spectrum reader.
Xenografts
Female, 4-week-old athymic nude mice (Animal
Research Center of Nanjing University; n=5 weight, 18 g) were
housed in a pathogen-free environment (37°C, 5% CO2,
12-h light-dark cycle) and had free access to water and food. The
protocol was approved by the Institutional Animal Care and Use
Committee of Suqian First Hospital (Suqian, China). A total of
2×106 cells overexpressing Cyto C or mock cells were
suspended in 50 µl PBS and subcutaneously implanted in the mice.
Tumors were detected via bioluminescence imaging using an in
vivo Image System Spectrum (PerkinElmer, Inc). The tumor size
was measured every 7 days for a month using a Vernier caliper and
the volume was calculated as follows: Shortest diameter2
× longest diameter/2. The tumor-bearing mice were sacrificed 4
weeks after implantation, and the tumors were collected and
weighed. The xenografts were fixed with 4% paraformaldehyde for 4 h
at 37°C and paraffin embedded. Sections (4 µm) from the paraffin
block were deparaffinized in xylene and rehydrated in a descending
gradient (100, 95, 85 and 75%) of ethanol. Sections were then
boiled in 10 mM citrate buffer, for 15 min in a microwave oven, and
endogenous peroxidase activity was blocked with 0.3% hydrogen
peroxide and 0.1% saponin dissolved in TBS for 30 min at 37°C.
Sections were incubated with ki67 antibody (1:100; cat. no.
ab15580; Abcam) at 4°C overnight, and incubated with
avidin-biotin-peroxidase (1:1; cat. no. COD-Nr5007; Dako; Agilent
Technologies, Inc) at 37°C for 40 min. Signal was visualized with
the 3′-diaminobenzidine visualization kit (Dako; Agilent
Technologies, Inc.) and images were captured with an optical
microscope (magnification, ×40).
Statistical analysis
Statistical analyses were conducted using SPSS
software (v19.0; IBM Corp.). The association between Cyto C
expression and the clinicopathological characteristics of patients
