Introduction
Renal cell carcinoma (RCC) is the most prevalent
type of malignant tumor of the adult kidney worldwide (1). In the United States of America, an
estimated ~64,000 individuals were diagnosed with RCC in 2017
(2). Clear cell (cc)RCC is the most
common and invasive histological subtype of RCC and accounts for
70–80% of all RCC cases (3,4). The 5-year survival of patients with
ccRCC diagnosed in the early stage is >90% (5). However, for patients diagnosed at the
advanced stage, the 5-year survival is as low as 12% (5). Furthermore, a large proportion of
patients have been diagnosed at the advanced stage (6). The high mortality rate of patients with
ccRCC in the advanced stage may be due to lack of typical symptoms
and biomarkers with high sensitivity and accuracy for diagnosis in
the early stage, and an absence of a reliable risk stratification
method for assessing prognosis. Therefore, there is an urgent
requirement to identify tumor-specific biomarkers and to develop a
nomogram for the precise prediction of prognosis, which may lead to
the development of an accurate risk stratification method and guide
the clinical diagnosis and treatment of ccRCC.
Long non-coding RNAs (lncRNAs), a class of
transcripts, are >200 nt long and lack any protein-coding
capacity (7). Previous studies have
indicated that lncRNAs are aberrantly expressed in diverse
malignant tumor types and serve as pivotal regulators of different
biological processes in tumors, including cell proliferation,
invasion, apoptosis and metastasis (8). For instance, Ning et al
(9) demonstrated that lncRNA nuclear
paraspeckle assembly transcript 1 (NEAT1) was overexpressed in
ccRCC tissue, which was significantly associated with poor
prognosis in patients with ccRCC and silencing of NEAT1 was able to
inhibit ccRCC cell proliferation and invasion. In addition, Yang
et al (10) demonstrated that
lncRNA PVT1 positively regulated ccRCC cell proliferation and
invasion by interacting with microRNA (miR)-200s through increasing
the expression of zinc finger E-box binding homeobox 1 (ZEB1), ZEB2
and polycomb complex protein BMI-1.
Advances in omics technology have created
opportunities to mechanistically elucidate, diagnose and treat
cancer in a systematic manner (11).
RNA-sequencing profiling has been developed, which may be utilized
for identifying novel molecular markers and mechanisms in numerous
tumor types (12). Furthermore, with
their advantage of the combination of independent prognostic
factors to consider their multiple effects on probability of
outcome, prognostic nomograms have been widely used as a powerful
model for risk evaluation in cancer (13).
In the present study, all original matrix files for
ccRCC and corresponding clinical information were downloaded from
The Cancer Genome Atlas (TCGA, http://cancergenome.nih.gov) to detect the
differentially expressed (DE)lncRNAs between ccRCC tissues and
normal renal tissues. The best survival-associated candidate
DElncRNAs were then selected to develop a prognostic nomogram.
Furthermore, functional enrichment analysis was performed to
predict the biological functions of the candidate DElncRNAs. The
present study may contribute to the determination of independent
prognostic lncRNAs in ccRCC and provide additional information
regarding the molecular mechanisms of ccRCC.
Materials and methods
Data retrieval and processing
The lncRNA expression profiles were obtained from
TCGA, an open database to identify novel biomarkers in cancer
research (version: April 5, 2018), and were then subjected to
background correction and normalization with Perl 5.0 (http://www.perl.org/). Patients with a follow-up time
of <30 days or lack of pathologic diagnosis and corresponding
clinical information were removed. The data of 574 tissue samples
were included in the present study, comprising 70 adjacent
non-tumor kidney tissues and 504 ccRCC tissue samples. Relevant
clinical characteristics of the 504 cancer cases were also obtained
and collated.
Patient cohort
The 504 patients with ccRCC were randomly divided
into two cohorts: The training cohort and the validation cohort.
The training cohort comprised 380 cancer cases and the remaining
cases were in the validation cohort. For categorical variables,
data were expressed as numbers and compared using χ2
tests or Fisher's test, whereas for continuous variables, data were
expressed as the mean ± standard deviation and compared using
Student's t-tests in SPSS 20.0 (IBM Corp.). P<0.05 was
considered to indicate a statistically significant difference.
Processing of lncRNA expression
data
The statistical software R (version 3.5.2;
http://www.R-project.org) and the Bioconductor
package ‘edgeR’ (http://www.bioconductor.org/packages/release/bioc/html/edgeR.html)
(14) were used to identify
DElncRNAs with the criterion of |log fold change (FC)|>2 and
adjusted P<0.01, as described previously (15).
Construction and evaluation of
DElncRNA-based prognostic nomogram
In the training cohort, a univariate regression
analysis was performed to select DElncRNAs that were highly
associated with the overall survival (OS) of patients with ccRCC.
Subsequently, a LASSO regression analysis was performed to
additionally screen out the set of independent prognostic candidate
DElncRNAs with the strongest predictive power. Next, the most
significant survival-associated candidate DElncRNAs were subjected
to a multivariate Cox regression analysis to develop a risk score
formula. On the basis of this risk score formula, the risk score of
each patient was calculated and patients were then stratified into
low- and high-risk groups (cut-off =0.8863). Ultimately, a
prognostic nomogram of candidate DElncRNAs was developed for
predicting 3- or 5-year OS probabilities. Furthermore, a
calibration plot with a bootstrapping set of 1,000 resamples and
the receiver operating characteristic curve (ROC) were generated to
appraise the predictive capacity of the prognostic nomogram by
calculating the area under the curve (AUC). In the same manner, a
ROC curve in the validation cohort and a ROC curve based on
clinical information were also generated to validate the prognostic
nomogram.
Kyoto Encyclopedia of Genes and
Genomes (KEGG) and Gene Ontology (GO) analyses
To date, the functions of the majority of lncRNAs
remain unknown. Therefore, to investigate the roles of candidate
DElncRNAs, Pearson correlation coefficients between candidate
DElncRNAs and mRNAs in the expression matrix were calculated using
R software. mRNAs with a Pearson correlation coefficient of
>0.45 and P<0.01 were selected and subjected to KEGG and GO
analysis with an adjusted P<0.01 set as the threshold.
Protein-protein interaction (PPI)
network construction and module selection
The Search Tool for the Retrieval of Interacting
Genes/Proteins (STRING) database (version 10.5; http://string-db.org) (16), comprising 9643,763 proteins from
2,031 organisms, was adopted to predict PPIs for ‘Homo
sapiens’, with a confidence score of >0.9 set as the cut-off
criterion. Data from the PPI network were processed by Cytoscape
and the hub genes (top 10 degree genes in the PPI network), which
were considered to be the most important targets of candidate
DElncRNAs, were selected. Subsequently, an online tool, UALCAN
(http://ualcan.path.uab.edu/index.html) (17), was used to assess the expression of
the hub mRNAs in ccRCC.
