Introduction
Gallbladder carcinoma (GBC) is the most common
malignancy of the bile duct, with its incidence ranking sixth in
gastrointestinal tumors, and its prevalence is increasing annually
(1). GBC is characterized by its
high malignancy, and metastasis can occur in the early stages of
disease. Traditional therapeutics, including surgery and
chemoradiotherapy have a poor effect on GBC, and the 5-year
survival rate is currently >5% (1,2).
Therefore, investigating the pathogenesis and identifying effective
treatments for GBC is an important area of research.
Receptor-mediated apoptosis serves an important role
in killing tumor cells (3). The
induction of apoptosis primarily occurs through a combination of
tumor necrosis factor superfamily members, including tumor necrosis
factor α (TNF-α), factor associated suicide legend (FAS-L) and
tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) and
their receptors on the cell membrane, therefore, activating
proteins involved in caspase activity, which then hydrolyse and
destroy the cellular structure and proteins (3). However, tumor cells often exhibit
anti-apoptotic capabilities, and the existence of cellular
Fas-associated death domain-like interleukin-1-converting enzyme
inhibitory protein (c-FLIP/CFLAR) results in the insensitivity and
tolerance of tumor cells towards TNF-α, Fas-L and TRAIL (4–7). The
overexpression of c-FLIP inhibits receptor-mediated apoptosis and
promotes the growth of tumors (5).
In colon cancer, prostate cancer, pancreatic cancer and melanoma,
c-FLIP has been identified to be highly expressed, and patients
expressing high levels of c-FLIP had a poor prognosis (7). Our previous research also confirmed
that c-FLIP was highly expressed in GBC (8). The knockdown of c-FLIP has been
hypothesized to significantly enhance TRAIL-induced apoptosis in
gallbladder cancer cells (8), and
the results of a previous study indicated that c-FLIP has become a
novel target for tumor therapy (9).
However, the regulation of the expression of c-FLIP in tumors and
its associated mechanism of action has not been completely
elucidated.
The mutations or heterotopic expressions of
microRNAs (miRs) are associated with multiple types of human cancer
and may serve an important role in the development of a number of
different types of cancer, including breast cancer, hepatocellular
carcinoma and lung cancer (10).
Aberrant expression of various miRs has been identified in a
variety of tumors, and are associated with certain clinical
features of tumors, including drug resistance and poor survival
(10,11). It has been reported that miR-512-3p
can inhibit the expression of c-FLIP in HepG2 hepatocellular
carcinoma cells in combination with the 3′untranslated regions
(3′UTR) of c-FLIP (12). Therefore,
the present study investigated whether the regulation of c-FLIP by
miR could affect the function of tumor cells and the development of
GBC.
Our previous research demonstrated that the c-FLIP
protein was significantly upregulated in GBC tissues (as shown by
immunohistochemistry), and inhibiting the expression of c-FLIP
could significantly enhance the effect of TRAIL-induced apoptosis
on gallbladder cancer cells (8).
miRs that regulated the expression of c-FLIP were subsequently
predicted through bioinformatic analysis, and a microarray to
detect microRNA expression profile in GBC and normal gallbladder
tissue samples were further utilized. The results of the present
study demonstrated that miR-125b could significantly inhibit the
growth and colony-forming ability of gallbladder cancer cells, and
miR-125b was observed to be significantly downregulated in
gallbladder cancer tissues. The regulatory effect of miR-125b on
c-FLIP 3′UTR was verified by a luciferase reporter gene experiment.
This confirmed that the low expression of miR-125b in GBC increased
the RNA expression of c-FLIP and promoted the growth of GBC cells.
Taken together, these results suggest that c-FLIP may be a novel
target of miR-125b. In addition, miR-125b may be used as a
candidate target for the clinical treatment of GBC.
Materials and methods
Human tissues sample
In the present study, a total of 23 patients with
GBC (including 13 males and 10 females) were enrolled between April
2015 and October 2016 at Huashan Hospital (Fudan University,
Shanghai, China). The median age of the patients was 60 years
(range, 32–85) at the time of surgery. The tumor and adjacent
normal tissues were obtained from patients following resection from
the primary neoplasms. The clinical features of patients are
presented in Table I. Tissues were
graded using the Tumor-Node-Metastasis grading system as described
previously (8). The present study
was approved by the Ethics Committee of Huashan Hospital, and
written informed consent was obtained from all patients.
 | Table I.Association between the expression of
miRNA-125b and clinicopathological factors in patients with
gallbladder carcinoma. |
Table I.
Association between the expression of
miRNA-125b and clinicopathological factors in patients with
gallbladder carcinoma.
|
| miR-125b
expression |
|
|
|---|
|
|
|
|
|
|---|
| Variables | High (n=12) | Low (n=11) | Total (n=23) | P-value |
|---|
| Age (years) |
|
|
| 0.4003 |
|
>60 | 6 | 8 | 14 |
|
|
≤60 | 6 | 3 | 9 |
|
| Sex |
|
|
| 0.4136 |
|
Male | 8 | 5 | 13 |
|
|
Female | 4 | 6 | 10 |
|
| TNM grade |
|
|
| 0.0361 |
|
I+II | 10 | 4 | 14 |
|
|
III+IV | 2 | 7 | 9 |
|
Cell culture and transfection
The gallbladder cancer cell lines, SGC-996 and
GBC-SD, were used in the present study. SGC-996 cell line was
provided by the Academy of Life Sciences, Tongji University
(Shanghai, China), and the GBC-SD cells were purchased from the
Shanghai Institute for Biological Sciences (Shanghai, China).