with CCRCC was assessed by the Pearson χ2 test. The data
were presented as the means ± standard deviation (n=3).
Kaplan-Meier analysis and log-rank test were used to explore the
prognostic relevance of Cyto C in univariate analysis. The
statistical software X-tile (version 3.6.1) was used to determine
the cutoff in the 150-cohort of CRCC (9). Student's t-test for two groups and
one-way ANOVA followed by Least Significant Difference test for
multiple groups were applied. P<0.05 was considered to indicate
a statistically significant difference.
Results
Cyto C expression is downregulated in
CCRCC tissues
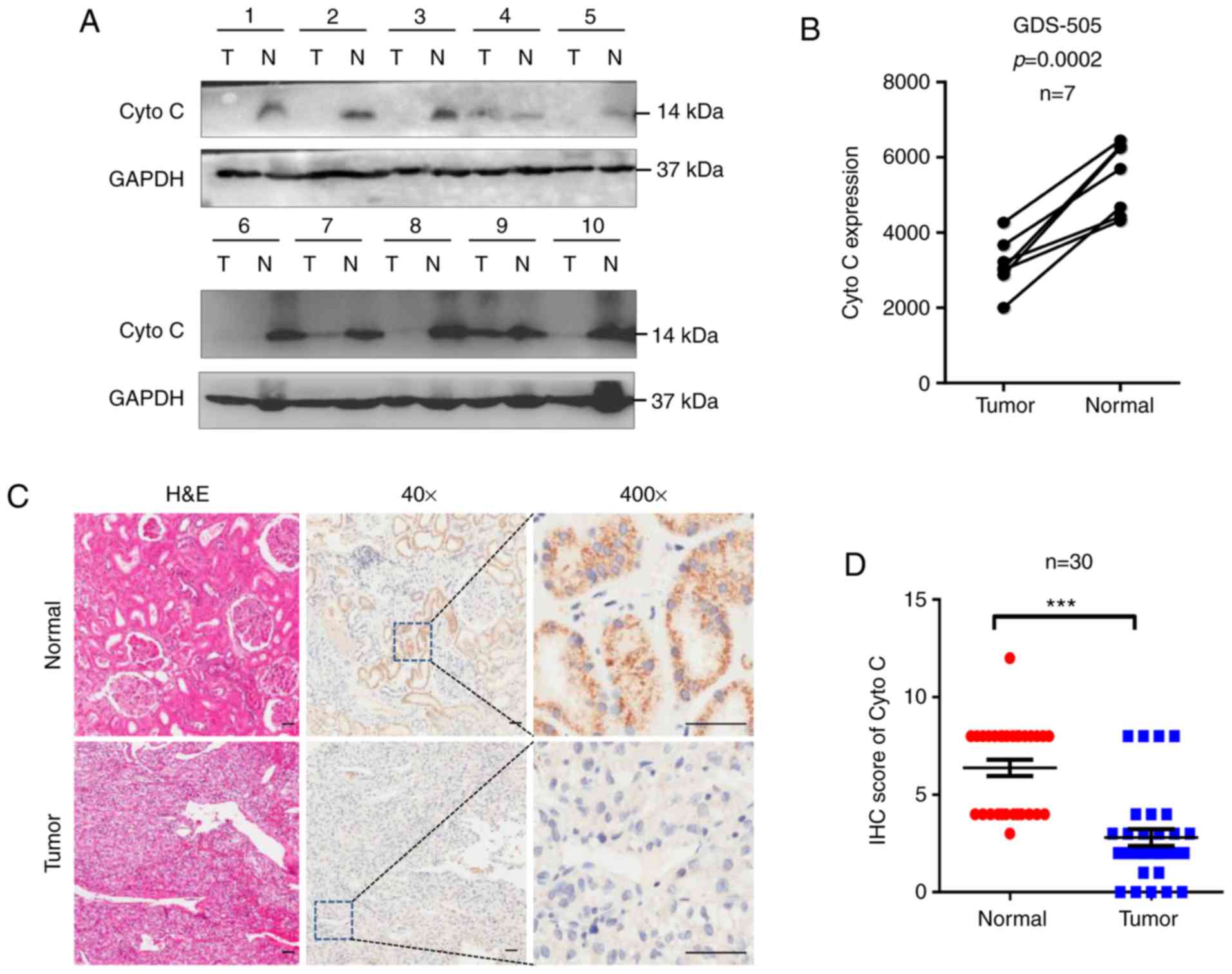
To determine the Cyto C status in CCRCC, 10 CCRCC
and corresponding normal tissues were examined using western
blotting. It was revealed that Cyto C protein expression levels
were decreased in CCRCC tissues compared with the corresponding
normal tissues (Fig. 1A). In
addition, analysis of the previously published GEO database GDS-505
revealed that Cyto C mRNA levels were significantly decreased in
CCRCC tissues compared with normal tissues (Fig. 1B). Furthermore, in the 30 paired
tissues from the cohort of 150 patients with CCRCC included in the
commercial tissue microarray used in the present study, Cyto C was
downregulated in the tumor tissues compared with their
corresponding normal tissues (Fig. 1C
and D). In addition, the proportion of Cyto Chigh
cells in CCRCC tissues (36.00%; 54/150) was significantly lower
compared with that in the corresponding normal tissues (96.67%;
29/30; Table I). Overall, these data
suggested that low expression levels of Cyto C may be associated
with CCRCC carcinogenesis.
 | Table I.Cyto C expression in renal carcinoma
and adjacent normal tissues from the tissue microarray
analysis. |
Table I.
Cyto C expression in renal carcinoma
and adjacent normal tissues from the tissue microarray
analysis.
