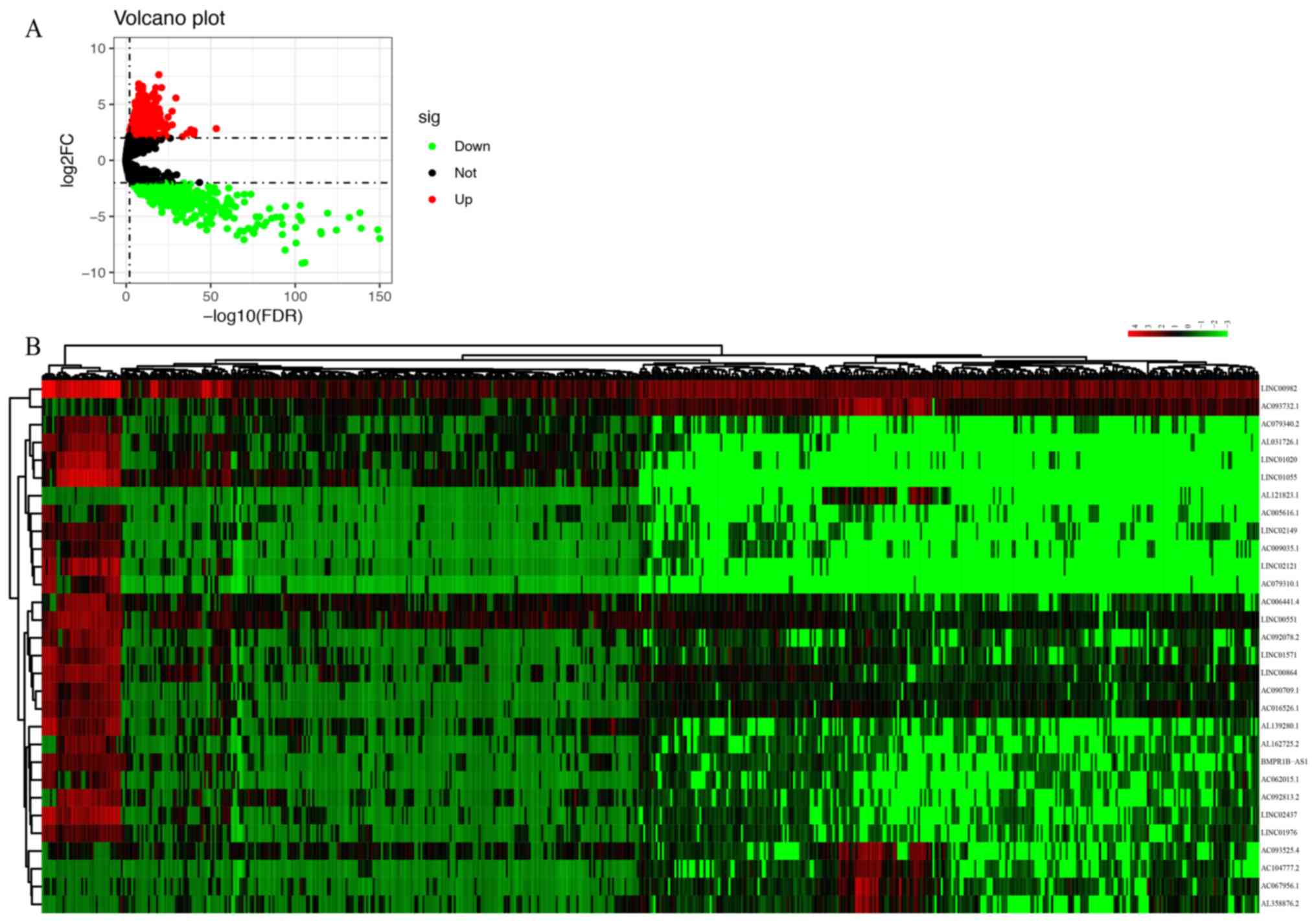
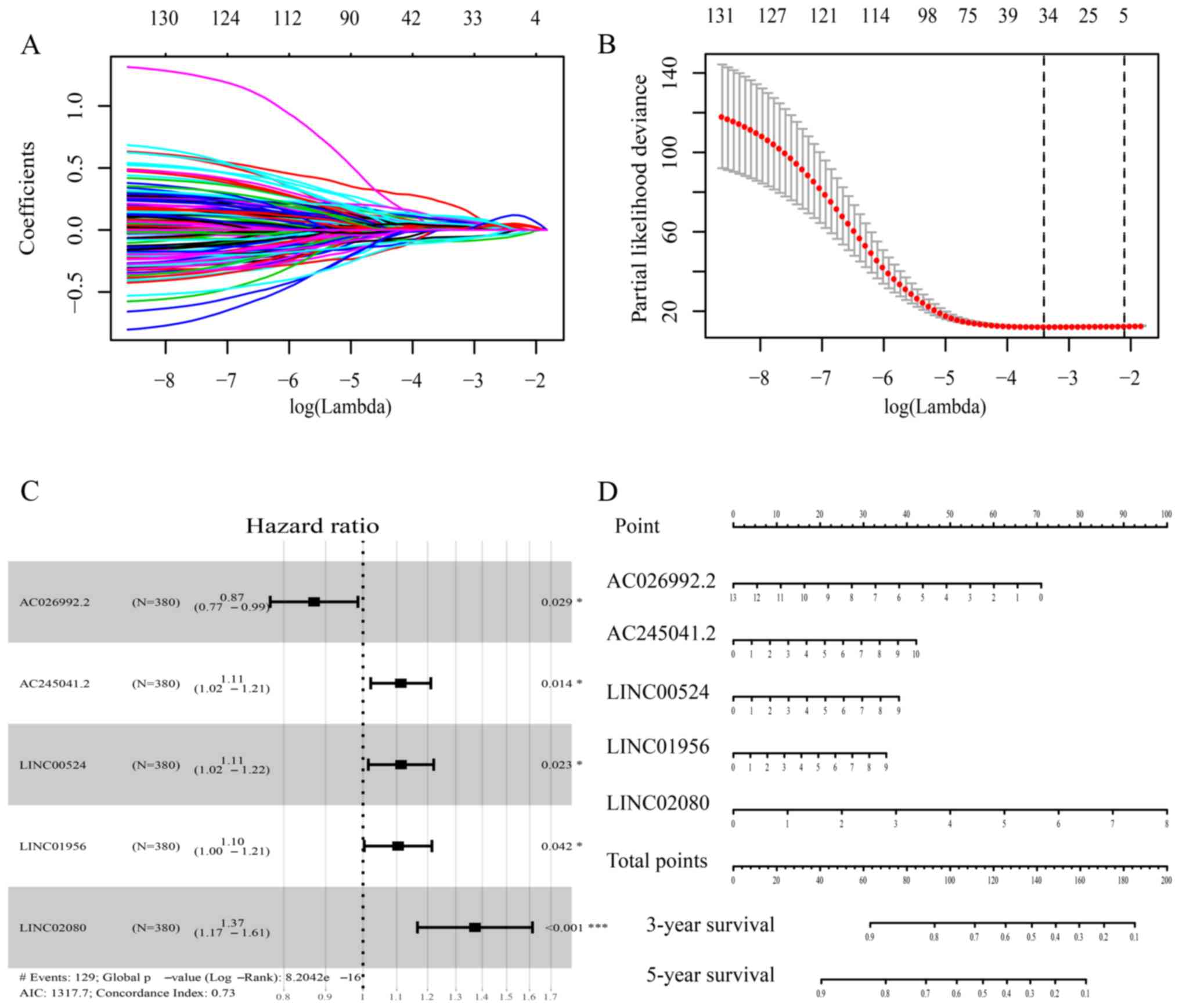
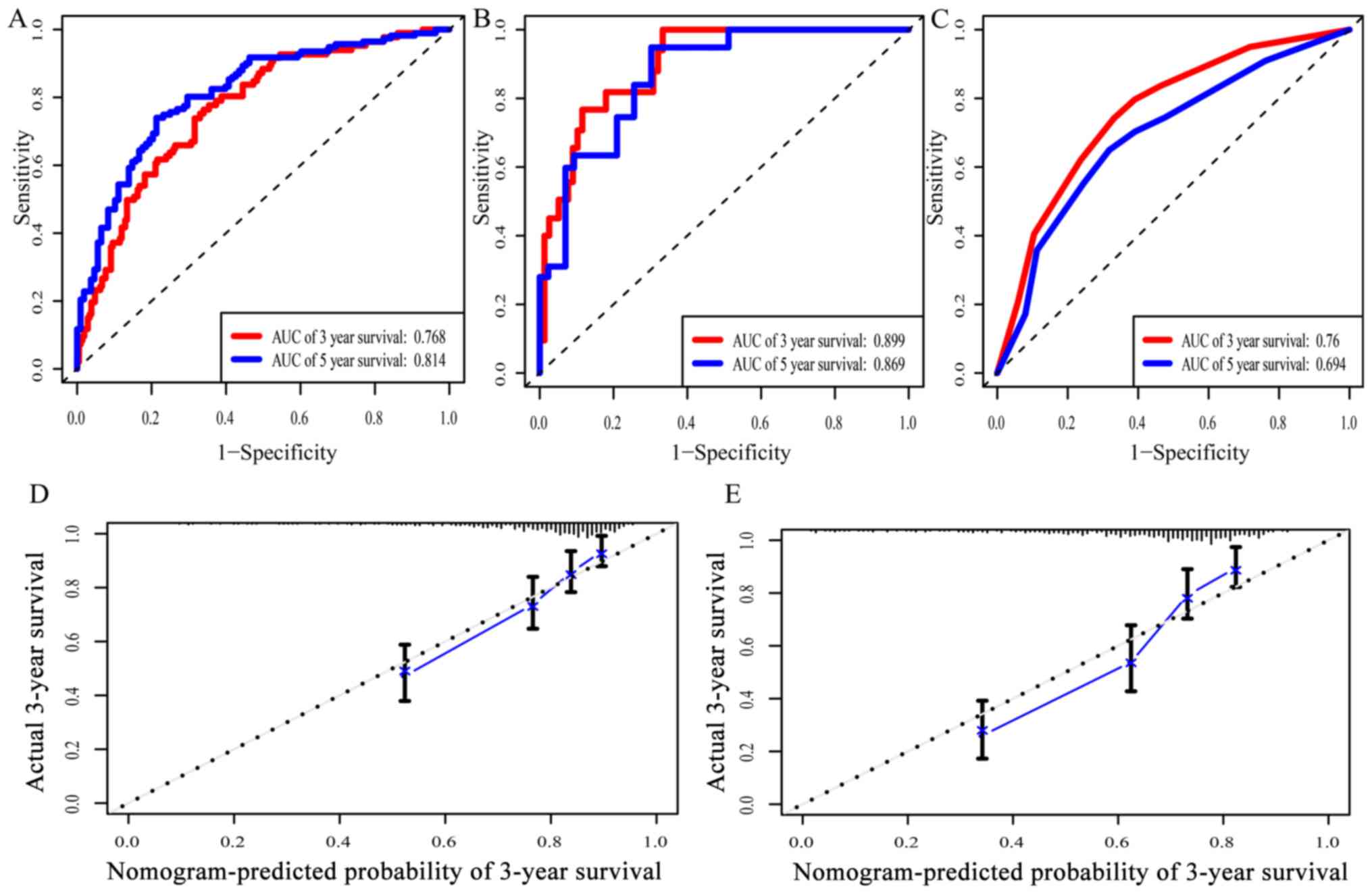
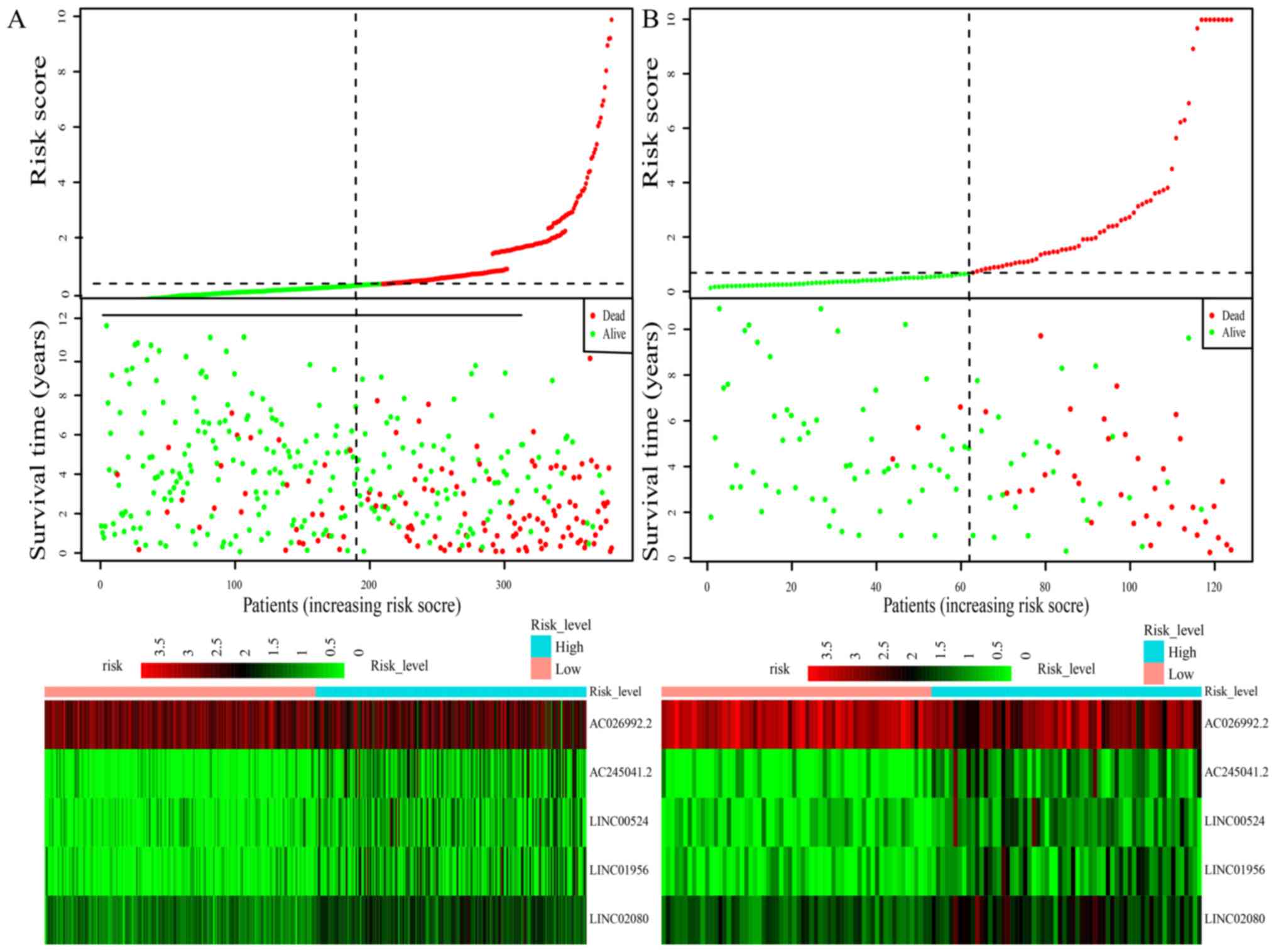
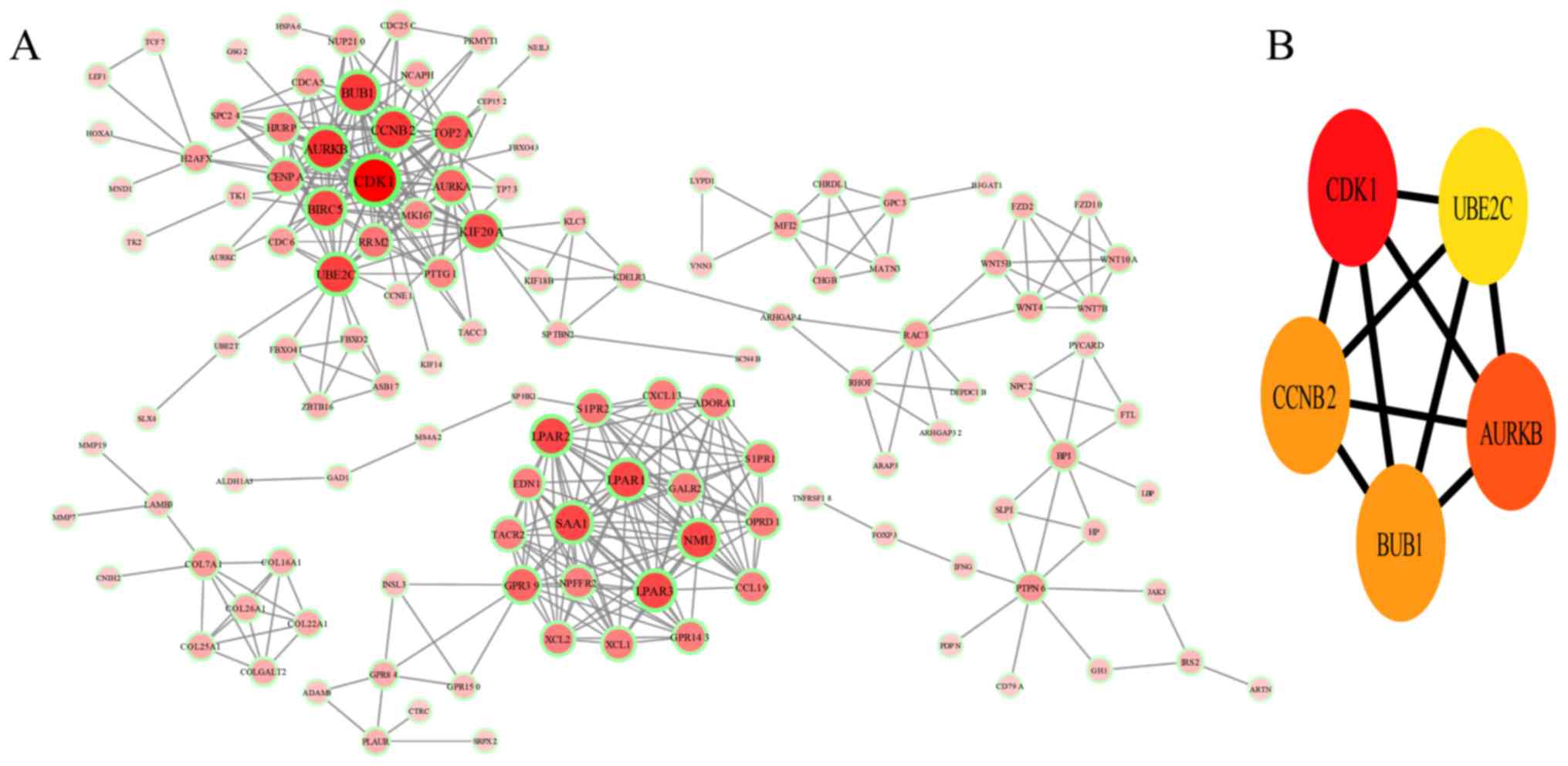
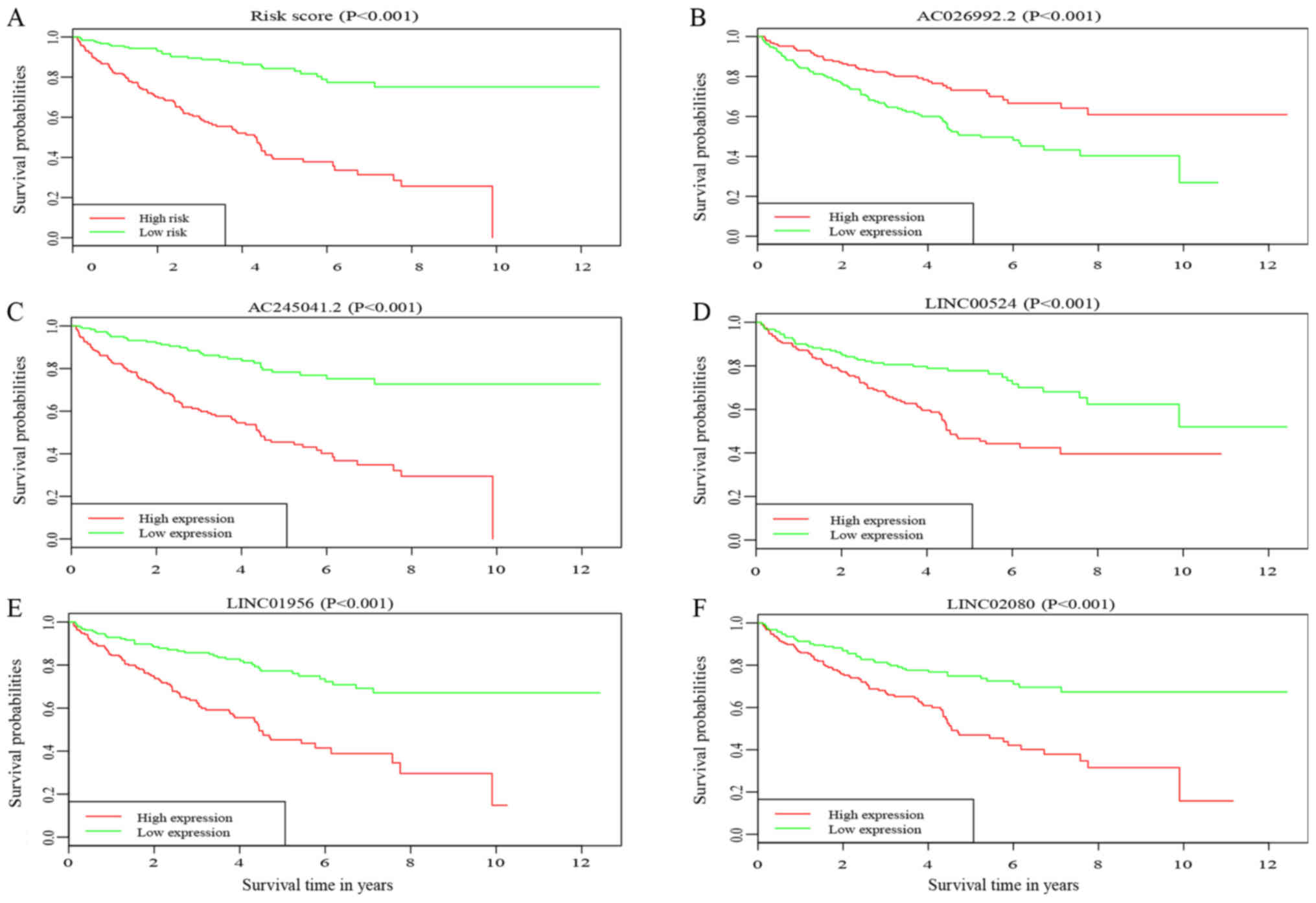
|
1
|
Yang K, Lu XF, Luo PC and Zhang J:
Identification of six potentially long noncoding RNAs as biomarkers
involved competitive endogenous RNA in clear cell renal cell
carcinoma. Biomed Res Int. 2018:93034862018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sanchez DJ and Simon MC: Genetic and
metabolic hallmarks of clear cell renal cell carcinoma. Biochim
Biophys Acta Rev Cancer. 1870:23–31. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Xing T and He H: Epigenomics of clear cell
renal cell carcinoma: Mechanisms and potential use in molecular
