Introduction
Multiple myeloma (MM) is a malignant plasma cell
(PC) disorder characterized by the presence of malignant PCs within
the bone marrow. MM accounts for ~1% of neoplastic diseases and 13%
of all hematological cancer types, with a 5-year survival rate of
only 40% (1). Although treatment
strategies have evolved from traditional chemotherapy and
autologous hematopoietic stem cell transplantation to novel
targeted drug therapies, patient outcomes have not improved notably
(2). Therefore, the need to
investigate novel functional molecular and therapeutic targets for
the treatment of MM is ever increasing.
Long noncoding RNAs (lncRNAs) are >200
nucleotides (nt) long and cannot be translated into proteins
(3). Previous studies have
demonstrated that lncRNAs are involved in tumor development and may
be used as diagnostic markers of cancers (4). Accumulating evidence has demonstrated
that lncRNAs [colon cancer associated transcript 1, metastasis
associated lung adenocarcinoma transcript 1 (MALAT1) and urothelial
cancer associated 1 (UCA1)] serve a role in MM, suggesting their
importance in MM progression (5–7).
However, the function of lncRNAs in MM malignancy and tumorigenesis
remains unclear.
The lncRNA IGF1R antisense imprinted non-protein
coding RNA (IRAIN), which is 5,359 nt in length, is located on
chromosome 15q26.3. Previous studies have suggested that IRAIN is
downregulated in prostate cancer and blood obtained from high-risk
acute myeloid leukemia (AML) patients (8,9). Kang
et al (10) reported that
IRAIN also acts as a tumor suppressor in breast cancer. However,
the functional role and underlying mechanisms of IRAIN in MM are
poorly understood. In the present study, IRAIN expression was
detected in MM tissues and cell lines and an initial analysis of
its molecular mechanisms of action was performed. This present
study provides novel insights into the function of IRAIN in MM
development, and suggests that this lncRNA may serve a role in MM
tumorigenesis.
Materials and methods
Sample collection
The specimens used in the present study were
obtained from 35 patients who were diagnosed according to the
National Comprehensive Cancer Network (NCCN) clinical practice
guidelines for MM at the First Affiliated Hospital of Nanchang
University (Nanchang, China), between October 2015 and May 2017.
All patients who received chemotherapy and/or biotherapy were
excluded and patients with other types of malignant tumors were
eliminated. A total of 20 plasma samples in the validation set from
healthy individuals were used as controls. Venous blood was
collected in EDTA tubes (BD Biosciences, Franklin Lakes, NJ, USA).
The plasma was transferred to a fresh tube and stored at −80°C
following snap-freezing in liquid nitrogen. The study was approved
by the Research Ethics Committee of Nanchang University and written
informed consent was obtained from all study subjects.
RNA isolation from human plasma and
reverse transcription-quantitative polymerase chain reaction
(RT-qPCR)
Total RNA was extracted from human plasma using the
mirVana PARIS RNA Isolation kit (Ambion; Thermo Fisher Scientific,
Inc., Waltham, MA, USA) following the manufacturer's protocol for
liquid samples. The concentration and purity of extracted RNA were
measured using 260 and 280 nm optical densities. cDNA was
synthesized from RNA via RT using gene-specific primers (Applied
Biosystems; Thermo Fisher Scientific, Inc.) using the Moloney
Murine Leukemia Virus RT kit (GeneCopoeia, Inc., Rockville, MD,
USA), according to the manufacturer's protocol: 42°C for 2–5 min,
42°C for 50 min, and 70°C for 15 min. To determine microRNA
(miR)-125b expression levels, qPCR was performed using SYBR Green
(Takara Bio, Inc., Otsu, Japan). U6 was used as the internal
control. PCR conditions were as follows: 40 cycles of 95°C for 5
min, 95°C for 45 sec, 55°C for 15 sec and 72°C for 50 sec. Samples
were analyzed in triplicate and gene expression was quantified by
normalizing target gene expression to that of the internal control
using the 2−ΔΔCq formula (11). The primer sequences used were as
follows: IRAIN forward, 5′-CGACACATGGTCCAATCACTGTT-3′ and reverse,
5′-AGACTCCCCTAGGACTGCCATCT-3′; miR-125b forward,
5′-TGCGCTAAAGTGCTTATAGTGC-3′ and reverse,
5′-CCAGTGCAGGGTCCGAGGTATT-3′; and U6 forward,
5′-CTCGCTTCGGCAGCACA-3′ and reverse,
5′-AACGCTTCACGAATTTGCGCTC-3′.