SGC-996 and GBC-SD cells were cultured in DMEM supplemented with
10% FBS (Gibco; Thermo Fisher Scientific, Inc.), 100 U/ml
penicillin and 100 µg/ml streptomycin, and incubated at 37°C with
5% CO2. The cells were detached with 0.25% trypsin and
were transmitted in a ratio of 1:2. The cells were then seeded in
six-well plates and transfected with miR-125b-5p mimics or a
negative control using Lipofectamine® 2000 transfection
reagent (Invitrogen; Thermo Fisher Scientific, Inc.), according to
the manufacturer's protocol. In brief, for each well, 5 µl 20 mM
mimics, inhibitors or siRNAs were added to 250 µl Opti-MEM (Gibco;
Thermo Fisher Scientific, Inc.) and Lipofectamine® 2000.
The mixture was added to cells and incubated for 6 h before
replacing the medium. miR-125b-5p mimics sequence:
5′-UCCCUGAGACCCUAACUUGUGA-3′; Negative control sequence:
5′-UUCUCCGAACGUGUCACGUTT-3′; miR-125b-5p mimics and negative
control were synthesized by Shanghai GenePharma Co, Ltd.
Cell proliferation assay
The cell growth curve was detected using the CCK-8
method. GBC-SD and SGC-996 gallbladder cancer cells were inoculated
in 96-well plates at a density of 800 cells/well, and were cultured
for 24 h. The experimental group was then transfected with
miR-125b-5p mimic, as described previously (8). At 24, 48, 72 and 96 h, the supernatant
was removed, and 10 µl CCK-8 solution and 90 µl culture medium was
added to each well, and the cells were cultured in the
aforementioned conditions for an additional 3 h. The medium was
analyzed at 450 nm on a microplate reader (Bio-Rad Laboratories,
Inc.).
RNA extraction and reverse
transcription-quantitative PCR (RT-qPCR)
Fresh tissues of unknown thickness were rinsed with
saline water and ground with liquid nitrogen. Total RNA was
isolated from GBC tissues, adjacent tissues and both cell lines
using TRIzol® reagent (Invitrogen; Thermo Fisher
Scientific, Inc.), according to the manufacturer's protocol. For
reverse transcription of total RNA containing miRNAs, the miScript
PCR system (Qiagen, Inc.) was used and the reverse transcription
temperature protocol was 37°C for 60 min and 95°C for 5 min. The
miScript II RT kit (Qiagen, Inc.) is part of the miScript PCR
system for miRNA detection and quantification. cDNA generated with
the miScript II RT kit was used as a template for real-time PCR,
with the expression quantified by PCR with specific primers and
probes using TaqMan microRNA assays (Applied Biosystems; Thermo
Fisher Scientific, Inc.), according to the manufacturer's
protocols. The thermocycling conditions were: 90°C For 20 sec;
followed by 40 cycles of 95°C for 1 sec and 60°C for 20 sec. The
primer sequences used for qPCR were the same as those in two
previous studies (13,14). The primer sequences used for qPCR
were as follows: miR-125b forward, 5′-GCTCCCTGAGACCCTAAC-3′ and
reverse, 5′-CAGTGCAGGGTCCGAGGT-3′; and U6 forward,
5′-CGCTTCGGCAGCACATATACTA-3′ and reverse,
5′-GCGAGCACAGAATTAATACGAC-3′. Relative expression was quantified
using the 2−ΔΔCq method (8).
Western blot analysis
miR-125b overexpression or control-treated GBC-SD
and SGC-996 cells were collected and lysed with
radioimmunoprecipitation assay buffer (Beyotime Institute of
Biotechnology) containing proteinase inhibitor cocktail (cat. no.
P8340; Sigma-Aldrich; Merck KGaA). The protein concentration was
measured using a bicinchoninic acid protein assay kit (Thermo
Fisher Scientific, Inc.). A total of 60 µg of protein was separated
by SDS-PAGE (12% gels) and transferred onto polyvinylidene
difluoride membranes. The membranes were subsequently blocked in 5%
fat-free milk at room temperature for 2 h and incubated with
anti-c-Flip (cat. no. ab8421; 1:1,000; rabbit polyclonal, Abcam)
and anti-GAPDH (cat. no. sc-32233; 1:1,000; mouse monoclonal; Santa
Cruz Biotechnology, Inc.) primary antibodies overnight at 4°C.
Following incubation with goat anti-mouse (cat. no. sc-2004;
1:5,000; Santa Cruz Biotechnology, Inc.) or goat anti-rabbit
IgG-HRP (cat. no. sc-2005; 1:5,000; Santa Cruz Biotechnology, Inc.)
secondary antibodies at room temperature for 2 h. The membranes
were visualized using the enhanced chemiluminescence-Plus kit (GE
Healthcare Life Sciences), according to the manufacturer's
protocols.