| Tissue | Cyto C (low), n | Cyto C (high), n | P-value |
|---|
| CCRCC cancer | 96 | 54 |
|
| Paracancerous | 1 | 29 | <0.001 |
Cyto C confers favorable prognosis in
CCRCC
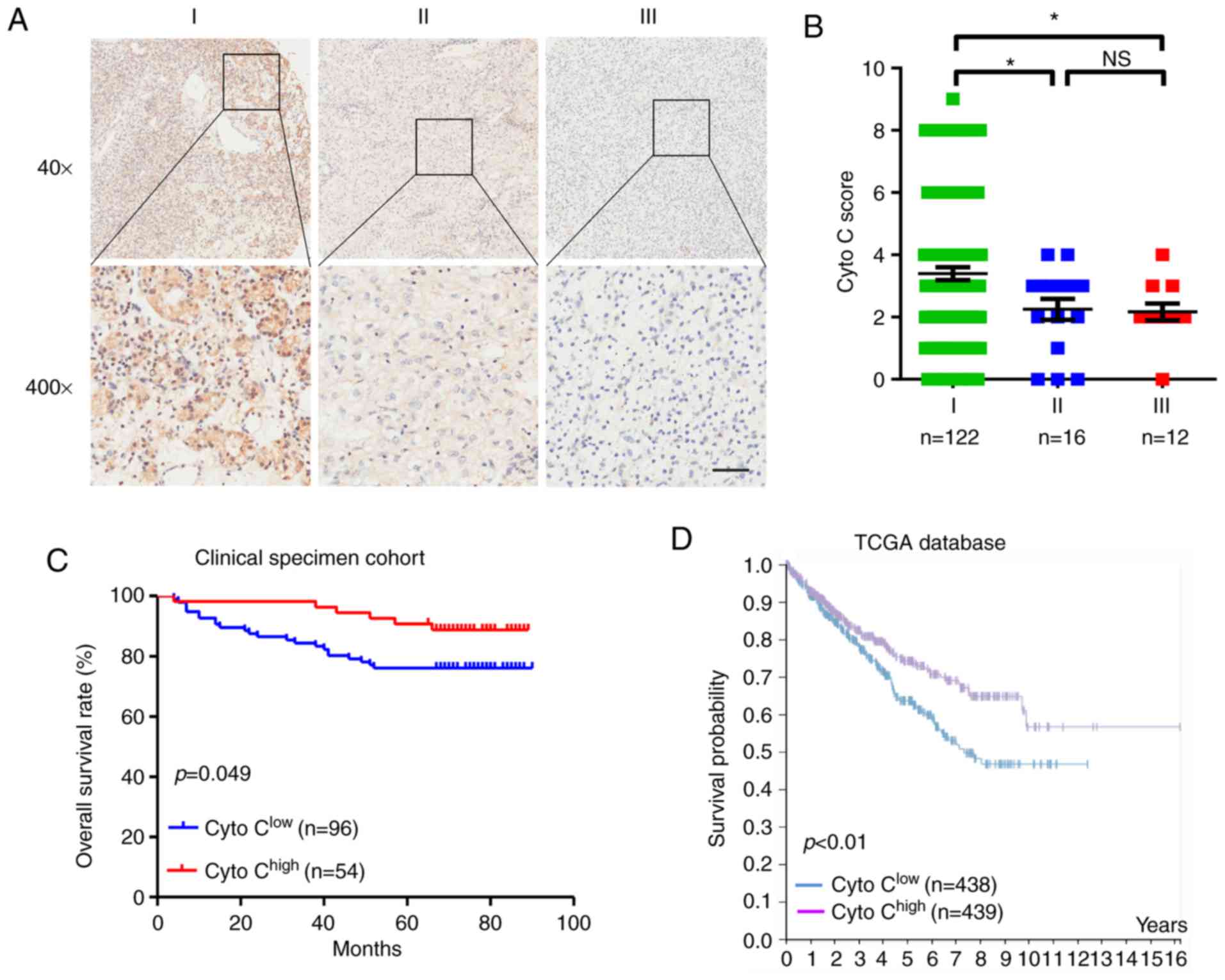
The association between Cyto C expression and
clinicopathological characteristics was investigated in patients
with CCRCC. It was revealed that low expression levels of Cyto C
were associated with worse TNM stage (P=0.001, Table II; and Fig. 2A and B) and tumor depth (P=0.008;
Table II). However, there was no
significant difference observed in Cyto C expression levels among
patients with well-, moderately- and poorly-differentiated tumors
(Table II). Furthermore, the
Kaplan-Meier survival analysis of the 150 CCRCC specimens from the
tissue microarray (P=0.049; Fig. 2C)
and TCGA database (P<0.01; Fig.