Cell lines and transfection
Human MM cell lines (MM.1S, U266 and RPMI-8226) and
normal PCs (nPCs) were cultured in RPMI-1640 (Gibco; Thermo Fisher
Scientific, Inc.) supplemented with 10% fetal bovine serum (FBS)
(Gibco; Thermo Fisher Scientific, Inc.) and 1%
penicillin/streptomycin at 37°C in a 5% CO2-containing
atmosphere. When the cells reached 80% confluence, cells in the
logarithmic growth phase were collected. The cell lines were
obtained from the American Type Culture Collection (Manassas, VA,
USA). nPCs in the control group were extracted from the plasma of
healthy volunteers and verified by flow cytometry (FITC-IgG, ab6755
and FITC-CD138, ab34164; Abcam, Cambridge, MA, USA) on a
FACSCalibur flow cytometer (BD Biosciences). When the cells reached
70–80% confluence, they were transfected with IRAIN-targeting small
interfering (si)RNA1, siRNA2, siRNA3 and negative control (NC)
using riboFECT™ CP transfection reagents (Guangzhou RiboBio Co.,
Ltd., Guangzhou, China), according to the manufacturer's
instructions. The sequences were: siRNA1 forward,
5′-GCGGGCACAUACUCACUUUTT-3′ and reverse,
5′-AAAGUGAGUAUGUGCCCGCTT-3′; siRNA2 forward,
5′-CCCUUAAUGUGGUCCGGUUTT-3′ and reverse,
5′-AACCGGACCACAUUAAGGGTT-3′; siRNA3 forward,
5′-GAGCGACACUGCUUAUUAATT-3′ and reverse,
5′-UUAAUAAGCAGUGUCGCUCTT-3′; si-NC forward,
5′-UUCUCCGAACGUGUCACGUTT-3′ and reverse,
5′-ACGUGACACGUUCGGAGAATT-3′. miR-125b inhibitor was purchased from
Shanghai GenePharma Co., Ltd. (Shanghai, China), with the following
sequence: miR-125b inhibitor forward, 5′-UCACAAGUUAGGGUCUCAGGGA-3′.
MiR-125b inhibitor was formulated in 0.9% NaCl to a final
concentration of 10 mg/ml. The cells were subjected to RT-qPCR to
measure the expression of IRAIN. As IRAIN siRNA3 was most effective
at knocking down IRAIN expression, this siRNA was used for all
subsequent experiments.
Cell proliferation and colony
formation assays
For the cell proliferation assays, cells were plated
in individual wells in a 96-well plate (1,500 cells/well) and
examined 48 h post-transfection using a Cell Counting Kit-8 (CCK8;
Dojindo Molecular Technologies, Inc., Kumamoto, Japan). Optical
density values were determined at 450 nm wavelength using a
microplate reader. For the survival rate assay, the numbers of
viable cells were counted using Trypan blue dye and a Countstar
Cell Counter (ALIT Life Sciences Co., Ltd., Shanghai, China),
according to the manufacturer's protocol. All assays were performed
in triplicate. For the colony formation assay, a total of 500 cells
were plated in a six-well plate and maintained in medium containing
10% FBS, which was replaced every 5 days. After 2 weeks, the cells
were fixed with methanol at 23°C for 1 h and stained with 0.1%
crystal violet at 37°C for 30 min. Visible colonies were manually
counted by visual inspection. Triplicate wells were measured in
each treatment group.
Apoptosis analysis
The cells in each group were collected at 24, 48 and
72 h following transfection and cold PBS was used to wash the cells
three times. The cells were resuspended in 500 µl pre-cooled
binding buffer at a concentration of 5×106 cells/ml. A
total of 100 µl of the cell suspension was added to flow cytometry
tubes and 5 µl Annexin V-fluorescein isothiocyanate (Beyotime
Institute of Biotechnology, Haimen, China) was added. Following
mixing, the samples were incubated at room temperature in the dark
for 15 min and at 5 min prior to the measurements, 5 µl of 10 mg/l
propidium iodide dye was added. Samples were immediately analyzed
via fluorescent-activated cell sorting and BD FACSuite™ software
(BD Biosciences), without washing or fixation. Each sample was
analyzed three times.
Luciferase reporter analysis
The pGL3-IRAIN-3′ untranslated region
(UTR)-wild-type/mutated vector (Promega Corporation, Madison, WI,
USA) was co-transfected with control plasmid or miR-125b-expressing
plasmid into 293T cells (CRL-3216™; American Type Culture
Collection, Manassas, VA, USA) using Lipofectamine® 2000
(Thermo Fisher Scientific, Inc.). Firefly and Renilla
luciferase activities were measured consecutively 24 h after
transfection using a Dual-Luciferase Reporter assay kit (Promega
Corporation).
Prediction of miR targets
Computational prediction of miR targets was
performed using the online database miRcode (www.mircode.org).
Statistical analysis
For data analysis, the SPSS 16.0 (SPSS, Inc.,
Chicago, IL, USA) and Microsoft Excel (Microsoft Office 2013 for
Windows; Microsoft Corporation. Redmond, WA, USA) were used. Each
experiment was performed at least three times and the values are
reported as the mean ± standard deviation. Differences between two
groups were evaluated by Student's t-tests. For multiple-groups
comparisons, one-way analysis of variance was used, followed by
post hoc Newman-Keuls tests. P<0.05 was considered to indicate a
statistically significant difference. For Pearson's correlation
coefficient analysis, GraphPad Prism software (version 5.01;
GraphPad Sofware, Inc., La Jolla, CA, USA) was used.
