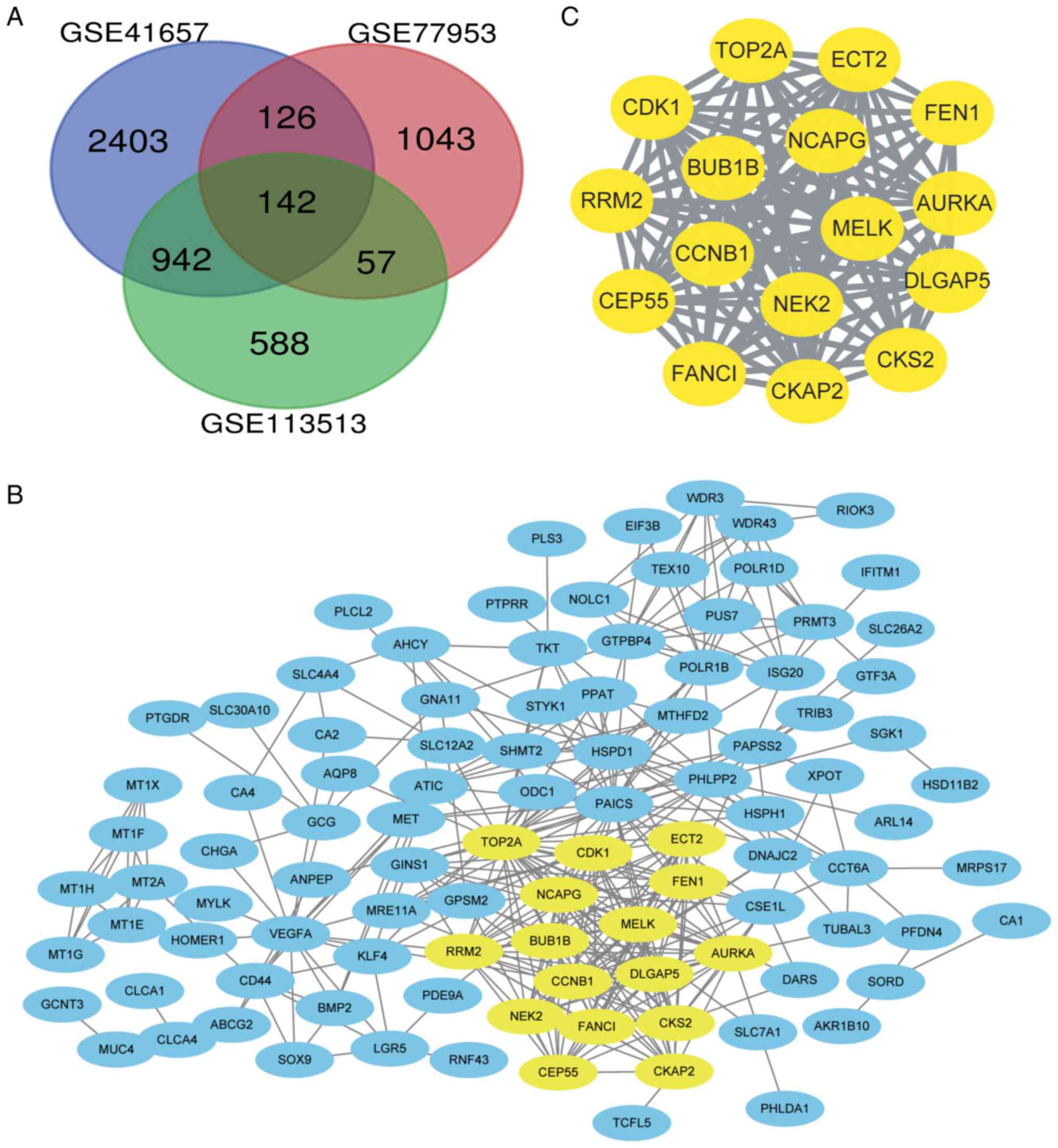
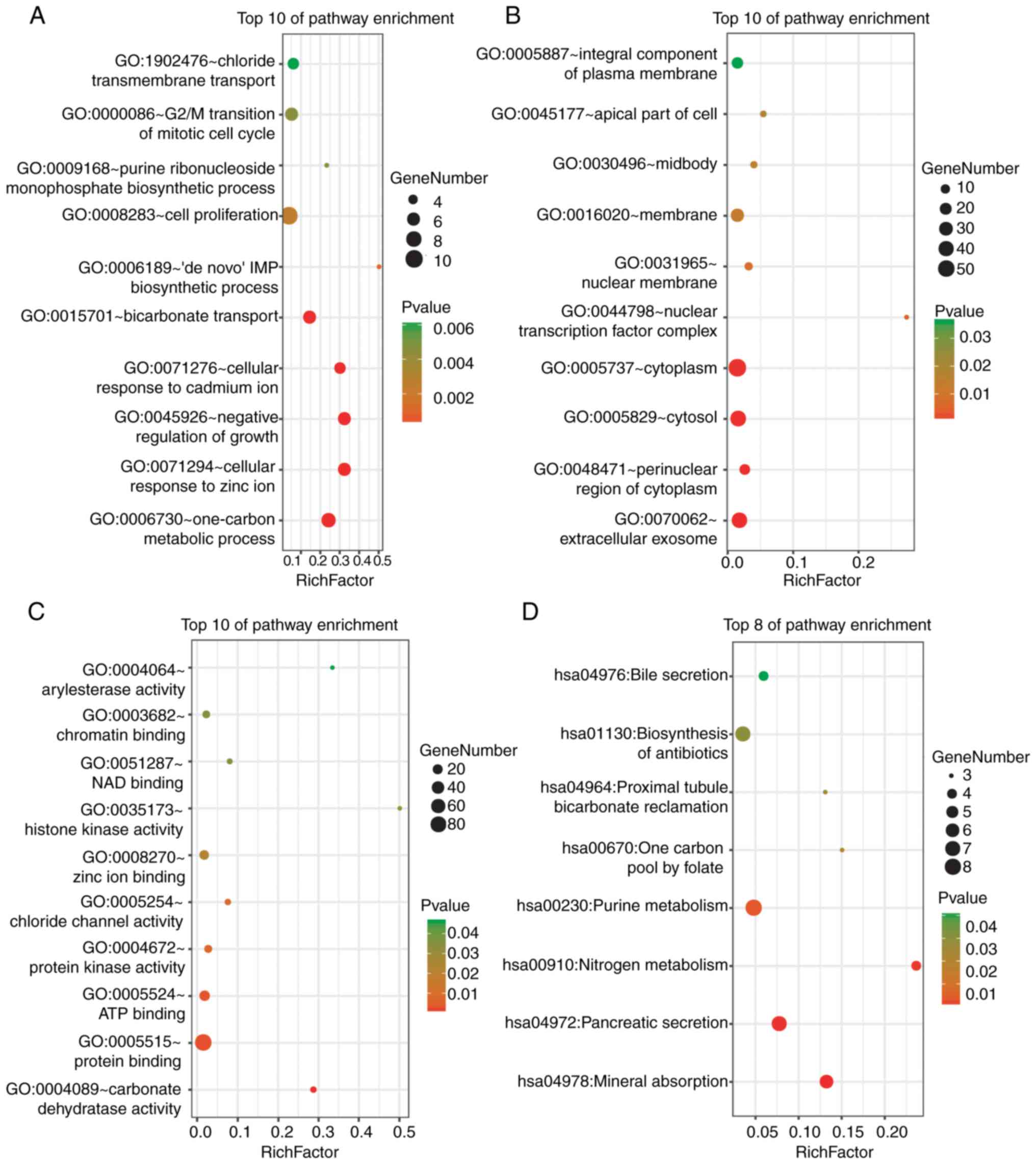
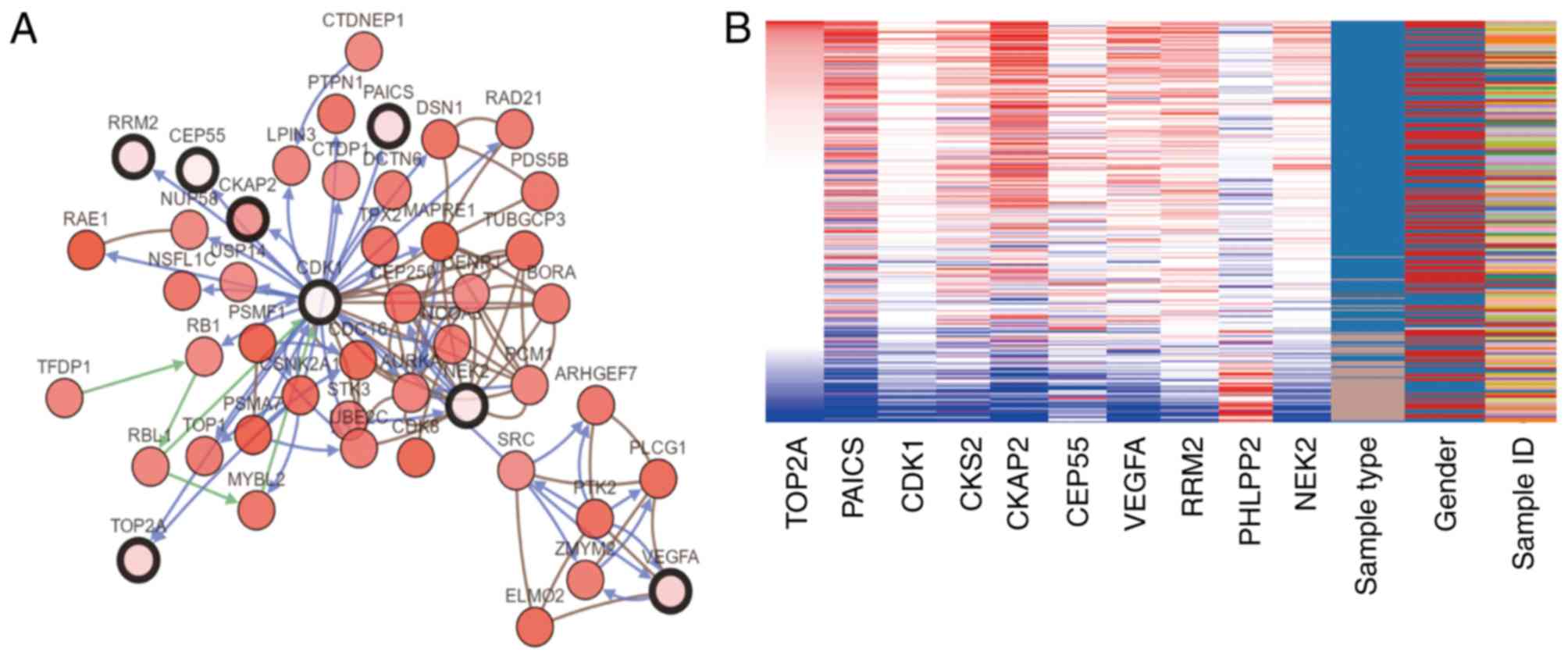
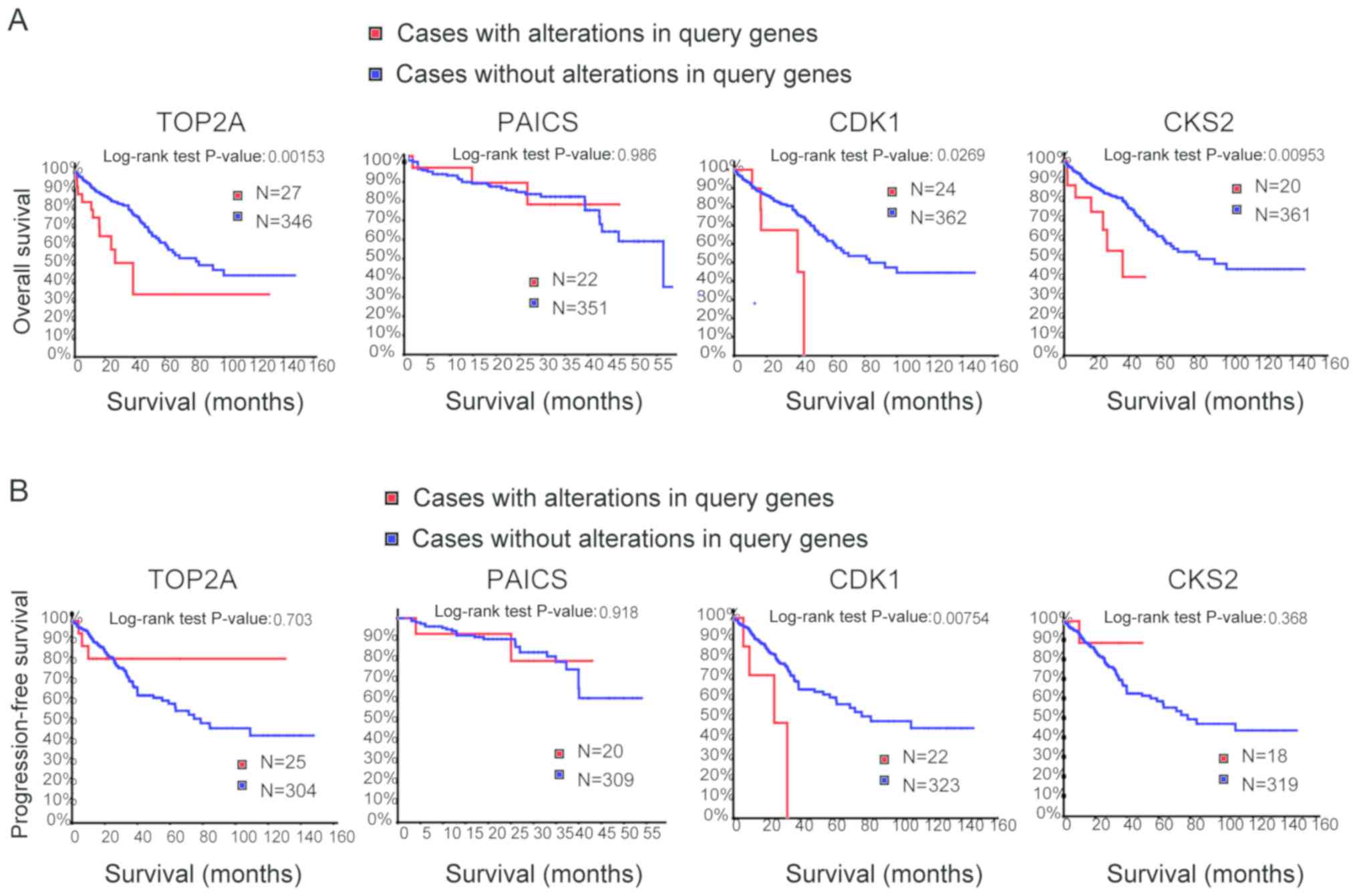
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Van Cutsem E, Lenz HJ, Kohne CH, Heinemann
V, Tejpar S, Melezinek I, Beier F, Stroh C, Rougier P, van Krieken
JH and Ciardiello F: Fluorouracil, leucovorin, and irinotecan plus
cetuximab treatment and RAS mutations in colorectal cancer. J Clin
Oncol. 33:692–700. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Tong K, Pellón-Cárdenas O, Sirihorachai
VR, Warder BN, Kothari OA, Perekatt AO, Fokas EE, Fullem RL, Zhou
A, Thackray JK, et al: Degree of tissue differentiation dictates
susceptibility to BRAF-driven colorectal cancer. Cell Rep.
21:3833–3845. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sanz-Garcia E, Argiles G, Elez E and
Tabernero J: BRAF mutant colorectal cancer: Prognosis, treatment,
and new perspectives. Ann Oncol. 28:2648–2657. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Corcoran RB, André T, Atreya CE, Schellens
JHM, Yoshino T, Bendell JC, Hollebecque A, McRee AJ, Siena S,
Middleton G, et al: Combined BRAF, EGFR, and MEK inhibition in
patients with BRAF(V600E)-mutant colorectal cancer. Cancer Discov.