2D) suggested that patients with low Cyto C expression
exhibited shorter overall survival rates compared with those with
high Cyto C levels. Univariate and multivariate analyses revealed
that decreased Cyto C expression was an independent prognostic risk
factor in CCRCC (hazard ratio, 0.539; 95% CI, 0.214–1.357; P=0.045;
Table III). Therefore, the data of
the present study indicated that Cyto C may be a novel prognostic
biomarker for CCRCC.
 | Table II.Association between Cyto C expression
and clinicopathological features of patients with RCC. |
Table II.
Association between Cyto C expression
and clinicopathological features of patients with RCC.
|
| Cyto C
expression |
|
|---|
|
|
|
|
|---|
| Feature | Low, n (n=96) | High, n (n=54) | P-value |
|---|
| Sex |
|
| 0.845 |
| Male | 69 | 38 |
|
|
Female | 27 | 16 |
|
| Age at diagnosis,
years |
|
| 0.290 |
|
<50 | 21 | 16 |
|
| ≥50 | 75 | 38 |
|
| Location |
|
| 0.859 |
| Left | 43 | 25 |
|
|
Right | 53 | 29 |
|
| T stage |
|
| 0.008 |
| T1 | 72 | 50 |
|
|
T2-T3 | 24 | 4 |
|
| N stage |
|
| 0.134 |
| N0 | 93 | 54 |
|
| N1-2 | 3 | 0 |
|
| TNM |
|
| 0.001 |
| I | 78 | 44 |
|
|
II+III | 18 | 10 |
|
| Histological
grade |
|
| 0.431 |
| Well | 41 | 21 |
|
|
Moderate | 44 | 25 |
|
|
Poor | 11 | 8 |
|
 | Table III.Univariate and multivariate analyses
for overall survival in clear renal cell carcinoma. |
Table III.
Univariate and multivariate analyses
for overall survival in clear renal cell carcinoma.
|
| Univariate | Multivariate |
|---|