Plasmid construction and luciferase
reporter experiment
In order to determine miRNA targeting of the 3′-UTR
region of c-FLIP, the full length of the c-FLIP 3′-UTR was
amplified and cloned into a pmiR-REPORT luciferase vector (Promega
Corporation). DNA was extracted from the tissue samples using the
PicoPure™ DNA Extraction kit (Applied Biosystems; Thermo Fisher
Scientific, Inc.), according to the manufacturer's protocols, and
was treated with DNA polymerase (Promega Corporation). The
temperature protocol was as follows: 95°C for 3 min, followed by 30
cycles of 94°C for 40 sec, 56°C for 35 sec and final extension at
72°C for 60 sec. QuickMutation™ Site-Directed Mutagenesis kit
(Beyotime Institute of Biotechnology) was then used to generate the
mutated vector by replacing the miR-9-5p binding site nucleotides,
according to the manufacturer's protocol.
The cells were inoculated into 24-well plates, which
were transfected with the expression plasmids. Cells were collected
at 24–48 h following transfection, and were washed with PBS.
Subsequently, 80 µl 1× Passive Lysis Buffer (Promega Corporation)
was added to each well, and placed on a shaker at room temperature
for 120 min. The cell lysates (8–10 µl/well) were used for
detection. The program was set with a 10-sec pre-read delay,
followed by a 30-sec measurement period. The supernatant was then
added to the plate, and the firefly luciferase reaction substrates
and Stop & Glo® reagents (Promega Corporation) were
added, including 30 µl of both LARII and Stop & Glo®
reagent/well. Firefly luciferase reaction substrates and Stop &
Glo® reagents were added to the fluorometer tube, and
the relative activity was measured.
Colony formation assay
The cells in the logarithmic growth phase following
transfection were detached using trypsin to form a single-cell
suspension, and inoculated into 6-well plates with 500 cells/well,
and cultured for 1–2 weeks at 37°C. When visible colonies appeared,
the culture was terminated. Following washing with PBS twice, cells
were treated with 4% paraformaldehyde for 15 min at 37°C. Cells
were subsequently stained with 0.25% crystal violet dye for 10–30
min at 37°C, and the residual crystal violet was removed with water
slowly, and the cultures were air-dried. The number of colonies
were counted in five randomly selected fields using a light
microscope at a ×4 magnification. The colony numbers were
calculated and analyzed statistically.
Lentivirus production and
transduction
The primary miRNA sequence was amplified from normal
genomic DNA and the open reading frame of c-Flip was amplified from
cDNA. Using these amplified sequences, the green fluorescent
protein fragment of the pWPXL mock vector was replaced to form the
lentivirus expression vector pWPXL-miR-125b and pWPXL-c-FLIP. After
transfecting pWPXL-miR-125b, pWPXL-c-FLIP or pWPXL-mock with the
envelope plasmid pMDG2 and the packaging plasmid psPAX2 into 293T
cells using Lipofectamine 2000 at 48 h, the virus particles were
subsequently harvested, and GBC-SD and SGC-996 cells were infected
with the recombinant lentivirus-transducing units and 6 µg/ml
polybrene.
Bioinformatics analysis
TargetScan (targetscan.org/vert_72/) was used to predict miRNAs
which regulated c-Flip.
Statistical analysis
The results of the present study were analyzed using
the STATA 8.0 statistical software (StataCorp LP). Differences
among groups were analyzed using Wilcoxon signed-rank test, a
one-way ANOVA with Bonferroni post hoc test, or a Student's t-test.
The association between miRNA expression and the
clinicopathological variables of the patients was analyzed using
the χ2 test or Fisher's exact test. P<0.05 was
considered to indicate a statistically significant difference.
Results
Expression of miR-125b in GBC tissues
is lower compared with the normal tissues
In order to identify miRs that regulated the
expression of c-FLIP, a bioinformatics analysis was performed and
it was identified that a series of miRs could target the c-FLIP
gene, including miR-125b, miR-93, miR-10a, miR-20a, miR-20b,
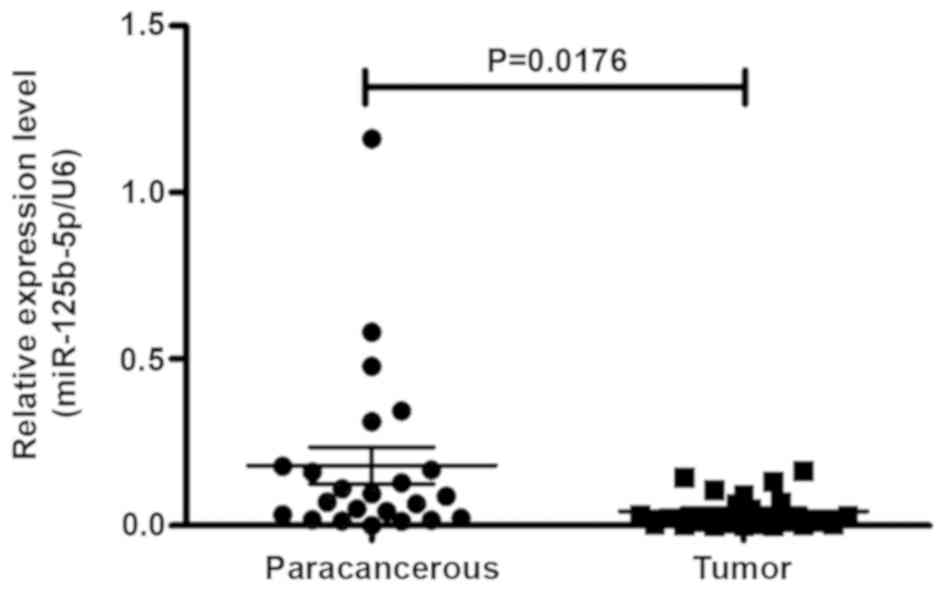
miR-143, miR-504, miR-150 and miR-149. The RT-qPCR results
demonstrated that the expression of miR-125b was significantly
decreased in GBC tissues when compared with 23 cases of matched
normal gallbladder tissues (Fig. 1).