Results
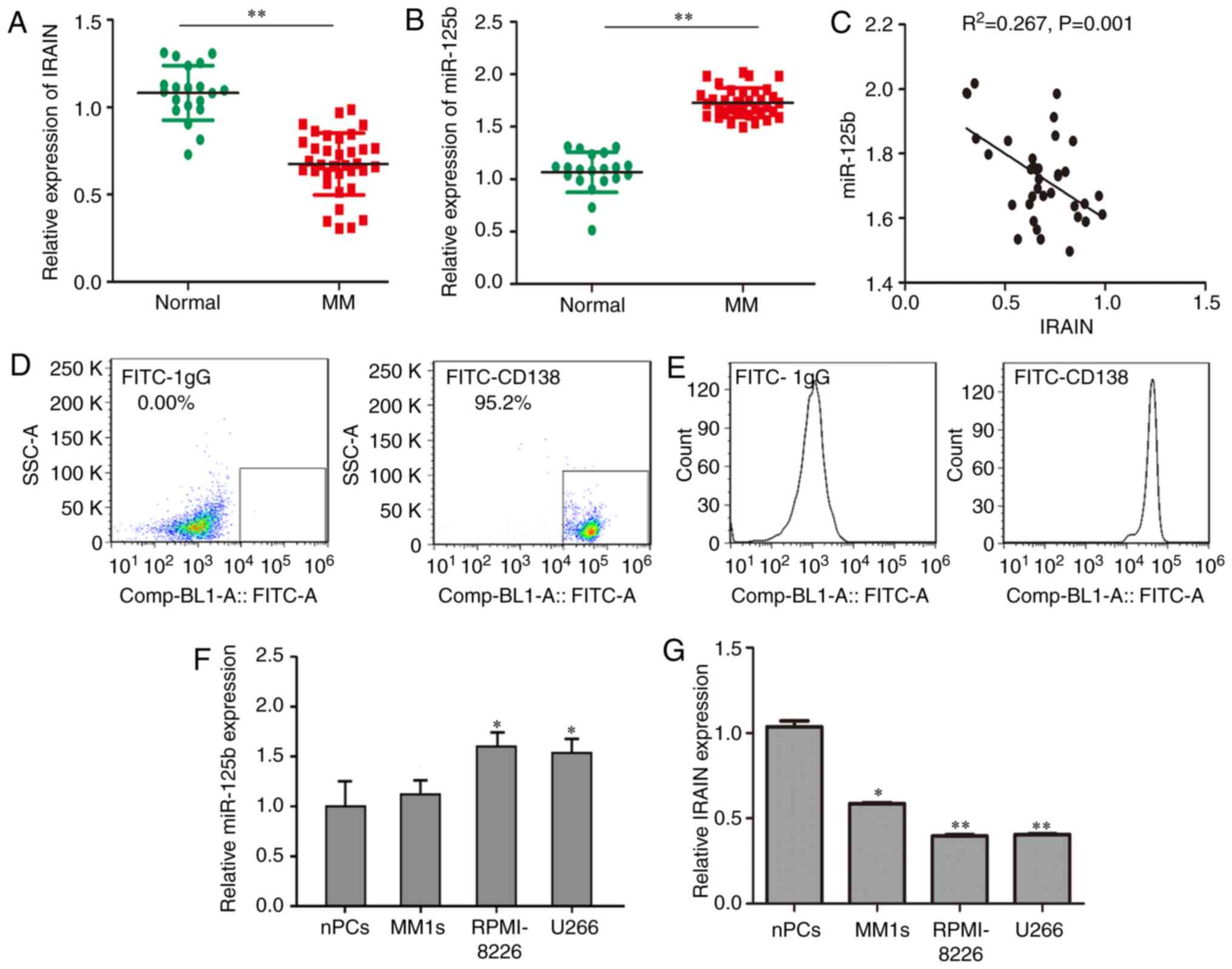
miR-125b is overexpressed and IRAIN is
downregulated in plasma from patients with MM and in MM cell
lines
RT-qPCR was performed in 35 plasma samples from
patients with MM and 20 samples from healthy individuals. The
expression level of IRAIN in patients with MM was significantly
lower compared to that in healthy individuals (Fig. 1A). By contrast, compared with that in
cells from healthy subjects, the level of circulating miR-125b was
upregulated in cells from patients with MM. The normalized miR-125b
expression levels in the patients with MM and healthy subjects were
1.69±0.03 and 1.30±0.08, respectively and miR-125b expression in MM
patients was significantly higher compared with that in healthy
individuals (Fig. 1B). An inverse
correlation between miR-125b and IRAIN was also observed in
patients with MM (P<0.01; R2=0.267) (Fig. 1C). Normal plasma cells in the control
group were cluster of differentiation 138-positive cells extracted
from the plasma of healthy volunteers and verified by flow
cytometry (Fig. 1D and E).
Consistent with these data, the RPMI-8226 cell line expressed the
highest level of miR-125b (Fig. 1F)
and the lowest level of IRAIN (Fig.
1G).