8:428–443. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hamzehzadeh L, Khadangi F, Ghayoor
Karimiani E, Pasdar A and Kerachian MA: Common KRAS and NRAS gene
mutations in sporadic colorectal cancer in Northeastern Iranian
patients. Curr Probl Cancer. 42:572–581. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Reggiani Bonetti L, Barresi V, Bettelli S,
Caprera C, Manfredini S and Maiorana A: Analysis of KRAS, NRAS,
PIK3CA, and BRAF mutational profile in poorly differentiated
clusters of KRAS-mutated colon cancer. Hum Pathol. 62:91–98. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Misale S, Yaeger R, Hobor S, Scala E,
Janakiraman M, Liska D, Valtorta E, Schiavo R, Buscarino M,
Siravegna G, et al: Emergence of KRAS mutations and acquired
resistance to anti-EGFR therapy in colorectal cancer. Nature.
486:532–536. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Cooks T, Pateras IS, Jenkins LM, Patel KM,
Robles AI, Morris J, Forshew T, Appella E, Gorgoulis VG and Harris
CC: Mutant p53 cancers reprogram macrophages to tumor supporting
macrophages via exosomal miR-1246. Nat Commun. 9:7712018.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Fu X, Huang Y, Fan X, Deng Y, Liu H, Zou
H, Wu P, Chen Z, Huang J, Wang J, et al: Demographic trends and
KRAS/BRAFV600E mutations in colorectal cancer patients
of South China: A single-site report. Int J Cancer. 144:2109–2117.
2019.PubMed/NCBI
|
|
11
|
O'Brien MJ, Yang S, Mack C, Xu H, Huang
CS, Mulcahy E, Amorosino M and Farraye FA: Comparison of
microsatellite instability, CpG island methylation phenotype, BRAF
and KRAS status in serrated polyps and traditional adenomas
indicates separate pathways to distinct colorectal carcinoma end
points. Am J Surg Pathol. 30:1491–1501. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Saito M, Momma T and Kono K: Targeted
therapy according to next generation sequencing-based panel
sequencing. Fukushima J Med Sci. 64:9–14. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Deshiere A, Berthet N, Lecouturier F,
Gaudaire D and Hans A: Molecular characterization of Equine
Infectious Anemia Viruses using targeted sequence enrichment and
next generation sequencing. Virology. 537:121–129. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Meng H, Wang L, You H, Huang C and Li J:
Circular RNA expression profile of liver tissues in an EtOH-induced
mouse model of alcoholic hepatitis. Eur J Pharmacol.
862:1726422019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Harada K, Okamoto W, Mimaki S, Kawamoto Y,
Bando H, Yamashita R, Yuki S, Yoshino T, Komatsu Y, Ohtsu A, et al:
Comparative sequence analysis of patient-matched primary colorectal
cancer, metastatic, and recurrent metastatic tumors after adjuvant
FOLFOX chemotherapy. BMC Cancer. 19:2552019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hu Y, He C, Liu JP, Li NS, Peng C, Yang-Ou
YB, Yang XY, Lu NH and Zhu Y: Analysis of key genes and signaling
pathways involved in Helicobacter pylori-associated gastric cancer
based on the cancer genome atlas database and RNA sequencing data.
Helicobacter. 23:e125302018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Nakagawa H and Fujita M: Whole genome
sequencing analysis for cancer genomics and precision medicine.
Cancer Sci. 109:513–522. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Barrett T, Wilhite SE, Ledoux P,
Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH,
Sherman PM, Holko M, et al: NCBI GEO: Archive for functional
genomics data sets-update. Nucleic Acids Res. 41:D991–D995. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Huang DW, Sherman BT, Tan Q, Collins JR,
Alvord WG, Roayaei J, Stephens R, Baseler MW, Lane HC and Lempicki
RA: The DAVID gene functional classification tool: A novel
biological module-centric algorithm to functionally analyze large
gene lists. Genome Biol. 8:R1832007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kanehisa M, Furumichi M, Tanabe M, Sato Y
and Morishima K: KEGG: New perspectives on genomes, pathways,
diseases and drugs. Nucleic Acids Res. 45:D353–D361. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: Tool for the unification of biology. The gene
ontology consortium. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Franceschini A, Szklarczyk D, Frankild S,
Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C
and Jensen LJ: STRING v9.1: Protein-protein interaction networks,
with increased coverage and integration. Nucleic Acids Res.