|
|
|
|
|---|
| Factors | HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| Sex | 2.594
(0.900–7.477) | 0.078 | 1.709
(0.570–5.217) | 0.339 |
| Age | 0.333
(0.101–1.031) | 0.071 | 0.413
(0.124–1.378) | 0.150 |
| Location | 1.084
(0.513–2.292) | 0.832 | 0.896
(0.416–1.928) | 0.778 |
| Cytochrome C
expression | 0.440
(0.178–1.085) | 0.049 | 0.539
(0.214–1.357) | 0.045 |
| Grade | 2.604
(1.516–4.472) | 0.001 | 1.764
(0.951–3.271) | 0.072 |
| TNM stage | 3.507
(2.313–5.316) | <0.001 | 3.546
(2.336–5.381) | <0.001 |
Cyto C decreases proliferation and
increases apoptosis of CCRCC cells in vitro
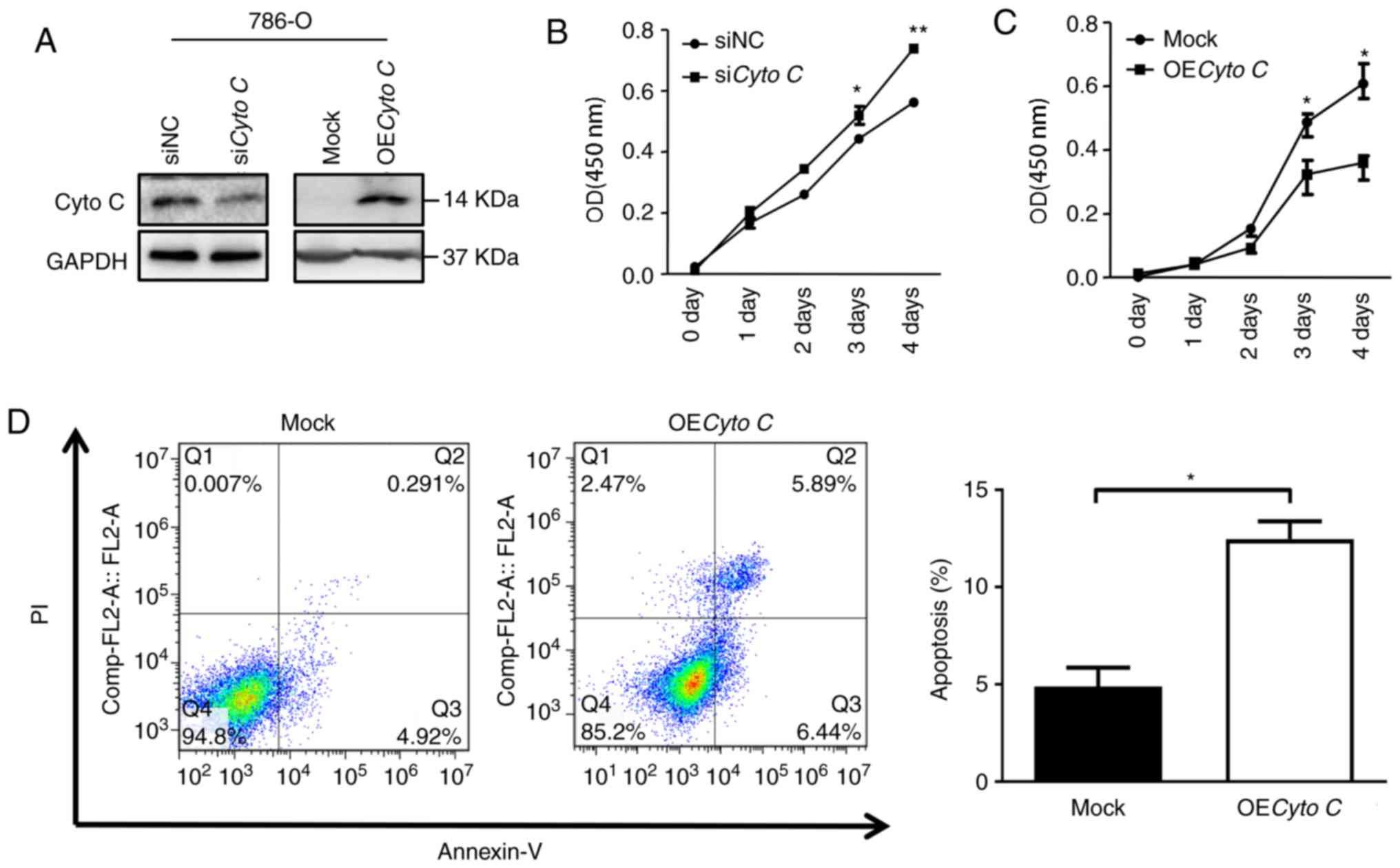
In order to explore the function of Cyto C in CCRCC
cells, the effects of Cyto C overexpression and knockdown on the
growth and apoptosis of CCRCC cells were investigated. Western
blotting confirmed that overexpression and knockdown of Cyto C
protein levels in 786-O cells were achieved (Fig. 3A). As a control, the expression
levels of COXIV, a house-keeping gene in mitochondria, were not
significantly altered following Cyto C-overexpression or knockdown
in 786-O cells (Fig. S2). It was
observed that the cell proliferation rate in OECyto C 786-O cells
was significantly decreased compared with the mock cells (Fig. 3C). By contrast, cell proliferation
was significantly increased following Cyto C knockdown by siRNA
(Fig. 3B). Furthermore, the cell
apoptosis rate was significantly increased in OECyto C 786-O cells
compared with mock cells (Fig.
3D).