This suggests that miR-125b functioned as a tumor suppressor gene
in GBC. We hypothesized that miR-125b may inhibit the expression of
c-FLIP. Therefore, the present study investigated whether miR-125b
expression was associated with the clinicopathological features of
patients with GBC. As presented in Table
I, the statistical analysis demonstrated that miR-125b
expression was significantly associated with the grade of GBC
(P=0.0361).
miR-125b inhibits the proliferation of
gallbladder cancer cells
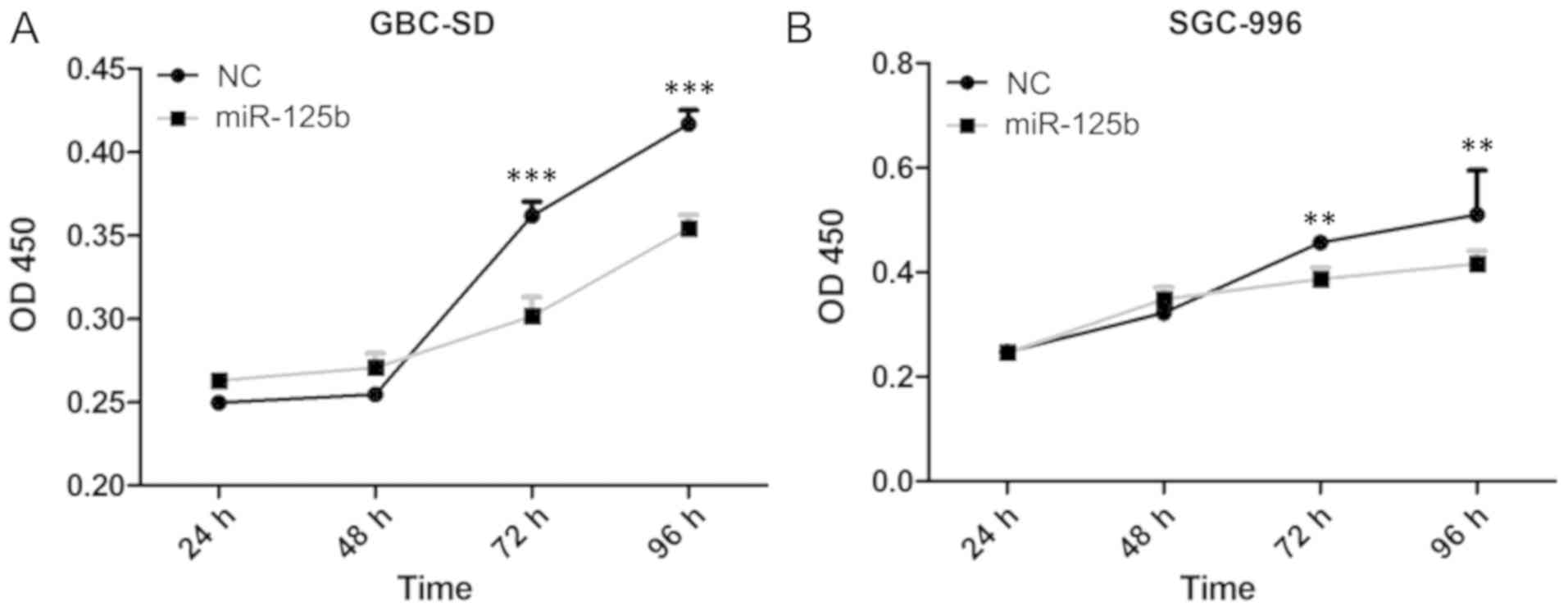
To examine the effects of miR-125b on the cell
growth of gallbladder cancer cells, the present study transfected
gallbladder cancer cells with miR-125b analogs (miR-125 mimic), and
detected the cellular proliferation rate of the gallbladder cancer
cells GBC-SD and SGC-966. The RT-qPCR results demonstrated that
miR-125b was efficiently overexpressed in gallbladder cancer cells
(Fig. S1A and B). miR-125b was
observed to have an inhibitory effect on the proliferation of
gallbladder cancer cells (Fig. 2A and
B). This indicated that increased expression of miR-125b in GBC
may reduce or delay cellular proliferation and tumor growth.