pathology. Chin J Cancer Res. 28:80–91. 2016.PubMed/NCBI
|
|
4
|
Godlewski J, Kiezun J, Krazinski BE,
Kozielec Z, Wierzbicki PM and Kmiec Z: The immunoexpression of YAP1
and LATS1 proteins in clear cell renal cell carcinoma: Impact on
Patients' survival. Biomed Res Int. 2018:26536232018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Atkins MB and Tannir NM: Current and
emerging therapies for first-line treatment of metastatic clear
cell renal cell carcinoma. Cancer Treat Rev. 70:127–137. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Cao Q, Ruan H, Wang K, Song Z, Bao L, Xu
T, Xiao H, Wang C, Cheng G, Tong J, et al: Overexpression of PLIN2
is a prognostic marker and attenuates tumor progression in clear
cell renal cell carcinoma. Int J Oncol. 53:137–147. 2018.PubMed/NCBI
|
|
7
|
Cui N, Liu J, Xia H and Xu D: LncRNA
SNHG20 contributes to cell proliferation and invasion by
upregulating ZFX expression sponging miR-495-3p in gastric cancer.
J Cell Biochem. 120:3114–3123. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Dong D, Mu Z, Wei N, Sun M, Wang W, Xin N,
Shao Y and Zhao C: Long non-coding RNA ZFAS1 promotes proliferation
and metastasis of clear cell renal cell carcinoma via targeting
miR-10a/SKA1 pathway. Biomed Pharmacother. 111:917–925. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ning L, Li Z, Wei D, Chen H and Yang C:
LncRNA, NEAT1 is a prognosis biomarker and regulates cancer
progression via epithelial-mesenchymal transition in clear cell