Survival analysis
A Kaplan-Meier plot and the log-rank test were used
to construct survival curves and assess significant differences in
OS between the low- and high-risk groups and the associations
between the expression levels of candidate DElncRNAs and OS in
patients with ccRCC. P<0.05 was set as a cutoff value.
Results
Patient characteristics
A total of 504 patients with ccRCC (380 cases in the
training cohort and 124 cases in the validation cohort) were
included in the present study. All patients had been pathologically
diagnosed with ccRCC. The detailed demographic and baseline
characteristics of the two cohorts are summarized in Table I. The mean age was 60.46±12.05 and
61.3±12.77 years in the training and validation cohorts,
respectively. There was no significant difference in any of the
clinicopathological parameters, including age, sex, ethnicity,
tumor stage and survival status between the two cohorts.
 | Table I.Baseline characteristics of all
patients with clear cell renal cell carcinoma. |
Table I.
Baseline characteristics of all
patients with clear cell renal cell carcinoma.
| Demographic
characteristics | Training cohort
(n=380) | Validation cohort
(n=124) | Total (n=504) | P-value |
|---|
| Age, years | 60.46±12.05 | 62.12±12.76 | 60.87±12.08 | 0.272 |
| Sex |
|
|
| 0.914 |
| Male | 247 | 133 | 329 |
|
|
Female | 82 | 42 | 175 |
|
| Ethnicity |
|
|
| 0.668 |
|
Caucasian | 338 | 108 | 446 |
|
| African
descent | 37 | 13 | 50 |
|
|
Asian | 5 | 3 | 8 |
|
| TNM stage |
|
|
| 0.264 |
| I | 180 | 68 | 254 |
|
| II | 40 | 14 | 54 |
|
|
III | 94 | 20 | 114 |
|
| IV | 60 | 22 | 82 |
|
| Survival
status |
|
|
| 0.661 |
|
Alive | 251 | 85 | 336 |
|
|
Dead | 129 | 39 | 168 |
|
Identification of DElncRNAs in
ccRCC
With the selection criteria set as |logFC|>2 and
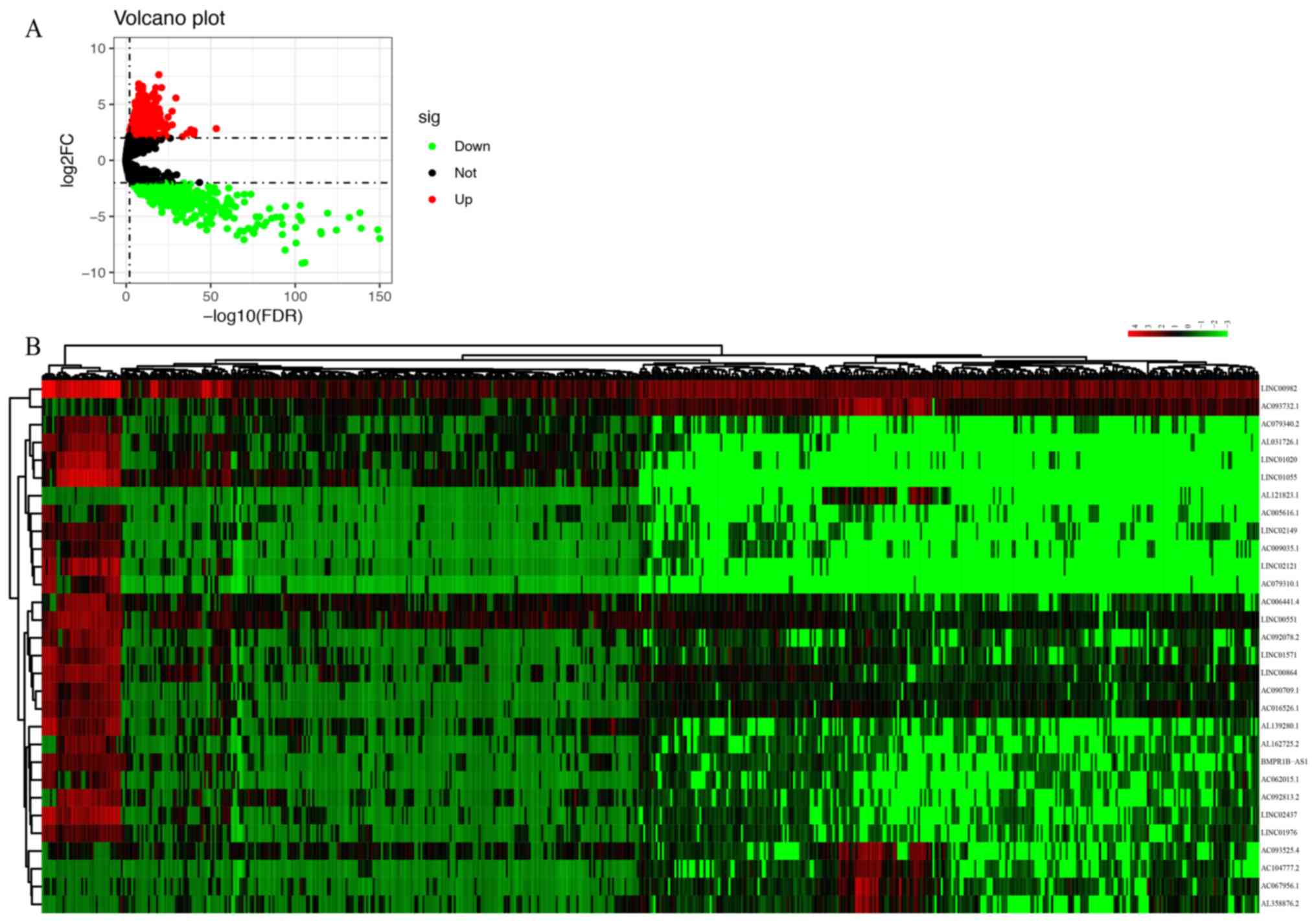
P<0.01, 1,064 upregulated and 489 downregulated DElncRNAs were
identified between ccRCC tissues and normal renal tissues (Fig. 1A). The heatmap of the top 30
DElncRNAs is provided in Fig.
1B.