Cyto C is crucial for CCRCC tumor
growth in vivo
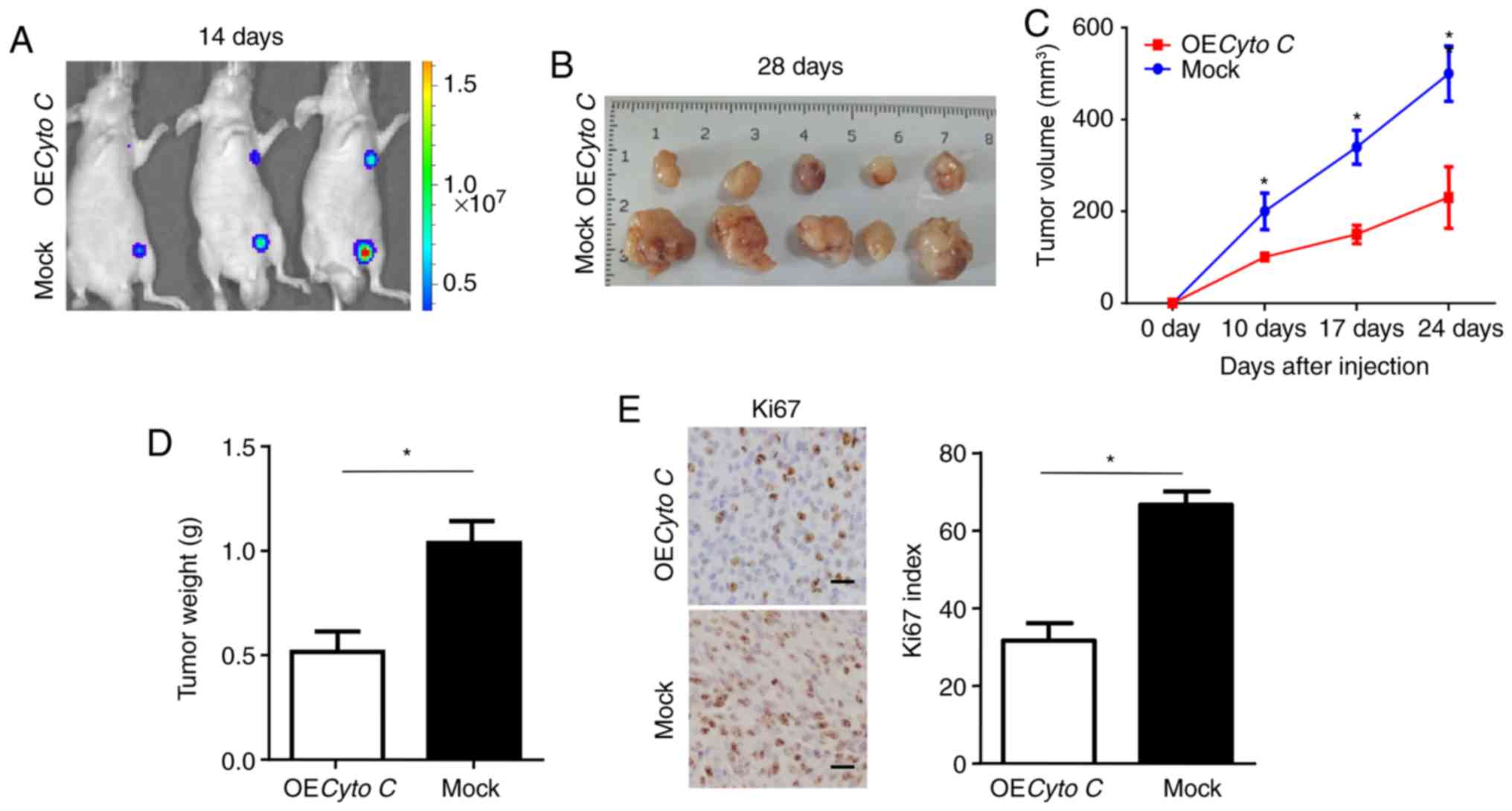
To further investigate the effect of Cyto C on CCRCC
cells in vivo, a subcutaneous xenograft model was
established. Bioluminescent imaging in nude mice detected smaller
tumors formed by OECyto C CCRCC cells compared with mock cells
after 14 days (Fig. 4A).
Furthermore, the xenograft tumors formed by OECyto C cells had a
slower growth rate and smaller average volume and weight compared
with those formed by mock cells (Fig.
4B-D). In addition, the xenografted tumors derived from the
OECyto C cells had decreased proliferation, as indicated by Ki67
staining (Fig. 4E). Overall, these
results indicated that overexpression of Cyto C inhibited the
tumorigenicity of CCRCC cells in vivo.
Discussion
Apoptosis or programmed cell death is vital for the
stability of cells, which is associated with several diseases,
including a number of different types of cancer, such as CCRCC
(10). The mitochondrial signaling
pathway, a major apoptosis signaling pathway, is involved in
releasing Cyto C and apoptosis-inducing factors from mitochondria
via the Bcl-2/Bax axis (11–13), thereby activating downstream
apoptotic executors. Therefore, Cyto C may be involved in cancer
initiation and progression. Although serum Cyto C has been shown to
be a precise indicator of cell death and decreased tumor size
following the first cycle of chemotherapy (14), to the best of our knowledge, the
relevance and function of Cyto C in CCRCC prior to chemotherapy
remains to be elucidated. In the present study, Cyto C expression
was demonstrated to be decreased at both the mRNA and protein
levels in CCRCC tissues, which suggested that decreased Cyto C may
be involved in CCRCC tumorigenesis.