renal cell carcinoma. Cancer Biomark. 19:75–83. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yang T, Zhou H, Liu P, Yan L, Yao W, Chen
K, Zeng J, Li H, Hu J, Xu H and Ye Z: lncRNA PVT1 and its splicing
variant function as competing endogenous RNA to regulate clear cell
renal cell carcinoma progression. Oncotarget. 8:85353–85367.
2017.PubMed/NCBI
|
|
11
|
Casuscelli J, Weinhold N, Gundem G, Wang
L, Zabor EC, Drill E, Wang PI, Nanjangud GJ, Redzematovic A,
Nargund AM, et al: Genomic landscape and evolution of metastatic
chromophobe renal cell carcinoma. JCI Insight. 2(pii): 926882017.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Feng Y, Shen Y, Chen H, Wang X, Zhang R,
Peng Y, Lei X, Liu T, Liu J, Gu L, et al: Expression profile
analysis of long non-coding RNA in acute myeloid leukemia by
microarray and bioinformatics. Cancer Sci. 109:340–353. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang C, Yang C, Wang W, Xia B, Li K, Sun F
and Hou Y: Prognostic nomogram for cervical cancer after surgery
from SEER database. J Cancer. 9:3923–3928. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Robinson MD, McCarthy DJ and Smyth GK:
edgeR: A Bioconductor package for differential expression analysis
of digital gene expression data. Bioinformatics. 26:139–140. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhu Q, Sun Y, Zhou Q, He Q and Qian H:
Identification of key genes and pathways by bioinformatics analysis
with TCGA RNA sequencing data in hepatocellular carcinoma. Mol Clin
Oncol. 9:597–606. 2018.PubMed/NCBI
|
|
16
|
Szklarczyk D, Gable AL, Lyon D, Junge A,
Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork
P, et al: STRING v11: Protein-protein association networks with
increased coverage, supporting functional discovery in genome-wide
experimental dataset. Nucleic Acids Res. 47:D607–D613. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chandrashekar DS, Bashel B, Balasubramanya
SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK and
Varambally S: UALCAN: A portal for facilitating tumor subgroup gene
expression and survival analyses. Neoplasia. 19:649–658. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Martínez-Salamanca JI, Huang WC, Millán I,
Bertini R, Bianco FJ, Carballido JA, Ciancio G, Hernández C,
Herranz F, Haferkamp A, et al: Prognostic impact of the 2009
UICC/AJCC TNM staging system for renal cell carcinoma with venous
extension. Eur Urol. 59:120–127. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Vecchio SJD and Ellis RJ: Cabozantinib for
the management of metastatic clear cell renal cell carcinoma. J
Kidney Cancer VHL. 5:1–5. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Majer W, Kluzek K, Bluyssen H and Wesoły
J: Potential approaches and recent advances in biomarker discovery
in clear-cell renal cell carcinoma. J Cancer. 6:1105–1113. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang L, Yin W, Yao L, Li X, Fang D, Ren
D, Zhang Z, Fan Y, He Q, Ci W, et al: Growth pattern of clear cell
renal cell carcinoma in patients with delayed surgical
intervention: Fast growth rate correlates with high grade and may
result in poor prognosis. Biomed Res Int.