Construction and evaluation of
DElncRNA-based prognostic nomogram
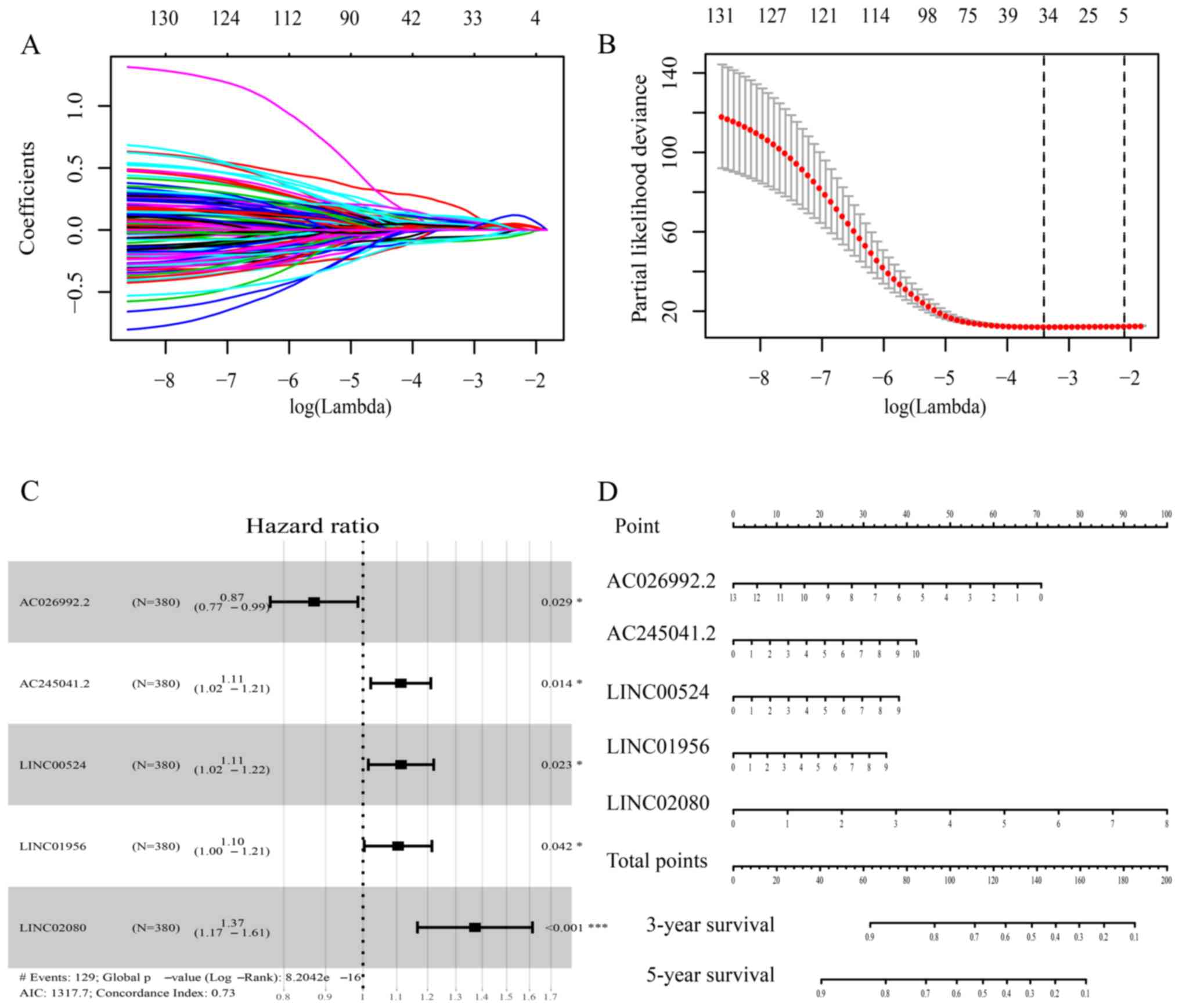
By using univariate regression analysis, 135
DElncRNAs significantly associated with OS of patients were
screened out and then additionally analyzed by LASSO regression
analysis in the training cohort (Fig. 2A
and B). A total of 5 candidate DElncRNAs (AC026992.2,
AC245041.2, LINC00524, LINC01956 and LINC02080) were selected to
develop a prognostic nomogram (Fig.
2D) and a risk score formula using multivariate Cox regression
analysis (Fig. 2C) as follows: Risk
score=(expression level of AC026992.2 ×-0.13804) + (expression
level of AC245041.2 ×0.10658) + (expression level of LINC00524
×0.10723) + (expression level of LINC01956 ×0.09903) + (expression
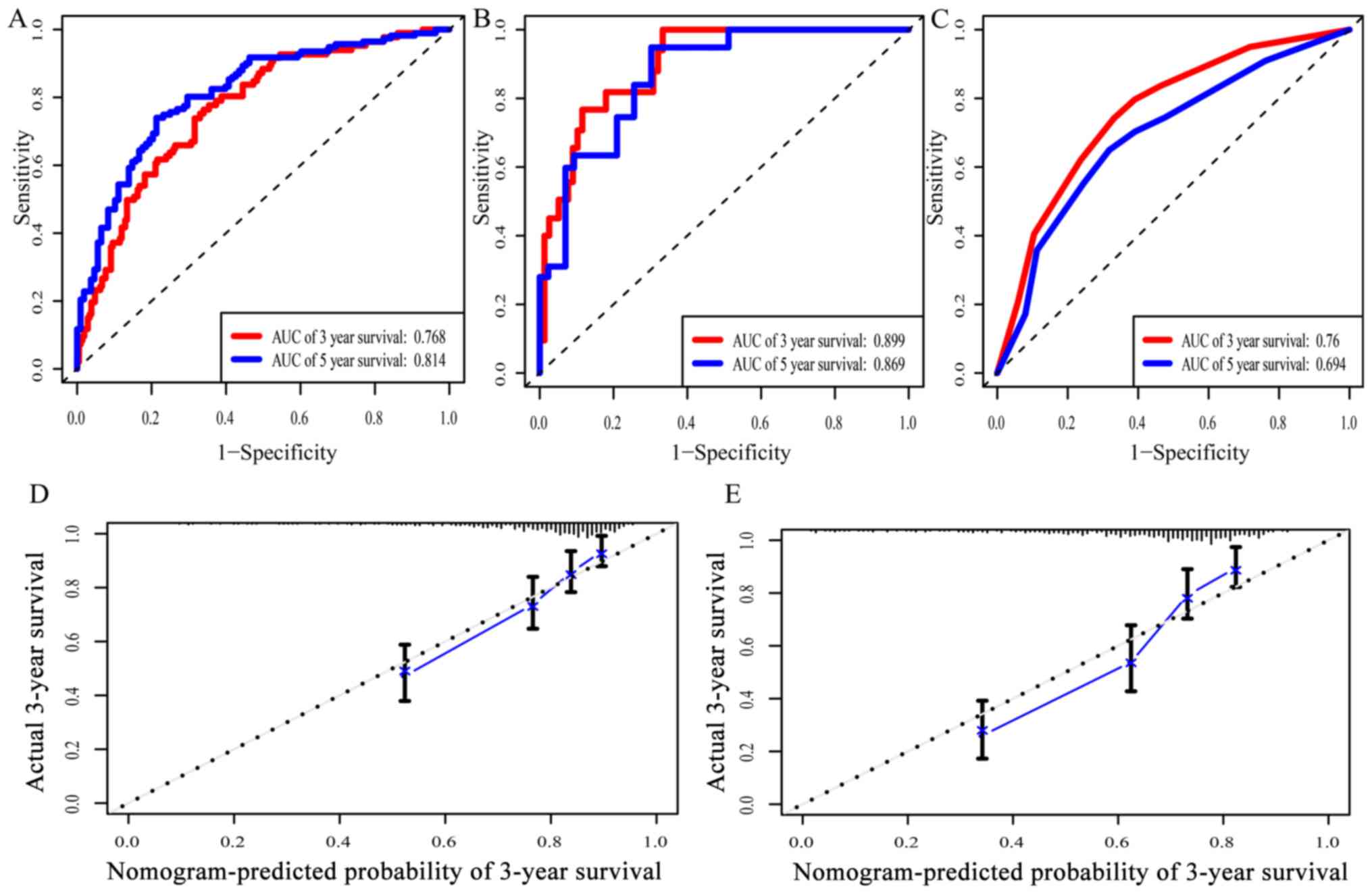
level of LINC02080 ×0.31598). The AUC of the prognostic nomogram
for 3- and 5-year OS in the training cohort was 0.768 and 0.814,
respectively, with a Harrell's concordance index (C-index) of 0.729
(Fig. 3A). The calibration plots for
3- or 5-year survival probabilities in the training cohort are