41:D808–D815. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Smoot ME, Ono K, Ruscheinski J, Wang PL
and Ideker T: Cytoscape 2.8: New features for data integration and
network visualization. Bioinformatics. 27:431–432. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Bandettini WP, Kellman P, Mancini C,
Booker OJ, Vasu S, Leung SW, Wilson JR, Shanbhag SM, Chen MY and
Arai AE: MultiContrast Delayed Enhancement (MCODE) improves
detection of subendocardial myocardial infarction by late
gadolinium enhancement cardiovascular magnetic resonance: A
clinical validation study. J Cardiovasc Magn Reson. 14:832012.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Cerami E, Gao J, Dogrusoz U, Gross BE,
Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et
al: The cBio cancer genomics portal: An open platform for exploring
multidimensional cancer genomics data. Cancer Discov. 2:401–404.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Haeussler M, Zweig AS, Tyner C, Speir ML,
Rosenbloom KR, Raney BJ, Lee CM, Lee BT, Hinrichs AS, Gonzalez JN,
et al: The UCSC genome browser database: 2019 update. Nucleic Acids
Res. 47:D853–D858. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Rhodes DR, Yu J, Shanker K, Deshpande N,
Varambally R, Ghosh D, Barrette T, Pandey A and Chinnaiyan AM:
ONCOMINE: A cancer microarray database and integrated data-mining
platform. Neoplasia. 6:1–6. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang ZZ, Yang J, Jiang BH, Di JB, Gao P,
Peng L and Su XQ: KIF14 promotes cell proliferation via activation
of Akt and is directly targeted by miR-200c in colorectal cancer.
Int J Oncol. 53:1939–1952. 2018.PubMed/NCBI
|
|
29
|
Wu J, Yi J, Wu Y, Chen X, Zeng J, Wu J and
Peng W: 3, 3′-dimethylquercetin inhibits the proliferation of human
colon cancer RKO cells through Inducing G2/M cell cycle arrest and
apoptosis. Anticancer Agents Med Chem. 19:402–409. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Cheng J, Dwyer M, Okolotowicz KJ, Mercola
M and Cashman JR: A novel inhibitor targets both wnt signaling and
ATM/p53 in colorectal cancer. Cancer Res. 78:5072–5083. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Li J, Liu YY, Yang XF, Shen DF, Sun HZ,
Huang KQ and Zheng HC: Effects and mechanism of STAT3 silencing on
the growth and apoptosis of colorectal cancer cells. Oncol Lett.
16:5575–5582. 2018.PubMed/NCBI
|
|
32
|
Chamberlain JA, Dugué PA, Bassett JK,
Hodge AM, Brinkman MT, Joo JE, Jung CH, Makalic E, Schmidt DF,
Hopper JL, et al: Dietary intake of one-carbon metabolism nutrients
and DNA methylation in peripheral blood. Am J Clin Nutr.
108:611–621. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ducker GS, Chen L, Morscher RJ,
Ghergurovich JM, Esposito M, Teng X, Kang Y and Rabinowitz JD:
Reversal of cytosolic one-carbon flux compensates for loss of the
mitochondrial folate pathway. Cell Metab. 23:1140–1153. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Nitiss JL: DNA topoisomerase II and its
growing repertoire of biological functions. Nat Rev Cancer.
9:327–337. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Pommier Y, Leo E, Zhang H and Marchand C:
DNA topoisomerases and their poisoning by anticancer and
antibacterial drugs. Chem Biol. 17:421–433. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Deweese JE and Osheroff N: The DNA
cleavage reaction of topoisomerase II: Wolf in sheep's clothing.
Nucleic Acids Res. 37:738–748. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
McLeod HL, Douglas F, Oates M, Symonds RP,
Prakash D, van der Zee AG, Kaye SB, Brown R and Keith WN:
Topoisomerase I and II activity in human breast, cervix, lung and
colon cancer. Int J Cancer. 59:607–611. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Shibao K, Takano H, Nakayama Y, Okazaki K,
Nagata N, Izumi H, Uchiumi T, Kuwano M, Kohno K and Itoh H:
Enhanced coexpression of YB-1 and DNA topoisomerase II alpha genes
in human colorectal carcinomas. Int J Cancer. 83:732–737. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Coss A, Tosetto M, Fox EJ, Sapetto-Rebow
B, Gorman S, Kennedy BN, Lloyd AT, Hyland JM, O'Donoghue DP,
Sheahan K, et al: Increased topoisomerase IIalpha expression in
colorectal cancer is associated with advanced disease and
chemotherapeutic resistance via inhibition of apoptosis. Cancer
Lett. 276:228–238. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Goswami MT, Chen G, Chakravarthi BV, Pathi
SS, Anand SK, Carskadon SL, Giordano TJ, Chinnaiyan AM, Thomas DG,
Palanisamy N, et al: Role and regulation of coordinately expressed
de novo purine biosynthetic enzymes PPAT and PAICS in lung cancer.