miR-125b weakens the colony formation
ability of gallbladder cancer cells
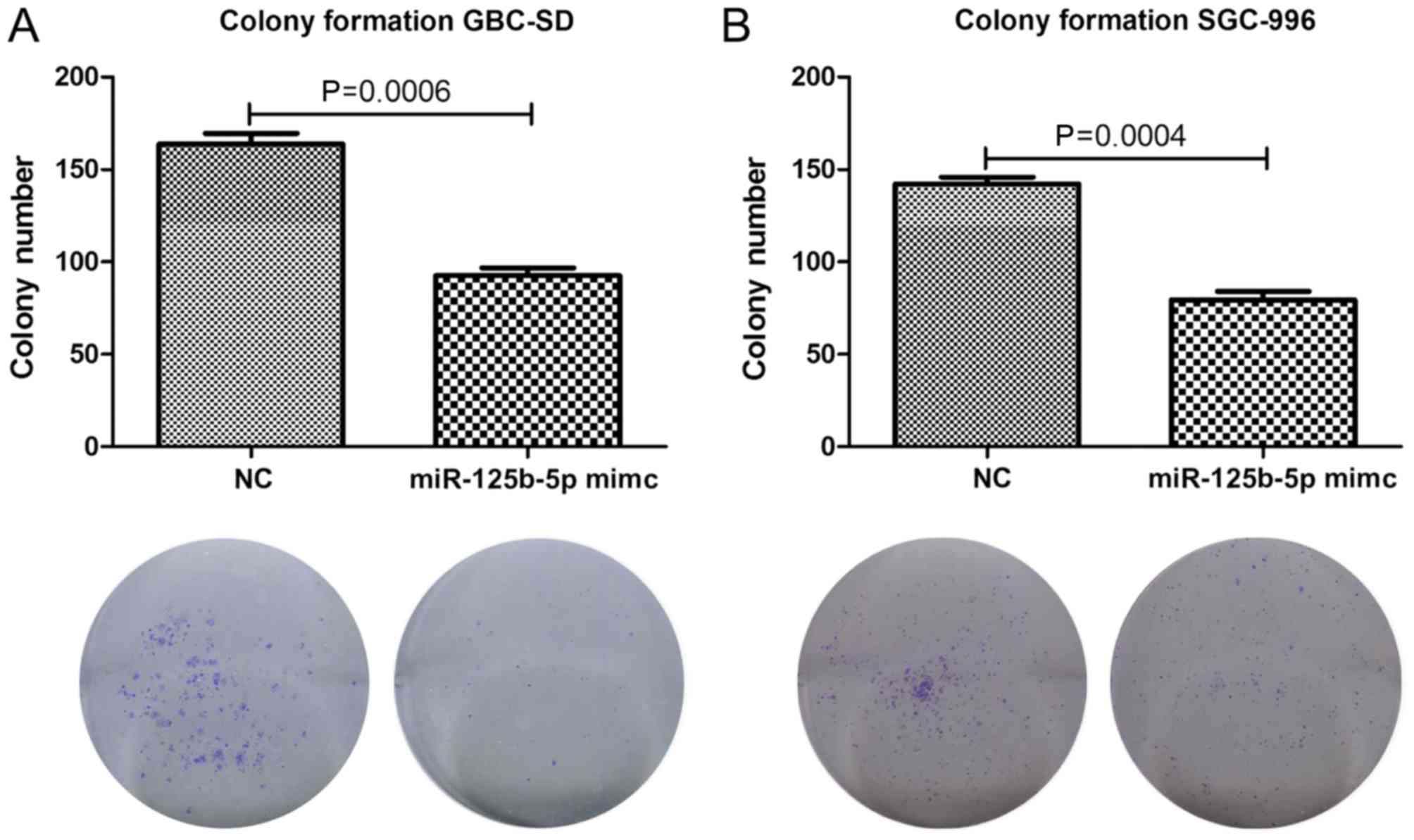
A colony formation assay was subsequently performed
in order to examine the influence of miR-125b on the colony forming
ability of gallbladder cancer cells. miR-125b was overexpressed in
the gallbladder cancer cell lines GBC-SD and SGC-966, and the
number of colonies formed was significantly decreased (Fig. 3A and B). The cell activity analysis
and the alteration of the colony formation ability of gallbladder
cancer cells suggested that miR-125b mimics significantly decrease
the colony formation ability of gallbladder cancer cells.
Therefore, these results suggest that miR-125b functions a
potential tumor suppressor and serves an important role in
inhibiting the initiation and development of GBC.
miR-125b suppresses the expression of
c-FLIP by interacting with the 3′UTR of the c-FLIP mRNA
It has been previously reported that the
overexpression of c-FLIP could inhibit the apoptosis of gallbladder
cancer cells (8). In order to
determine whether miR-125b inhibited the growth of gallbladder
cancer cells by regulating the expression of c-FLIP, the present
study initially predicted the binding sites of miR-125b and the
3′UTR of c-FLIP mRNA (Fig. 4A). By
cloning the c-FLIP mRNA 3′UTR and c-FLIP mRNA 3′ UTR mutation into
the luciferase report plasmid vector, the present study examined
the change of luciferase activity following the overexpression of
miR-125b, and found that miR-125b inhibited the luciferase
activity. In addition, this inhibitory effect was not observed when
the binding sites were mutated (Fig.
4B). Furthermore, the overexpression of miR-125b significantly
inhibited the protein expression of c-FLIP (Fig. 4C). This indicated that miR-125b may
inhibit the expression of c-FLIP through direct binding with the
3′UTR of c-FLIP mRNA, and further demonstrated that miR-125b may
suppress the proliferation of gallbladder cancer cells through
reducing the expression of c-FLIP.