presented in Fig. 3D and E,
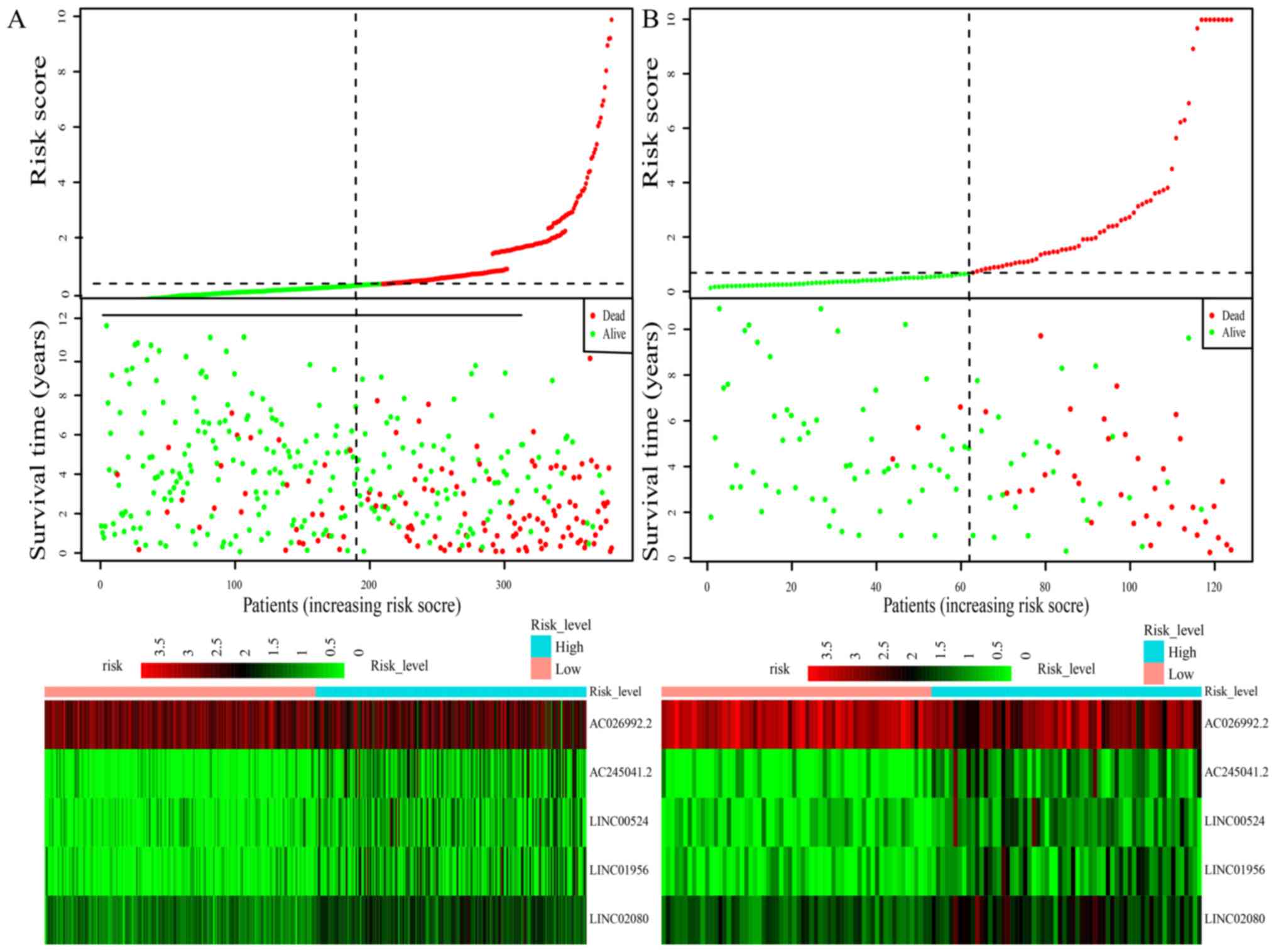
respectively. The distribution of risk score, survival status and
expression profile of the 5 prognostic DElncRNAs for the training
cohort are depicted in Fig. 4A.
Verification of the candidate DElncRNA
nomogram
To confirm the accuracy of the prediction of 3- or
5-year OS by the nomogram established in the training cohort, it
was validated in the 124 patients with ccRCC of the validation
cohort. Similar to the procedure in the training cohort, the 124
patients in the validation cohort were stratified into high- and
low-risk groups according to their risk score. The AUC for
predicting 3- and 5-year OS in the validation cohort was 0.899 and
0.869, respectively, with a C-index of 0.88 (Fig. 3B). The AUC for predicting 3- and
5-year OS based on the TNM staging (AJCC 7th edition, 2010)
(18) was 0.760 and 0.694,
respectively (Fig. 3C). The
distribution of the risk score, survival status and expression
profile of the 5 candidate DElncRNAs in the validation cohort are
demonstrated in Fig. 4B.