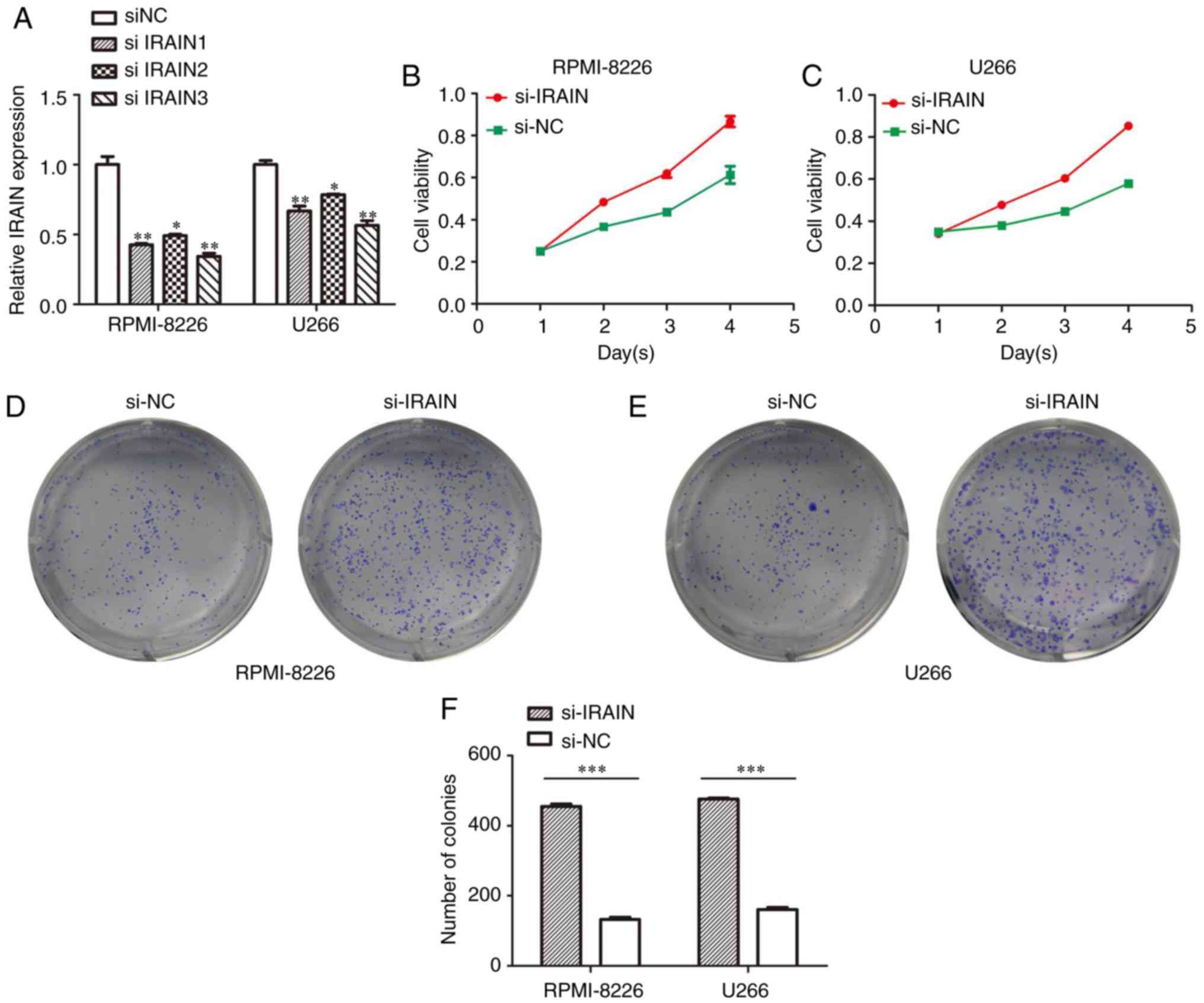
Downregulation of IRAIN promotes cell
proliferation
To investigate the biological role of IRAIN in MM
cell lines, MM cells were transfected with IRAIN-targeting siRNA.
IRAIN siRNA 3 possessed the most potent ability to knock down IRAIN
(Fig. 2A). The CCK8 assay results
demonstrated that the viability of U266 and RPMI-8226 cells was
increased by transfection with si-IRAIN (Fig. 2B and C). Consistent with the CCK8
assay results, the colony numbers in IRAIN-silenced U266 and
RPMI-8226 cells exhibited a marked increase compared with
si-NC-transfected cells (Fig. 2D and
E). The data from the colony formation assay are presented in
Fig. 2F.