Discussion
The treatment of GBC is challenging for
hepatobiliary surgeons in a clinical setting; however, the
combination of surgery with chemotherapy and radiotherapy serves as
the principal means of treatment for patients with GBC (15,16). As
the majority of tumors are at an advanced stage of the disease at
the time of diagnosis, only 10–30% of patients with GBC can undergo
radical surgery (15–17). The mutation rate of the P53 gene in
GBC is high (18–20) and the multidrug-resistance gene
MDR1 has also been demonstrated to be expressed in GBC
(21), therefore, contributing to
the insensitivity of GBC to radiotherapy and chemotherapy. Previous
studies have reported that the response rate of gemcitabine and
platinum compounds and 5-FU, when used alone or in combination was
>30% (22–24). The overall treatment efficacy of GBC
is poor, with 5-year overall survival rate of only 2–5% (1,2). In
addition to early detection and surgical treatment, it is
imperative to identify novel and more effective methods for the
treatment of GBC.
c-FLIP is an inhibitory protein of cellular
apoptosis, which was initially reported by Irmler et al
(25). c-FLIP is an analog of
procaspase-8 and procaspase-10, which can bind to FADD, interfering
with the recruitment of caspase-8/caspase-10, thereby blocking the
conduction of death receptor signals and inhibiting apoptosis
(26). c-FLIP contains two
sequential terminal domains, which are termed the death effector
domains (5). In c-FLIP, there is an
extending domain from the C-terminal that contains a caspase-like
domain; however, the two amino acid residues, Cys and His, in this
caspase-like domain are replaced by Tyr, which results in the
proteolytic enzyme activity of c-FLIP being lost (27). At present, the high expression of
c-FLIP has been detected in various tumors, including colon cancer,
gastric cancer, pancreatic cancer and melanoma. In colon cancer,
the expression level of c-FLIP is one of the independent factors
that affect the prognosis of patients (4). The high expression of c-FLIP has an
antagonistic effect in anti-androgen receptor drugs in patients
with prostate cancer that are ineffective in the treatment of
castration (28). Further studies
have confirmed that inhibiting the expression of c-FLIP in
melanoma, and in prostate and liver cancer can enhance the
apoptosis induced by a tumor in TRAIL and CD95L (29–31). In
addition, the sensitivity of tumor cells to chemotherapeutic drugs,
including taxol, cisplatin and 5-FU is elevated in tumors such as
colon and cervical cancer (32,33). The
results of the present study indicated that c-FLIP may function as
an oncogene and its high expression may be associated with tumor
growth. The present study demonstrated that c-FLIP is highly
expressed in GBC, and may suppress cellular apoptosis through the
regulation of TRAIL.
miR is a type of single-stranded non-coding RNA of
~19-25 nucleotides in length. It can bind to the mRNA coding region
or UTR, which directly results in the degradation of target genes
or inhibits the translation process of target genes, thereby
influencing their expression at the post-transcriptional level
(34,35). We have previously demonstrated that a
series of miRNAs could target the c-FLIP gene, including miR-125b,
miR-93, miR-10a, miR-20a, miR-20b, miR-143, miR-504, miR-150,
miR-149 and miR-125b was associated with the occurrence,
development, metastasis and prognosis of human tumors (36). Shi et al (37) revealed that miR-125b inhibited the
proliferation of glioma stem cells by blocking the cell cycle at
the G1/S phase. A previous study also demonstrated that
miR-125b suppressed the growth of hepatocellular carcinoma cells by
inhibiting the phosphorylation of Akt and the cell cycle,
additionally, the survival rate was higher in patients with cancer
expressing elevated levels of miR-125b (38). It has also been reported that
miR-125b could attenuate the growth of melanoma by directly
regulating the expression of the c-Jun protein (39). Studies have reported that miR-125b is
an important prognostic indicator in colon cancer, and patients
with a high expression of miR-125b had a larger tumor volume,
exhibited high tumor invasion and had a poor prognosis (40,41).
Furthermore, miR-125a and miR-125b could act on tumor necrosis
factor α-induced protein 3, and promote the activation of the NF-κB
pathway in diffuse large B-cell lymphoma (42). In conclusion, the results of these
studies suggest that miR-125b serves an important role in a variety
of tumors. We hypothesize that miR-125b could target c-FLIP in GBC;
however, to the best of our knowledge, the regulation of miR-125b
on c-FLIP has not yet been investigated.
Using bioinformatics analysis, the present study
demonstrated that miR-125b can bind to the 3′UTR of c-FLIP mRNA.
Furthermore, the use of a luciferase reporter gene assay confirmed
that miR-125b can inhibit the expression of c-FLIP, therefore,
significantly inhibiting the protein expression of c-FLIP. In
addition, it was observed that miR-125b was lowly expressed in GBC
tissues, and the overexpression of miR-125b could increase the
proliferation and colony formation capacity of gallbladder cancer
cells. This suggests that miR-125b may function as a potential
tumor suppressor. However, whether miR-125b may also inhibit tumor
cell growth in other tumors remains to be further studied. The
results of the present study revealed the important role of the
miR-125b-c-FLIP signal in the growth of gallbladder cancer cells,
providing a novel strategy for clinical treatment of GBC.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by Shanghai
Municipal Commission of Health and Family Planning (grant no.
201540191).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
YX and GW conceived the study, carried out the
experimental design and data interpretation, and prepared and
revised the manuscript. HJZ and HDZ performed the experiments. All
authors have read and approved the final version of this
manuscript.