2015:5981342015.PubMed/NCBI
|
|
22
|
Qiao F, Li N and Li W: Integrative
bioinformatics analysis reveals potential long non-coding RNA
biomarkers and analysis of function in Non-smoking females with
lung cancer. Med Sci Monit. 24:5771–5778. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yan J, Zhou C, Guo K, Li Q and Wang Z: A
novel seven-lncRNA signature for prognosis prediction in
hepatocellular carcinoma. J Cell Biochem. 120:213–223. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Shi D, Qu Q, Chang Q, Wang Y, Gui Y and
Dong D: A five-long non-coding RNA signature to improve prognosis
prediction of clear cell renal cell carcinoma. Oncotarget.
8:58699–58708. 2017.PubMed/NCBI
|
|
25
|
Qu L, Wang ZL, Chen Q, Li YM, He HW, Hsieh
JJ, Xue S, Wu ZJ, Liu B, Tang H, et al: Prognostic value of a long
Non-coding RNA signature in localized clear cell renal cell
carcinoma. Eur Urol. 74:756–763. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Taciak B, Pruszynska I, Kiraga L, Bialasek
M and Krol M: Wnt signaling pathway in development and cancer. J
Physiol Pharmacol. 69:185–196. 2018.
|
|
27
|
Huang JL, Liao Y, Qiu MX, Li J and An Y:
Long non-coding RNA CCAT2 promotes cell proliferation and invasion
through regulating Wnt/β-catenin signaling pathway in clear cell
renal cell carcinoma. Tumour Biol. 39:10104283177113142017.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Tian T, Li X and Zhang J: mTOR signaling
in cancer and mTOR inhibitors in solid tumor targeting therapy. Int
J Mol Sci. 20(pii): E7552019. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Liu G, Zhao X, Zhou J, Cheng X, Ye Z and
Ji Z: LncRNA TP73-AS1 promotes cell proliferation and inhibits cell
apoptosis in clear cell renal cell carcinoma through repressing
KISS1 expression and inactivation of PI3K/Akt/mTOR signaling
pathway. Cell Physiol Biochem. 48:371–384. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Li Y and Burridge K: Cell-cycle-dependent
regulation of cell adhesions: Adhering to the schedule: Three
papers reveal unexpected properties of adhesion structures as cells
progress through the cell cycle. Bioessays. 41:e18001652019.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Hongo F, Takaha N, Oishi M, Ueda T,
Nakamura T, Naitoh Y, Naya Y, Kamoi K, Okihara K, Matsushima T, et
al: CDK1 and CDK2 activity is a strong predictor of renal cell
carcinoma recurrence. Urol Oncol. 32:1240–1246. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Li Y, Quan J, Chen F, Pan X, Zhuang C,
Xiong T, Zhuang C, Li J, Huang X, Ye J, et al: MiR-31-5p acts as a
tumor suppressor in renal cell carcinoma by targeting
cyclin-dependent kinase 1 (CDK1). Biomed Pharmacother. 111:517–526.
2019. View Article : Google Scholar : PubMed/NCBI
|