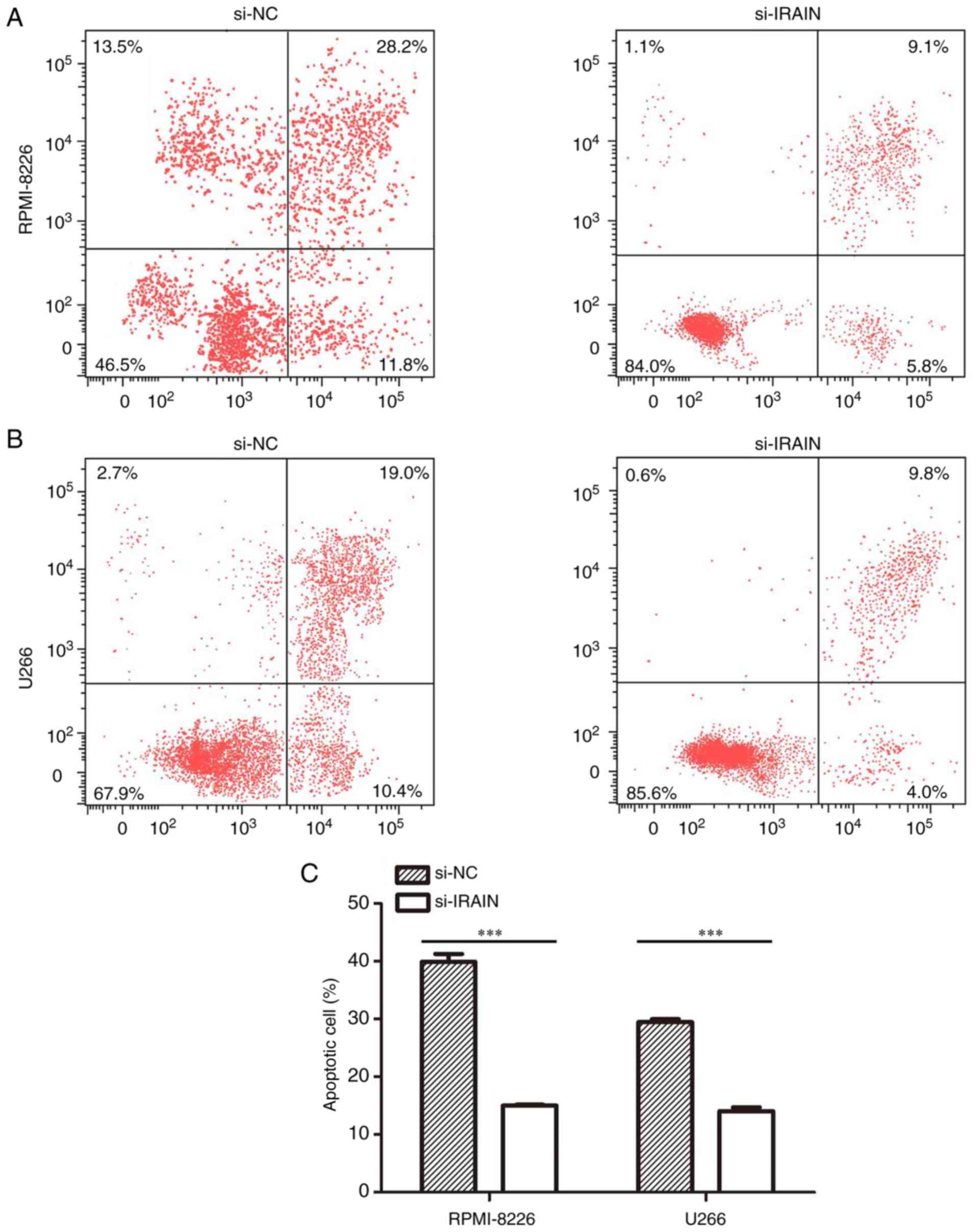
Knockdown of IRAIN inhibits
apoptosis
The effect of IRAIN knockdown on apoptosis was
analyzed by flow cytometry. Compared with that in the si-NC group,
the apoptosis of MM cells was inhibited in the si-IRAIN group. The
apoptotic rates were 13.8% in the U266 group and 14.9% in the
RPMI-8226 group (Fig. 3A and B).
Compared with si-NC transfection, si-IRAIN transfection inhibited
apoptosis in MM cells (P<0.001, Fig.
3C).
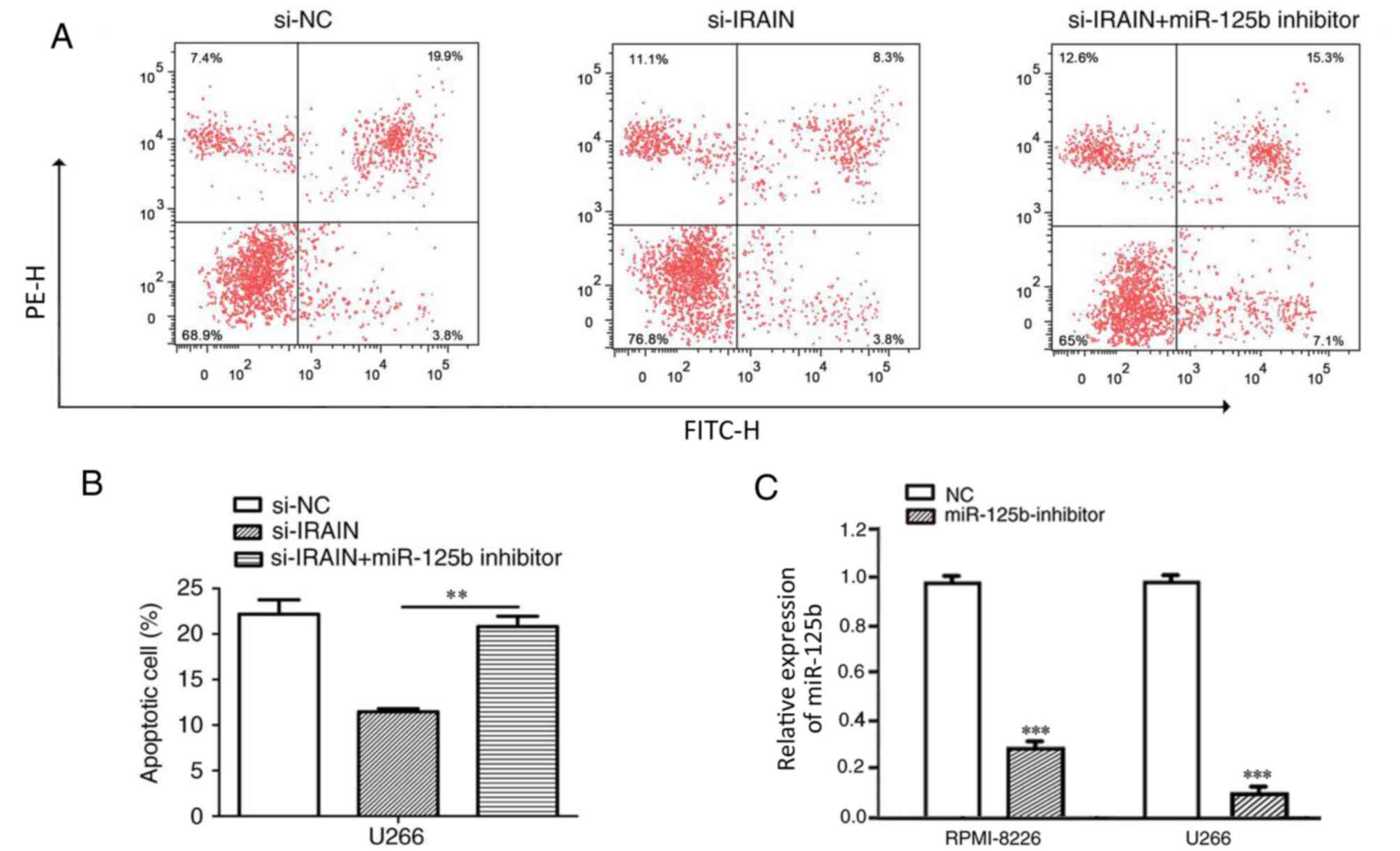
miR-125b reverses the effect of IRAIN
on MM cell apoptosis
To further investigate whether IRAIN exerts
biological functions through miR-125b, rescue experiments were
performed by inhibiting miR-125b expression in si-IRAIN cells
(U266). Flow cytometry demonstrated that cell apoptosis was reduced
in si-IRAIN cells, whereas the addition of an miR-125b inhibitor
partially reversed this effect (Fig.
4; P<0.01).
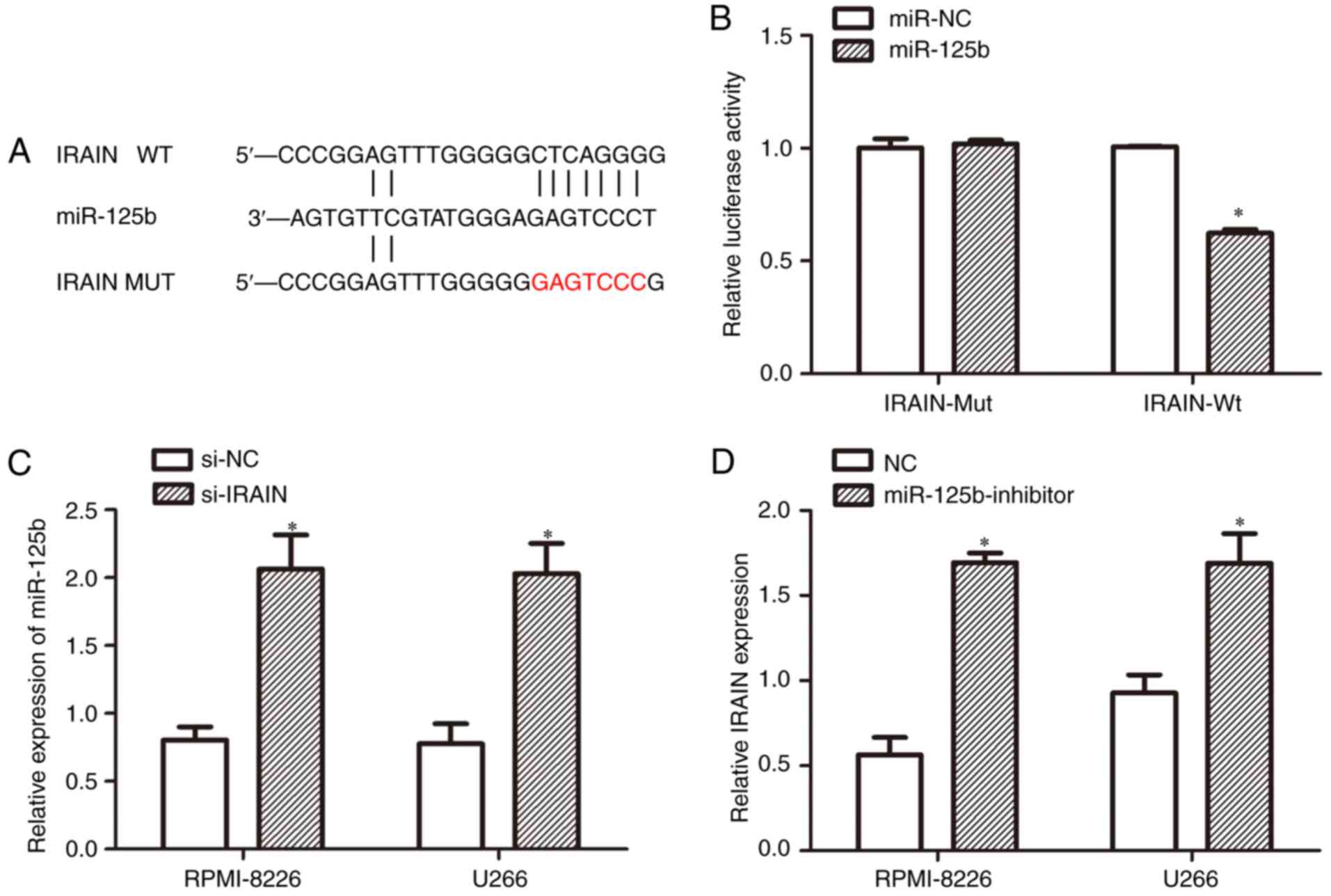
IRAIN is a target of miR-125b
In the present study it was discovered that the
tumor suppressor IRAIN was a target of miR-125b as predicted by
miRcode (Fig. 5A). To determine
whether miR-125b bound to the 3′-UTR of IRAIN, luciferase reporter
assays were performed. As expected, miR-125b overexpression
inhibited luciferase activity. By contrast, cells with mutant IRAIN
3′-UTRs displayed increased luciferase activity (Fig. 5B). The results also demonstrated that
IRAIN knockdown increased miR-125b expression levels (Fig. 5C). At the same time, treatment with
the miR-125b inhibitor enhanced IRAIN expression (Fig. 5D).
Discussion
MM is a neoplasm of terminally differentiated B