Functional enrichment analysis
A total of 521 mRNAs, the expression levels of which
were associated with the 5 candidate DElncRNAs (Pearson correlation
coefficient >0.45 and P<0.01), were identified. The GO
analysis revealed that the 521 mRNAs were enriched in 17 terms,
including receptor ligand activity and channel activity (Fig. 5A). The KEGG analysis indicated that
the 521 mRNAs were primarily involved in 25 pathways, among which a
few pathways were highly associated with oncogenesis, progression
and metastasis of neoplasm, including the Wnt, p53 and mTOR
signaling pathways (Fig. 5B).
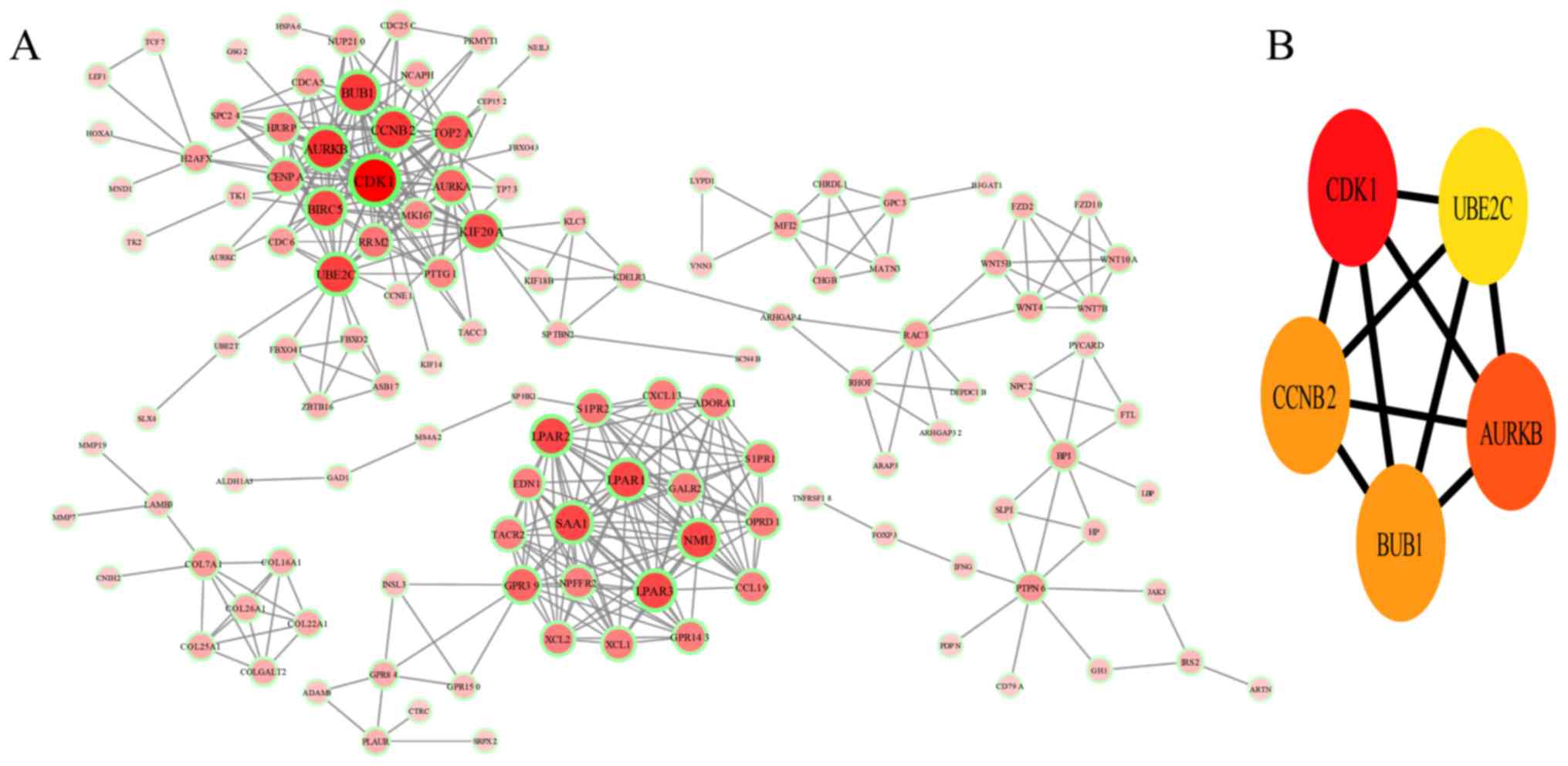
PPI network construction and module
selection
A PPI network was constructed using the online
database STRING and then processed with Cytoscape software
(Fig. 6A). 5 hub mRNAs were selected
as the most crucial targets of the candidate DElncRNAs, comprising
cyclin D kinase 1 (CDK1; degree of connectivity =27), aurora kinase
B (AURKB; degree of connectivity =21), cyclin B1 (CCNB2; degree of
connectivity =20), BUB1 mitotic checkpoint serine/threonine kinase
(BUB1; degree of connectivity =20) and ubiquitin conjugating enzyme
E2 C (UBE2C; degree of connectivity =20) (Fig. 6B). The result obtained with the
UALCAN tool demonstrated that the 5 mRNAs were expressed at
increased levels in ccRCC compared with that in normal tissues
(Fig. 6C).