Ethics approval and consent to
participate
The study was approved by the Ethics Committee of
Huashan Hospital (Fudan University) and written informed consent
was obtained from all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
GBC
|
gallbladder carcinoma
|
|
c-FLIP
|
cellular Fas-associated death
domain-like interleukin-1-converting enzyme inhibitory protein
|
|
3′UTR
|
3′untranslated regions
|
|
TNF-α
|
tumor necrosis factor α
|
|
FAS-L
|
factor associated suicide legend
|
|
TRAIL
|
tumor necrosis factor-related
apoptosis-inducing ligand
|
References
|
1
|
Chen YL, Huang ZQ, Zhou NX, Zhang WZ,
Huang XQ, Duan WD, Liu R and Liu Y: Clinical analysis of 110
patients with primary gallbladder carcinoma. Zhonghua Zhong Liu Za
Zhi. 29:704–706. 2007.(In Chinese). PubMed/NCBI
|
|
2
|
de Groen PC, Gores GJ, LaRusso NF,
Gunderson LL and Nagorney DM: Biliary tract cancers. N Engl J Med.
341:1368–1378. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ashkenazi A: Targeting the extrinsic
apoptosis pathway in cancer. Cytokine Growth Factor Rev.
19:325–331. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Korkolopoulou P, Saetta AA, Levidou G,
Gigelou F, Lazaris A, Thymara I, Scliri M, Bousboukea K,
Michalopoulos NV, Apostolikas N, et al: c-FLIP expression in
colorectal carcinomas: Association with Fas/FasL expression and
prognostic implications. Histopathology. 51:150–156. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhang X, Li W and Olumi AF: Overcoming
resistance to trail-induced apoptosis in prostate cancer by
regulation of c-FLIP. Methods Enzymol. 446:333–349. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Haag C, Stadel D, Zhou S, Bachem MG,
Moller P, Debatin KM and Fulda S: Identification of c-FLIP(L) and
c-FLIP(S) as critical regulators of death receptor-induced
apoptosis in pancreatic cancer cells. Gut. 60:225–237. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Tian F, Lu JJ, Wang L, Li L, Yang J, Li Y,
Liu YQ, Shen GX, Tu YT and Tao J: Expression of c-FLIP in malignant
melanoma, and its relationship with the clinicopathological
features of the disease. Clin Exp Dermatol. 37:259–265. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zong H, Yin B, Chen J, Ma B, Cai D and He
X: Over-expression of c-FLIP confers the resistance to
TRAIL-induced apoptosis on gallbladder carcinoma. Tohoku J Exp Med.
217:203–208. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Shirley S and Micheau O: Targeting c-FLIP
in cancer. Cancer Lett. 332:141–150. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lin S and Gregory RI: MicroRNA biogenesis
pathways in cancer. Nat Rev Cancer. 15:321–333. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Rupaimoole R and Slack FJ: MicroRNA
therapeutics: Towards a new era for the management of cancer and
other diseases. Nat Rev Drug Discov. 16:203–222. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chen F, Zhu HH, Zhou LF, Wu SS, Wang J and
Chen Z: Inhibition of c-FLIP expression by miR-512-3p contributes
to taxol-induced apoptosis in hepatocellular carcinoma cells. Oncol
Rep. 23:1457–1462. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Jia K, Shi P, Han X, Chen T, Tang H and
Wang J: Diagnostic value of miR-30d-5p and miR-125b-5p in acute
myocardial infarction. Mol Med Rep. 14:184–194. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang XL, Zhao YY, Sun L, Shi Y, Li ZQ,
Zhao XD, Xu CG, Ji HG, Wang M, Xu WR and Zhu W: Exosomes derived
from human umbilical cord mesenchymal stem cells improve myocardial
repair via upregulation of Smad7. Int J Mol Med. 41:3063–3072.
2018.PubMed/NCBI
|
|
15
|
Benoist S, Panis Y and Fagniez PL:
Long-term results after curative resection for carcinoma of the
gallbladder. French University Association for Surgical Research.
Am J Surg. 175:118–122. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Gourgiotis S, Kocher HM, Solaini L,
Yarollahi A, Tsiambas E and Salemis NS: Gallbladder cancer. Am J
Surg. 196:252–264. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Misra S, Chaturvedi A, Misra NC and Sharma
ID: Carcinoma of the gallbladder. Lancet Oncol. 4:167–176. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Saetta AA: K-ras, p53 mutations, and
microsatellite instability (MSI) in gallbladder cancer. J Surg
Oncol. 93:644–649. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Takagawa M, Muguruma N, Oguri K, Imoto Y,
Okamoto K, Ii K and Ito S: Prediction of prognosis in gallbladder
carcinoma by mucin and p53 immunohistochemistry. Dig Dis Sci.
50:1410–1413. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Masuhara S, Kasuya K, Aoki T, Yoshimatsu
A, Tsuchida A and Koyanagi Y: Relation between K-ras codon 12
mutation and p53 protein overexpression in gallbladder cancer and
biliary ductal epithelia in patients with pancreaticobiliary
maljunction. J Hepatobiliary Pancreat Surg. 7:198–205. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang BL, Zhai HY, Chen BY, Zhai SP, Yang
HY, Chen XP, Zhao WT and Meng L: Clinical relationship between MDR1
gene and gallbladder cancer. Hepatobiliary Pancreat Dis Int.