cells (PCs), accounting for ~0.8% of all new cancer cases (12). Dysregulation of lncRNA expression
serves an important role in cancer development and lncRNAs are
becoming potential prognostic biomarkers in cancer (13). An increasing number of studies have
provided evidence to suggest that the dysregulation of lncRNAs may
contribute to MM progression. For example, Sedlarikova et al
(14) reported that lncRNA UCA1 was
downregulated in MM. Furthermore, Cho et al (15) demonstrated that the lncRNA MALAT1 is
overexpressed in MM and may serve as a marker to predict disease
progression. However, the function and underlying mechanism of
lncRNAs in MM remain unclear. Understanding the roles of lncRNAs as
tumor suppressors or oncogenes may help to identify novel potential
biomarkers for early diagnosis and new epigenetic molecular targets
for MM patients.
IRAIN is an antisense noncoding RNA that was first
identified in hematopoietic malignancies, with a pattern of
decreased expression in AML (9).
Previous studies have indicated that IRAIN is associated with
breast cancer (10), non-small cell
lung cancer (16) and pancreatic
cancer (17). However, there is
limited evidence to suggest a link between IRAIN and MM. Notably
miR-125b is a member of the miR-17-92 cluster and has been
demonstrated to function as an oncomir in numerous human cancer
types. A study performed by Wang et al (18) demonstrated that miR-125b was highly
expressed in breast cancer and Shen et al (19) reported that miR-125b expression was
markedly increased in type 2 diabetes mellitus. Previous research
has also demonstrated that high levels of miR-125b was associated
with shortened progression-free survival times (20). In the current study, it was