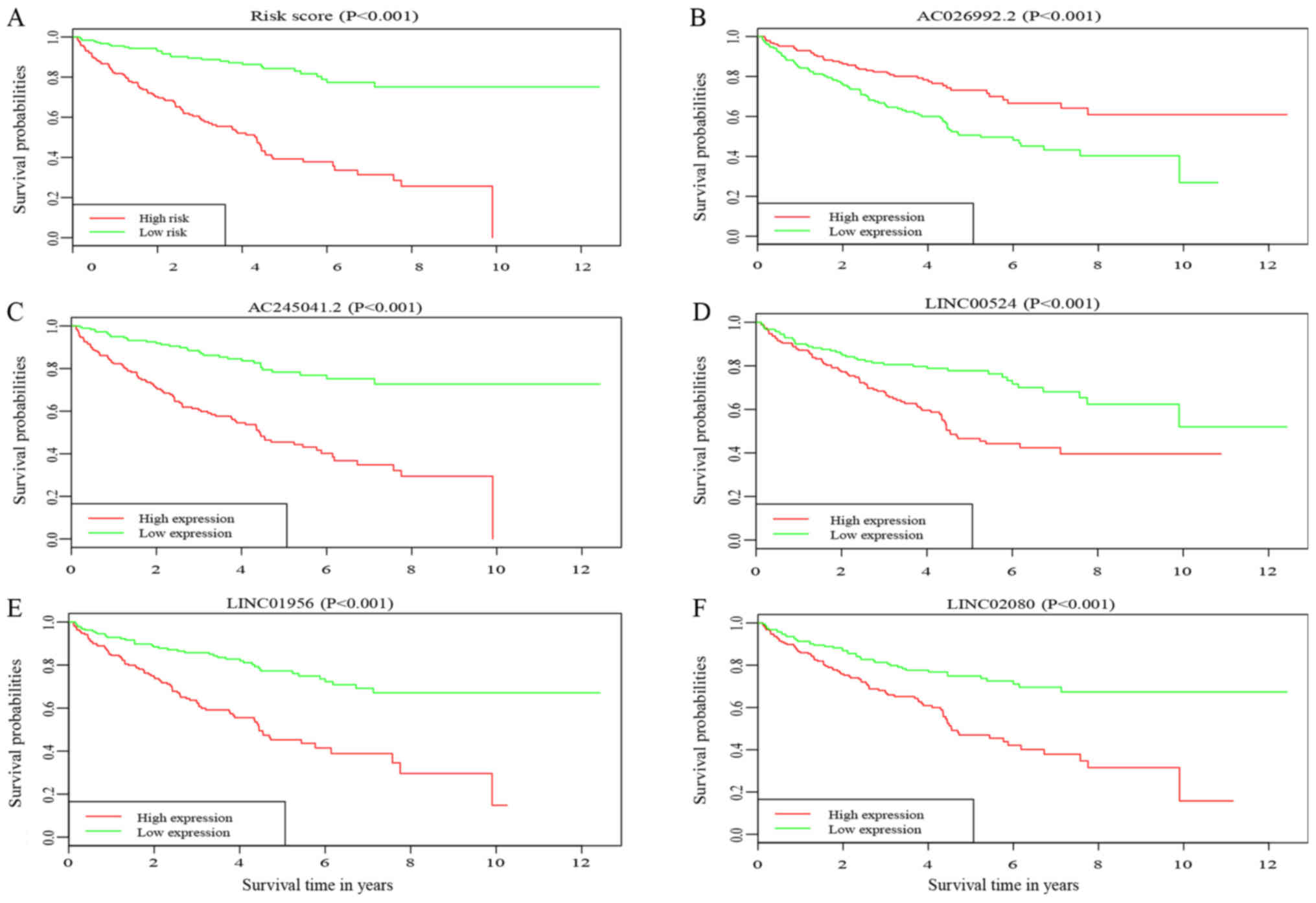
Survival analysis
Kaplan-Meier analysis was performed to determine the
association between OS of patients with ccRCC and risk score, and
the expression levels of 5 candidate DElncRNAs. The result of the
survival analysis demonstrated that the prognosis of the patients
with ccRCC in the high-risk group was poorer compared with that in
the low-risk group (Fig. 7A).
Furthermore, high expression of AC245041.2 (P<0.001), LINC00524
(P<0.001), LINC01956 (P<0.001) and LINC02080 (P<0.001),
along with low expression level of AC026992.2 (P<0.001), was
associated with a poor outcome for patients with ccRCC (Fig. 7B-F).
Discussion
ccRCC ranks first in incidence amongst all
histological types of kidney malignancies (19), and accounts for nearly 3% of all
types of adult malignancies (20).
Although the 5-year survival rate of patients with ccRCC at the
early stage is >90%, it decreases to 10% in patients with
advanced ccRCC (5), and >100,000
patients succumb to ccRCC per year worldwide (21). Therefore, it is imperative to
identify tumor-specific markers for risk stratification that may be
utilized for assessing the prognosis of patients, and which may
facilitate the development of novel strategies for the diagnosis
and therapy of ccRCC. lncRNAs, a class of non-coding RNAs of
>200 nt in length, have been demonstrated to have a significant
role in transcriptional and post-transcriptional regulation, and
deregulation of certain lncRNAs is involved in the initiation and
progression of various cancer types (22). The roles of lncRNAs have become
active areas of research, which will undoubtedly be propitious for
the elucidation of the functions of lncRNAs in cancer. However, to
date, the functions of the majority of lncRNAs remain elusive
(23).
In the present study, a comprehensive analysis of
lncRNA expression matrix files and corresponding clinical
information of patients with ccRCC was performed. The patients were
randomly assigned to training or validation cohorts. In the
training cohort, a total of 1,553 DElncRNAs (1,064 upregulated and
489 downregulated DElncRNAs) were identified. Following screening
by univariate regression analysis and LASSO regression analysis,
the top 5 candidate DElncRNAs (AC026992.2, AC245041.2, LINC00524,
LINC01956 and LINC02080) associated with survival were selected and
used to develop a prognostic nomogram. In the training cohort, the
AUC of the prognostic nomogram for 3- and 5-year OS was 0.768 and
0.814, respectively, and in the validation cohort, it was 0.899 and
0.869, respectively, demonstrating an excellent predictive accuracy
for the probability of survival at 3 or 5 years. The AUC for
predicting 3-year OS based on the clinical index (0.760) was
similar to that in the training cohort. However, the AUC for
predicting 5-year OS based on the clinical index (0.694) was
markedly decreased compared with that in training cohort,
indicating that the accuracy of the prognostic nomogram was
improved compared with that of the clinical index for predicting
5-year OS. Previously, Shi et al (24) used 5 lncRNAs (ENSG00000229178,
ENSG00000236453, ENSG00000245060, ENSG00000258789 and
ENSG00000272558) for predicting 3-year OS of patients with ccRCC.
In addition, Qu et al (25)
also built a prognostic lncRNA signature for predicting 5-year OS
in localized ccRCC with 4 lncRNAs (ENSG00000255774,
ENSG00000248323, ENSG00000260911 and ENSG00000231666). In the
present study, 5 lncRNAs, which were completely different from
lncRNAs used in the studies of Shi et al (24) and Qu et al (25), were used to develop a prognostic
nomogram. The AUC for 3-year OS in the training cohort and
validation cohort, 0.768 and 0.899, respectively, was superior to
that of Shi et al (24),
0.703 and 0.630, respectively. Furthermore, in the study by Qu
et al (25), the AUC for
5-year OS in the training and validation cohort was 0.690 and
0.663, respectively, which was lower compared with those of the
present study. In addition, the calibration plot for 3- or 5-year
OS demonstrated an outstanding consistency between the prediction
by the prognostic nomogram and the actual outcome. All of the
results suggested that the prognostic nomogram established is
suitable for estimating the probability of OS of patients with
ccRCC at 3 and 5 years.