3:296–299. 2004.PubMed/NCBI
|
|
22
|
Meyerhardt JA, Zhu AX, Stuart K, Ryan DP,
Blaszkowsky L, Lehman N, Earle CC, Kulke MH, Bhargava P and Fuchs
CS: Phase-II study of gemcitabine and cisplatin in patients with
metastatic biliary and gallbladder cancer. Dig Dis Sci. 53:564–570.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Iyer RV, Gibbs J, Kuvshinoff B, Fakih M,
Kepner J, Soehnlein N, Lawrence D and Javle MM: A phase II study of
gemcitabine and capecitabine in advanced cholangiocarcinoma and
carcinoma of the gallbladder: A single-institution prospective
study. Ann Surg Oncol. 14:3202–3209. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hong YS, Lee J, Lee SC, Hwang IG, Choi SH,
Heo JS, Park JO, Park YS, Lim HY and Kang WK: Phase II study of
capecitabine and cisplatin in previously untreated advanced biliary
tract cancer. Cancer Chemother Pharmacol. 60:321–328. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Irmler M, Thome M, Hahne M, Schneider P,
Hofmann K, Steiner V, Bodmer JL, Schroter M, Burns K, Mattmann C,
et al: Inhibition of death receptor signals by cellular FLIP.
Nature. 388:190–195. 1997. View
Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhang X, Jin TG, Yang H, DeWolf WC,
Khosravi-Far R and Olumi AF: Persistent c-FLIP(L) expression is
necessary and sufficient to maintain resistance to tumor necrosis
factor-related apoptosis-inducing ligand-mediated apoptosis in
prostate cancer. Cancer Res. 64:7086–7091. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Safa AR: c-FLIP, a master anti-apoptotic
regulator. Exp Oncol. 34:176–184. 2012.PubMed/NCBI
|
|
28
|
McCourt C, Maxwell P, Mazzucchelli R,
Montironi R, Scarpelli M, Salto-Tellez M, O'Sullivan JM, Longley DB
and Waugh DJ: Elevation of c-FLIP in castrate-resistant prostate
cancer antagonizes therapeutic response to androgen
receptor-targeted therapy. Clin Cancer Res. 18:3822–3833. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Geserick P, Drewniok C, Hupe M, Haas TL,
Diessenbacher P, Sprick MR, Schon MP, Henkler F, Gollnick H,
Walczak H and Leverkus M: Suppression of cFLIP is sufficient to
sensitize human melanoma cells to TRAIL-and CD95L-mediated
apoptosis. Oncogene. 27:3211–3220. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
White SJ, Lu P, Keller GM and
Voelkel-Johnson C: Targeting the short form of cFLIP by RNA
interference is sufficient to enhance TRAIL sensitivity in PC3
prostate carcinoma cells. Cancer Biol Ther. 5:1618–1623. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Du X, Bao G, He X, Zhao H, Yu F, Qiao Q,
Lu J and Ma Q: Expression and biological significance of c-FLIP in
human hepatocellular carcinomas. J Exp Clin Cancer Res. 28:242009.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Longley DB, Wilson TR, McEwan M, Allen WL,
McDermott U, Galligan L and Johnston PG: c-FLIP inhibits
chemotherapy-induced colorectal cancer cell death. Oncogene.
25:838–848. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Luo A, Wang W, Sima N, Lu Y, Zhou J, Xu G,
Yu H, Wang S and Ma D: Short hairpin RNA targeting c-FLIP
sensitizes human cervical adenocarcinoma Hela cells to chemotherapy
and radiotherapy. Cancer Lett. 271:323–332. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Brennecke J, Hipfner DR, Stark A, Russell
RB and Cohen SM: Bantam encodes a developmentally regulated
microRNA that controls cell proliferation and regulates the
proapoptotic gene hid in Drosophila. Cell. 113:25–36. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Sun YM, Lin KY and Chen YQ: Diverse
functions of miR-125 family in different cell contexts. J Hematol
Oncol. 6:62013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Shi L, Zhang J, Pan T, Zhou J, Gong W, Liu
N, Fu Z and You Y: MiR-125b is critical for the suppression of
human U251 glioma stem cell proliferation. Brain Res. 1312:120–126.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Li W, Xie L, He X, Li J, Tu K, Wei L, Wu
J, Guo Y, Ma X, Zhang P, et al: Diagnostic and prognostic
implications of microRNAs in human hepatocellular carcinoma. Int J
Cancer. 123:1616–1622. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Kappelmann M, Kuphal S, Meister G,
Vardimon L and Bosserhoff AK: MicroRNA miR-125b controls melanoma
progression by direct regulation of c-Jun protein expression.
Oncogene. 32:2984–2991. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Nishida N, Yokobori T, Mimori K, Sudo T,
Tanaka F, Shibata K, Ishii H, Doki Y, Kuwano H and Mori M: MicroRNA
miR-125b is a prognostic marker in human colorectal cancer. Int J
Oncol. 38:1437–1443. 2011.PubMed/NCBI
|
|
41
|
Yamada A, Horimatsu T, Okugawa Y, Nishida
N, Honjo H, Ida H, Kou T, Kusaka T, Sasaki Y, Yagi M, et al: Serum
miR-21, miR-29a, and miR-125b are promising biomarkers for the
early detection of colorectal neoplasia. Clin Cancer Res.
21:4234–4242. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Kim SW, Ramasamy K, Bouamar H, Lin AP,
Jiang D and Aguiar RC: MicroRNAs miR-125a and miR-125b
constitutively activate the NF-KB pathway by targeting the tumor
necrosis factor alpha-induced protein 3 (TNFAIP3, A20). Proc Natl
Acad Sci USA. 109:7865–7870. 2012. View Article : Google Scholar : PubMed/NCBI
|