One of the main features of cancer is to evade
apoptosis. Oncogenic events, such as downregulated tumor suppressor
genes or upregulated oncogenes that disrupt apoptosis, cause tumor
initiation, progression and metastasis (15). In the process of apoptosis, Bcl-2, an
anti-apoptotic factor and oncoprotein, prevents Cyto C release into
the cytoplasm from the mitochondrial inner membrane (16). The release of Cyto C triggers the
formation of an apoptosome and cell death cascade via binding to
the apoptosis regulator apoptotic peptidase activating factor 1
(16). The results of the present
study revealed that Cyto C may be involved in tumor growth by
altering cell apoptosis. Previous studies have demonstrated that
serum Cyto C levels are lower prior to treatment compared with
after chemotherapy and radiotherapy (14). However, another study has
demonstrated that serum Cyto C (17), which is decreased in patients with
non-small cell lung carcinoma (NSCLC) prior to treatment compared
with healthy individuals, is associated with low TNM stage and a
favorable prognosis in NSCLC. The results of the present study
demonstrated that low expression levels of Cyto C were associated
with advanced TNM stage and poor prognosis, indicating that Cyto C
was a tumor suppressor gene, and may be a prognostic biomarker in
CCRCC. Together with the results from previous studies (14,17),
this provides improved knowledge of the association between Cyto C
expression and tumor progression.
Unlimited cell proliferation and cell apoptosis are
two major characteristics of various types of cancer (18). In the present study, Cyto C acted as
a tumor suppressor gene to decrease the proliferation of 786-O
in vitro and in xenograft tumors in vivo.
Overexpression of Cyto C in CCRCC cells resulted in decreased
growth and increased cancer cell apoptosis in vitro. These
data indicated that Cyto C could serve as a therapeutic target for
the treatment of cancer. Furthermore, considering that Cyto C is a
protein with high selectivity and specificity (19), it is an attractive substitute as a
cytotoxic drug. In addition, a previous study revealed that Cyto C
can be packed with nano-sized smart drug delivery systems to induce
apoptosis as a method of cancer treatment (20). The results from the present study
provided evidence to further suggest that Cyto C may be a useful
and selective therapeutic drug.
In summary, the present study indicated an important
function of Cyto C in controlling the malignant behavior of CCRCC
cells, as well as its prognostic value in CCRCC. Therefore, Cyto C
may be a promising therapeutic drug for clinical treatment of
CCRCC.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by Suqian Guiding
Science and Technology Project (grant no. Z2018174) and Jiangsu
Youth Medical Talents Project (grant no. QNRC2016480).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
ZL and XZ performed experiments and contributed to
the conception of the study. LZ and BP analyzed the data and wrote
the manuscript. XZ revised the article. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethical
Committee of Suqian First Hospital (Suqian Branch Jiangsu Province
Hospital). The requirement to obtain patient consent for
participation was waived.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Moch H, Montironi R, Lopez-Beltran A,
Cheng L and Mischo A: Oncotargets in different renal cancer
subtypes. Curr Drug Targets. 16:125–135. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Mijuskovic M, Stanojevic I, Milovic N,
Cerovic S, Petrovic D, Maksic D, Kovacevic B, Andjelic T, Aleksic
P, Terzic B, et al: Tissue and urinary KIM-1 relate to tumor
characteristics in patients with clear renal cell carcinoma. Int
Urol Nephrol. 50:63–70. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Manickam P, Kaushik A, Karunakaran C and
Bhansali S: Recent advances in cytochrome c biosensing
technologies. Biosens Bioelectron. 87:654–668. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Renz A, Berdel WE, Kreuter M, Belka C,
Schulze-Osthoff K and Los M: Rapid extracellular release of
cytochrome c is specific for apoptosis and marks cell death in
vivo. Blood. 98:1542–1548. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wen Q, Zhang X, Cai J and Yang PH: A novel
strategy for real-time and in situ detection of cytochrome c and
caspase-9 in Hela cells during apoptosis. Analyst. 139:2499–2506.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Barczyk K, Kreuter M, Pryjma J, Booy EP,
Maddika S, Ghavami S, Berdel WE, Roth J and Los M: Serum cytochrome
c indicates in vivo apoptosis and can serve as a prognostic marker
during cancer therapy. Int J Cancer. 116:167–173. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Osaka A, Hasegawa H, Tsuruda K, Inokuchi
N, Yanagihara K, Yamada Y, Aoyama M, Sawada T and Kamihira S: Serum
cytochrome c to indicate the extent of ongoing tumor cell death.