demonstrated that IRAIN expression was substantially downregulated
in MM plasma and cell lines. Furthermore, an increased level of
miR-125b expression in plasma from samples was observed. It was
demonstrated that miR-125b was also upregulated in MM cell lines.
IRAIN depletion promoted MM cell proliferation and led to the
inhibition of MM cell apoptosis. Furthermore, the present study
reported that IRAIN was a direct target of miR-125b. These data
suggest that IRAIN may be a tumor suppressor lncRNA and that the
upregulation of miR-125b in MM may be associated with the
development and progression of the disease.
Increasing evidence suggests that lncRNAs may act as
endogenous miRNA sponges by binding to miRNAs (21,22).
Zhang et al (23)
demonstrated that lncRNA UCA1 promoted cancer progression by acting
as a competing endogenous RNA of activating transcription factor 2
in prostate cancer. Additionally, Xia et al (24) demonstrated that lncRNA Fer-1 like
family member 4 suppressed cancer cell growth by acting as a
competing endogenous RNA and regulating phosphatase and tensin
homolog expression. However, further investigations are required to
determine whether the relationship between IRAIN and miR-125b is
similar in MM.
In summary, the present study demonstrated that
IRAIN may act as a tumor suppressor in MM, whilst miR-125b may act
as a tumor promoter. A negative correlation between IRAIN and
miR-125b was observed in MM plasma. Downregulation of IRAIN
significantly increased the expression of miR-125b. Silencing of
IRAIN promoted cell growth and inhibited cell apoptosis, and
miR-125b reversed the effect of IRAIN on MM cell apoptosis.