Oncotarget. 6:23445–23461. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Meng M, Chen Y, Jia J, Li L and Yang S:
Knockdown of PAICS inhibits malignant proliferation of human breast
cancer cell lines. Biol Res. 51:242018. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Chakravarthi BVSK, Rodriguez Pena MDC,
Agarwal S, Chandrashekar DS, Hodigere Balasubramanya SA, Jabboure
FJ, Matoso A, Bivalacqua TJ, Rezaei K, Chaux A, et al: A role for
de novo purine metabolic enzyme PAICS in bladder cancer
progression. Neoplasia. 20:894–904. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Santamaría D, Barrière C, Cerqueira A,
Hunt S, Tardy C, Newton K, Cáceres JF, Dubus P, Malumbres M and
Barbacid M: Cdk1 is sufficient to drive the mammalian cell cycle.
Nature. 448:811–815. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Brown NR, Korolchuk S, Martin MP, Stanley
WA, Moukhametzianov R, Noble MEM and Endicott JA: CDK1 structures
reveal conserved and unique features of the essential cell cycle
CDK. Nat Commun. 6:67692015. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Asghar U, Witkiewicz AK, Turner NC and
Knudsen ES: The history and future of targeting cyclin-dependent
kinases in cancer therapy. Nat Rev Drug Discov. 14:130–146. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Goga A, Yang D, Tward AD, Morgan DO and
Bishop JM: Inhibition of CDK1 as a potential therapy for tumors
over-expressing MYC. Nat Med. 13:820–827. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Martinsson-Ahlzén HS, Liberal V,
Grünenfelder B, Chaves SR, Spruck CH and Reed SI: Cyclin-dependent
kinase-associated proteins Cks1 and Cks2 are essential during early
embryogenesis and for cell cycle progression in somatic cells. Mol
Cell Biol. 28:5698–5709. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Li M, Lin YM, Hasegawa S, Shimokawa T,
Murata K, Kameyama M, Ishikawa O, Katagiri T, Tsunoda T, Nakamura Y
and Furukawa Y: Genes associated with liver metastasis of colon
cancer, identified by genome-wide cDNA microarray. Int J Oncol.
24:305–312. 2004.PubMed/NCBI
|
|
49
|
Claesson-Welsh L and Welsh M: VEGFA and
tumour angiogenesis. J Intern Med. 273:114–127. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Ferrara N and Adamis AP: Ten years of
anti-vascular endothelial growth factor therapy. Nat Rev Drug
Discov. 15:385–403. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Terme M, Pernot S, Marcheteau E, Sandoval
F, Benhamouda N, Colussi O, Dubreuil O, Carpentier AF, Tartour E
and Taieb J: VEGFA-VEGFR pathway blockade inhibits tumor-induced
regulatory T-cell proliferation in colorectal cancer. Cancer Res.
73:539–549. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Seki A and Fang G: CKAP2 is a
spindle-associated protein degraded by APC/C-Cdh1 during mitotic
exit. J Biol Chem. 282:15103–15113. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Tsuchihara K, Lapin V, Bakal C, Okada H,
Brown L, Hirota-Tsuchihara M, Zaugg K, Ho A, Itie-Youten A,
Harris-Brandts M, et al: Ckap2 regulates aneuploidy, cell cycling,
and cell death in a p53-dependent manner. Cancer Res. 65:6685–6691.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Weinberger P, Ponny SR, Xu H, Bai S,
Smallridge R, Copland J and Sharma A: Cell cycle M-phase genes are
highly upregulated in anaplastic thyroid carcinoma. Thyroid.