Int J Lab Hematol. 31:307–314. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Medeiros LJ, Jones EC, Aizawa S, Aldape
HC, Cheville JC, Goldstein NS, Lubensky IA, Ro J, Shanks J, Pacelli
A and Jung SH: Grading of renal cell carcinom a: Workgroup No. 2.
union internationale contre le cancer and the American Joint
Committee on cancer (AJCC). Cancer. Sep. 80:990–991. 1997.
|
|
9
|
Lenburg ME, Liou LS, Gerry NP, Frampton
GM, Cohen HT and Christman MF: Previously unidentified changes in
renal cell carcinoma gene expression identified by parametric
analysis of microarray data. BMC Cancer. 3:312003. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Camp RL, Dolled-Filhart M and Rimm DL:
X-tile: A new bio-informatics tool for biomarker assessment and
outcome-based cut-point optimization. Clin Cancer Res.
10:7252–7259. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kao SJ, Lee WJ, Chang JH, Chow JM, Chung
CL, Hung WY and Chien MH: Suppression of reactive oxygen
species-mediated ERK and JNK activation sensitizes
dihydromyricetin-induced mitochondrial apoptosis in human non-small
cell lung cancer. Environ Toxicol. 32:1426–1438. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Irizarry Rovira AR, Bennet BM, Bolon B,
Braendli-Baiocco A, Chandra S, Fleurance R, Garman R, Hutto D, Lane
J, Romeike A, et al: Scientific and regulatory policy committee
points to consider: Histopathologic evaluation in safety assessment
studies for PEGylated pharmaceutical products. Toxicol Pathol.
46:616–635. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Trotta AP and Chipuk JE: Mitochondrial
dynamics as regulators of cancer biology. Cell Mol Life Sci.
74:1999–2017. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kadam CY and Abhang SA: Serum levels of
soluble Fas ligand, granzyme B and cytochrome c during adjuvant
chemotherapy of breast cancer. Clin Chim Acta. 438:98–102. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Vladimirov YA, Sarisozen C, Vladimirov GK,
Filipczak N, Polimova AM and Torchilin VP: The cytotoxic action of
cytochrome C/Cardiolipin nanocomplex (Cyt-CL) on cancer cells in
culture. Pharm Res. 34:1264–1275. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hockenbery D, Nunez G, Milliman C,
Schreiber RD and Korsmeyer SJ: Bcl-2 is an inner mitochondrial
membrane protein that blocks programmed cell death. Nature.
348:334–336. 1990. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Javid J1, Mir R, Julka PK, Ray PC and
Saxena A: Extracellular cytochrome c as a biomarker for monitoring
therapeutic efficacy and prognosis of non-small cell lung cancer
patients. Tumour Biol. 36:4253–4260. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chen Q, Weng HY, Tang XP, Lin Y, Yuan Y,
Li Q, Tang Z, Wu HB, Yang S, Li Y, et al: ARL4C stabilized by
AKT/mTOR pathway promotes the invasion of PTEN-deficient primary
human glioblastoma. J Pathol. 247:266–278. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Loo JF, Lau PM, Kong SK and Ho HP: An
assay using localized surface plasmon resonance and gold nanorods
functionalized with aptamers to sense the cytochrome-c released
from apoptotic cancer cells for anti-cancer drug effect
determination. micromachines (Basel). 8(pii): E3382017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kim ST, Lee YJ, Hwang YS and Lee S: Study
on aggregation behavior of Cytochrome C-conjugated silver
nanoparticles using asymmetrical flow field-flow fractionation.
Talanta. 132:939–944. 2015. View Article : Google Scholar : PubMed/NCBI
|