Compared with normal renal tissues, the 5 candidate
DElncRNAs were aberrantly expressed in ccRCC tissues. Survival
analysis for low- and high-risk groups indicated that the high-risk
group exhibited a poorer prognosis compared with the low-risk
group. Furthermore, high expression levels of AC245041.2,
LINC00524, LINC01956 and LINC02080 (P<0.001 for each) along with
low expression levels of AC026992.2 (P<0.001) were highly
negatively associated with OS of patients with ccRCC.
However, at present, little is known regarding the
biological functions of the 5 candidate DElncRNAs. Therefore, to
additionally explore the biological roles of the 5 candidate
DElncRNAs, 521 mRNAs, the expression levels of which were highly
associated with the expression of the 5 candidate DElncRNAs, were
selected to perform functional enrichment analysis and a PPI
network was constructed. According to the GO analysis, the mRNAs
were primarily involved in 17 terms, including ‘receptor ligand
activity’ and ‘channel activity’, and the KEGG analysis revealed
enrichment in 25 pathways. Several of these pathways are known to
be associated with oncogenesis, progression and metastasis of
cancer. For example, the Wnt signaling pathway is generally
involved in cell proliferation and division via controlling the
β-catenin degradation complex (26).
Through activation of the Wnt signaling pathway, the lncRNA colon
cancer-associated transcript 2 improved the proliferation and
invasion of ccRCC cells (27). In
addition, the mTOR signaling pathway serves vital roles in
modulating diverse biological behaviors, including cell growth,
metabolism, protein synthesis and autophagy (28). Liu et al (29) identified that inactivation of the
mTOR signaling pathway inhibited apoptosis and promoted cell
proliferation in ccRCC. The results of the GO and KEGG analyses
conducted in the present study predicted that the 5 candidate
DElncRNAs had an important effect on the oncogenesis and
progression of ccRCC by affecting a series of biological pathways
and processes. The PPI network was constructed to determine the
interaction among 521 mRNAs, and 5 hub mRNAs (CDK1, AURKB, CCNB,
BUB1 and UBE2C) were identified as the most important targets. All
of the 5 top degree mRNAs were overexpressed in ccRCC. CDK1 is
essential for the eukaryotic cell cycle by regulating the onset of
mitosis and the centrosome cycle (30). A previous study indicated that CDK1
expression was highly associated with the prognosis of patients
with RCC (31). Furthermore, Li
et al (32) indicated that
through targeting CDK1, miR-31-5p inhibited the proliferation,
migration and invasion of ccRCC cells. At present, little is known
about the functions of AURKB, CCNB, BUB1 and UBE2C in ccRCC.
Therefore, future studies investigating the functions of AURKB,
CCNB, BUB1 and UBE2C in ccRCC re required.
Although the prognostic nomogram established in the
present study demonstrated good predictive accuracy for patients
with ccRCC, there are a few limitations that should be addressed.
Firstly, as all cases were retrieved from TCGA database, the risk
of selection bias could not be excluded. Furthermore, to date, no
experimental studies have been performed to examine the functions
of 4 of the identified lncRNAs in cancer. Therefore, further in
vitro and in vivo studies are required to confirm the
results of the present study.
In conclusion, 5 candidate DElncRNAs (AC026992.2,
AC245041.2, LINC00524, LINC01956 and LINC02080) were identified in
the present study, which were independent prognostic factors for
patients with ccRCC, and exhibited potential utility as powerful
molecular biomarkers for prognosis and risk assessment. A novel and
convenient prognostic nomogram was then developed for predicting 3-
and 5-year OS for patients with ccRCC based on lncRNAs. The results
of the present study may contribute to an improved understanding of
ccRCC at the molecular level. However, additional experimental
research concerning lncRNAs in vitro and in vivo is
required to verify the results of the present study.
Acknowledgements
Not applicable.
Funding
The present study was supported by Ke-qun Chai
Inheritance Studio of National Prominent Chinese Medicine Doctor,
State Administration of Traditional Chinese Medicine (grant no.
2A21533).
Availability of data and materials
The datasets analyzed during the present study are
available in the TCGA repository, https://portal.gdc.cancer.gov/.
Authors' contributions
SW conceived and designed the study, performed the
experiment, and wrote the manuscript. KC performed statistical
analysis, reviewed and edited the manuscript. JC conceived and
performed the experiments and reviewed the manuscript. All authors
approved the manuscript and agreed to be accountable for all
aspects of the research and for ensuring that the accuracy or
integrity of any part of the work were appropriately investigated
and resolved.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
All authors confirm that they have no competing
interests.
References
|
1
|
Yang K, Lu XF, Luo PC and Zhang J:
Identification of six potentially long noncoding RNAs as biomarkers
involved competitive endogenous RNA in clear cell renal cell
carcinoma. Biomed Res Int. 2018:93034862018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sanchez DJ and Simon MC: Genetic and
metabolic hallmarks of clear cell renal cell carcinoma. Biochim
Biophys Acta Rev Cancer. 1870:23–31. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Xing T and He H: Epigenomics of clear cell
renal cell carcinoma: Mechanisms and potential use in molecular
pathology. Chin J Cancer Res. 28:80–91. 2016.PubMed/NCBI
|
|
4
|
Godlewski J, Kiezun J, Krazinski BE,
Kozielec Z, Wierzbicki PM and Kmiec Z: The immunoexpression of YAP1
and LATS1 proteins in clear cell renal cell carcinoma: Impact on
Patients' survival. Biomed Res Int. 2018:26536232018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Atkins MB and Tannir NM: Current and
emerging therapies for first-line treatment of metastatic clear
cell renal cell carcinoma. Cancer Treat Rev. 70:127–137. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Cao Q, Ruan H, Wang K, Song Z, Bao L, Xu
T, Xiao H, Wang C, Cheng G, Tong J, et al: Overexpression of PLIN2
is a prognostic marker and attenuates tumor progression in clear
cell renal cell carcinoma. Int J Oncol. 53:137–147. 2018.PubMed/NCBI
|
|
7
|
Cui N, Liu J, Xia H and Xu D: LncRNA
SNHG20 contributes to cell proliferation and invasion by
upregulating ZFX expression sponging miR-495-3p in gastric cancer.