27:236–252. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Wang Y, Jin T, Dai X and Xu J:
Lentivirus-mediated knockdown of CEP55 suppresses cell
proliferation of breast cancer cells. Biosci Trends. 10:67–73.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Jiang W, Wang Z, Chen G and Jia Y:
Prognostic significance of centrosomal protein 55 in stage I
pulmonary adenocarcinoma after radical resection. Thorac Cancer.
7:316–322. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
D'Angiolella V, Donato V, Forrester FM,
Jeong YT, Pellacani C, Kudo Y, Saraf A, Florens L, Washburn MP and
Pagano M: Cyclin F-mediated degradation of ribonucleotide reductase
M2 controls genome integrity and DNA repair. Cell. 149:1023–1034.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Brognard J and Newton AC: PHLiPPing the
switch on Akt and protein kinase C signaling. Trends Endocrinol
Metab. 19:223–230. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Brognard J, Sierecki E, Gao T and Newton
AC: PHLPP and a second isoform, PHLPP2, differentially attenuate
the amplitude of Akt signaling by regulating distinct Akt isoforms.
Mol Cell. 25:917–931. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Liao WT, Li TT, Wang ZG, Wang SY, He MR,
Ye YP, Qi L, Cui YM, Wu P, Jiao HL, et al: microRNA-224 promotes
cell proliferation and tumor growth in human colorectal cancer by
repressing PHLPP1 and PHLPP2. Clin Cancer Res. 19:4662–4672. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Cai J, Fang L, Huang Y, Li R, Yuan J, Yang
Y, Zhu X, Chen B, Wu J and Li M: miR-205 targets PTEN and PHLPP2 to
augment AKT signaling and drive malignant phenotypes in non-small
cell lung cancer. Cancer Res. 73:5402–5415. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Santarpia L, Qi Y, Stemke-Hale K, Wang B,
Young EJ, Booser DJ, Holmes FA, O'Shaughnessy J, Hellerstedt B,
Pippen J, et al: Mutation profiling identifies numerous rare drug
targets and distinct mutation patterns in different clinical
subtypes of breast cancers. Breast Cancer Res Treat. 134:333–343.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Hu CM, Zhu J, Guo XE, Chen W, Qiu XL, Ngo
B, Chien R, Wang YV, Tsai CY, Wu G, et al: Novel small molecules
disrupting Hec1/Nek2 interaction ablate tumor progression by
triggering Nek2 degradation through a death-trap mechanism.
Oncogene. 34:1220–1230. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Hayward DG, Clarke RB, Faragher AJ, Pillai
MR, Hagan IM and Fry AM: The centrosomal kinase Nek2 displays
elevated levels of protein expression in human breast cancer.
Cancer Res. 64:7370–7376. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Hawkins SM, Loomans HA, Wan YW,
Ghosh-Choudhury T, Coffey D, Xiao W, Liu Z, Sangi-Haghpeykar H and
Anderson ML: Expression and functional pathway analysis of nuclear
receptor NR2F2 in ovarian cancer. J Clin Endocrinol Metab.
98:E1152–E1162. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Neal CP, Fry AM, Moreman C, McGregor A,
Garcea G, Berry DP and Manson MM: Overexpression of the Nek2 kinase
in colorectal cancer correlates with beta-catenin relocalization
and shortened cancer-specific survival. J Surg Oncol. 110:828–838.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Lu L, Zhai X and Yuan R: Clinical
significance and prognostic value of Nek2 protein expression in
colon cancer. Int J Clin Exp Pathol. 8:15467–15473. 2015.PubMed/NCBI
|
|
68
|
Xu H, Zeng L, Guan Y, Feng X, Zhu Y, Lu Y,
Shi C, Chen S, Xia J, Guo J, et al: High NEK2 confers to poor
prognosis and contributes to cisplatin-based chemotherapy
resistance in nasopharyngeal carcinoma. J Cell Biochem.
120:3547–3558. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Zhang Y, Wang W, Wang Y, Huang X, Zhang Z,
Chen B, Xie W, Li S, Shen S and Peng B: NEK2 promotes
hepatocellular carcinoma migration and invasion through modulation
of the epithelial-mesenchymal transition. Oncol Rep. 39:1023–1033.
2018.PubMed/NCBI
|