J Cell Biochem. 120:3114–3123. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Dong D, Mu Z, Wei N, Sun M, Wang W, Xin N,
Shao Y and Zhao C: Long non-coding RNA ZFAS1 promotes proliferation
and metastasis of clear cell renal cell carcinoma via targeting
miR-10a/SKA1 pathway. Biomed Pharmacother. 111:917–925. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ning L, Li Z, Wei D, Chen H and Yang C:
LncRNA, NEAT1 is a prognosis biomarker and regulates cancer
progression via epithelial-mesenchymal transition in clear cell
renal cell carcinoma. Cancer Biomark. 19:75–83. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yang T, Zhou H, Liu P, Yan L, Yao W, Chen
K, Zeng J, Li H, Hu J, Xu H and Ye Z: lncRNA PVT1 and its splicing
variant function as competing endogenous RNA to regulate clear cell
renal cell carcinoma progression. Oncotarget. 8:85353–85367.
2017.PubMed/NCBI
|
|
11
|
Casuscelli J, Weinhold N, Gundem G, Wang
L, Zabor EC, Drill E, Wang PI, Nanjangud GJ, Redzematovic A,
Nargund AM, et al: Genomic landscape and evolution of metastatic
chromophobe renal cell carcinoma. JCI Insight. 2(pii): 926882017.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Feng Y, Shen Y, Chen H, Wang X, Zhang R,
Peng Y, Lei X, Liu T, Liu J, Gu L, et al: Expression profile
analysis of long non-coding RNA in acute myeloid leukemia by
microarray and bioinformatics. Cancer Sci. 109:340–353. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang C, Yang C, Wang W, Xia B, Li K, Sun F
and Hou Y: Prognostic nomogram for cervical cancer after surgery
from SEER database. J Cancer. 9:3923–3928. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Robinson MD, McCarthy DJ and Smyth GK:
edgeR: A Bioconductor package for differential expression analysis
of digital gene expression data. Bioinformatics. 26:139–140. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhu Q, Sun Y, Zhou Q, He Q and Qian H:
Identification of key genes and pathways by bioinformatics analysis
with TCGA RNA sequencing data in hepatocellular carcinoma. Mol Clin
Oncol. 9:597–606. 2018.PubMed/NCBI
|
|
16
|
Szklarczyk D, Gable AL, Lyon D, Junge A,
Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork
P, et al: STRING v11: Protein-protein association networks with
increased coverage, supporting functional discovery in genome-wide
experimental dataset. Nucleic Acids Res. 47:D607–D613. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chandrashekar DS, Bashel B, Balasubramanya
SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK and
Varambally S: UALCAN: A portal for facilitating tumor subgroup gene
expression and survival analyses. Neoplasia. 19:649–658. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Martínez-Salamanca JI, Huang WC, Millán I,
Bertini R, Bianco FJ, Carballido JA, Ciancio G, Hernández C,
Herranz F, Haferkamp A, et al: Prognostic impact of the 2009
UICC/AJCC TNM staging system for renal cell carcinoma with venous
extension. Eur Urol. 59:120–127. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Vecchio SJD and Ellis RJ: Cabozantinib for
the management of metastatic clear cell renal cell carcinoma. J
Kidney Cancer VHL. 5:1–5. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Majer W, Kluzek K, Bluyssen H and Wesoły
J: Potential approaches and recent advances in biomarker discovery
in clear-cell renal cell carcinoma. J Cancer. 6:1105–1113. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang L, Yin W, Yao L, Li X, Fang D, Ren
D, Zhang Z, Fan Y, He Q, Ci W, et al: Growth pattern of clear cell
renal cell carcinoma in patients with delayed surgical
intervention: Fast growth rate correlates with high grade and may
result in poor prognosis. Biomed Res Int.
2015:5981342015.PubMed/NCBI
|
|
22
|
Qiao F, Li N and Li W: Integrative
bioinformatics analysis reveals potential long non-coding RNA
biomarkers and analysis of function in Non-smoking females with
lung cancer. Med Sci Monit. 24:5771–5778. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yan J, Zhou C, Guo K, Li Q and Wang Z: A
novel seven-lncRNA signature for prognosis prediction in
hepatocellular carcinoma. J Cell Biochem. 120:213–223. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Shi D, Qu Q, Chang Q, Wang Y, Gui Y and
Dong D: A five-long non-coding RNA signature to improve prognosis
prediction of clear cell renal cell carcinoma. Oncotarget.
8:58699–58708. 2017.PubMed/NCBI
|
|
25
|
Qu L, Wang ZL, Chen Q, Li YM, He HW, Hsieh
JJ, Xue S, Wu ZJ, Liu B, Tang H, et al: Prognostic value of a long
Non-coding RNA signature in localized clear cell renal cell
carcinoma. Eur Urol. 74:756–763. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Taciak B, Pruszynska I, Kiraga L, Bialasek
M and Krol M: Wnt signaling pathway in development and cancer. J
Physiol Pharmacol. 69:185–196. 2018.
|
|
27
|
Huang JL, Liao Y, Qiu MX, Li J and An Y:
Long non-coding RNA CCAT2 promotes cell proliferation and invasion
through regulating Wnt/β-catenin signaling pathway in clear cell
renal cell carcinoma. Tumour Biol. 39:10104283177113142017.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Tian T, Li X and Zhang J: mTOR signaling
in cancer and mTOR inhibitors in solid tumor targeting therapy. Int
J Mol Sci. 20(pii): E7552019. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Liu G, Zhao X, Zhou J, Cheng X, Ye Z and
Ji Z: LncRNA TP73-AS1 promotes cell proliferation and inhibits cell
apoptosis in clear cell renal cell carcinoma through repressing
KISS1 expression and inactivation of PI3K/Akt/mTOR signaling
pathway. Cell Physiol Biochem. 48:371–384. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Li Y and Burridge K: Cell-cycle-dependent
regulation of cell adhesions: Adhering to the schedule: Three
papers reveal unexpected properties of adhesion structures as cells
progress through the cell cycle. Bioessays. 41:e18001652019.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Hongo F, Takaha N, Oishi M, Ueda T,
Nakamura T, Naitoh Y, Naya Y, Kamoi K, Okihara K, Matsushima T, et
al: CDK1 and CDK2 activity is a strong predictor of renal cell
carcinoma recurrence. Urol Oncol. 32:1240–1246. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Li Y, Quan J, Chen F, Pan X, Zhuang C,
Xiong T, Zhuang C, Li J, Huang X, Ye J, et al: MiR-31-5p acts as a
tumor suppressor in renal cell carcinoma by targeting
cyclin-dependent kinase 1 (CDK1). Biomed Pharmacother. 111:517–526.
2019. View Article : Google Scholar : PubMed/NCBI
|