Furthermore, by using dual-luciferase reporter assays, IRAIN was
identified as a target of miR-125b. Knockdown of IRAIN suppressed
MM cell proliferation in vitro. Therefore, the present study
highlights the importance of the miRNA-lncRNA interaction in
tumorigenesis. Consequently IRAIN may be used to predict the
clinical prognosis of MM patients and may be a novel therapeutic
target for treating MM.
Acknowledgements
Not applicable.
Funding
The present study was supported by the International
Collaboration Fund from the National Science and Technology
Committee of China (grant no. 2011DFA32820), Innovation Fund
Project in Jiangxi Province (grant no. YC2016-B018), and the
National Natural Science Fund Project (grant no. 81460037).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
YJ performed the experiments, analyzed the data and
wrote the manuscript. JC performed flow cytometry. GC conceived and
designed the experiments. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The Research Ethics Committee of Nanchang University
(Nanchang, China) approved this study and written informed consent
was obtained from all study subjects.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sonneveld P, Avet-Loiseau H, Lonial S,
Usmani S, Siegel D, Anderson KC, Chng WJ, Moreau P, Attal M, Kyle
RA, et al: Treatment of multiple myeloma with high-risk
cytogenetics: A consensus of the International Myeloma Working
Group. Blood. 127:2955–2962. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Mimura N, Hideshima T and Anderson KC:
Novel therapeutic strategies for multiple myeloma. Exp Hematol.
43:732–741. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Gao JR, Qin XJ, Jiang H, Gao YC, Guo MF
and Jiang NN: Potential role of lncRNAs in contributing to
pathogenesis of chronic glomerulonephritis based on microarray
data. Gene. 643:46–54. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Duguang L, Jin H, Xiaowei Q, Peng X,
Xiaodong W, Zhennan L, Jianjun Q and Jie Y: The involvement of
lncRNAs in the development and progression of pancreatic cancer.
Cancer Biol Ther. 18:927–936. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Meng YB, He X, Huang YF, Wu QN, Zhou YC
and Hao DJ: Long noncoding RNA CRNDE promotes multiple myeloma cell
growth by suppressing miR-451. Oncol Res. 25:1207–1214. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Li B, Shi C, Zhao J and Li B: Long
noncoding RNA CCAT1 functions as a ceRNA to antagonize the effect
of miR-410 on the down-regulation of ITPKB in human HCT-116 and
HCT-8 cells. Oncotarget. 8:92855–92863. 2017.PubMed/NCBI
|
|
7
|
Gu Y, Xiao X and Yang S: LncRNA MALAT1
acts as an oncogene in multiple myeloma through sponging miR-509-5p
to modulate FOXP1 expression. Oncotarget. 8:101984–101993. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Niu XB, Fu GB, Wang L, Ge X, Liu WT, Wen
YY, Sun HR, Liu LZ, Wang ZJ and Jiang BH: Insulin-like growth
factor-I induces chemoresistence to docetaxel by inhibiting miR-143
in human prostate cancer. Oncotarget. 8:107157–107166. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sun J, Li W, Sun Y, Yu D, Wen X, Wang H,
Cui J, Wang G, Hoffman AR and Hu JF: A novel antisense long
noncoding RNA within the IGF1R gene locus is imprinted in
hematopoietic malignancies. Nucleic Acids Res. 42:9588–9601. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kang L, Sun J, Wen X, Cui J, Wang G,
Hoffman AR, Hu JF and Li W: Aberrant allele-switch imprinting of a
novel IGF1R intragenic antisense non-coding RNA in breast cancers.
Eur J Cancer. 51:260–270. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Merz M, Jauch A, Hielscher T, Bochtler T,
Schönland SO, Seckinger A, Hose D, Bertsch U, Neben K, Raab MS, et
al: Prognostic significance of cytogenetic heterogeneity in
patients with newly diagnosed multiple myeloma. Blood Adv. 2:1–9.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Shen X, Zhang Y, Wu X, Guo Y, Shi W, Qi J,
Cong H, Wang X, Wu X and Ju S: Upregulated lncRNA-PCAT1 is closely
related to clinical diagnosis of multiple myeloma as a predictive
biomarker in serum. Cancer Biomark. 18:257–263. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sedlarikova L, Gromesova B, Kubaczkova V,
Radova L, Filipova J, Jarkovsky J, Brozova L, Velichova R, Almasi
M, Penka M, et al: Deregulated expression of long non-coding RNA
UCA1 in multiple myeloma. Eur J Haematol. 99:223–233. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Cho SF, Chang YC, Chang CS, Lin SF, Liu
YC, Hsiao HH, Chang JG and Liu TC: MALAT1 long non-coding RNA is
overexpressed in multiple myeloma and may serve as a marker to
predict disease progression. BMC Cancer. 14:8092014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Feng J, Sun Y, Zhang EB, Lu XY, Jin SD and
Guo RH: A novel long noncoding RNA IRAIN regulates cell
proliferation in non small cell lung cancer. Int J Clin Exp Pathol.
8:12268–12275. 2015.PubMed/NCBI
|
|
17
|
Lian Y, Wang J, Feng J, Ding J, Ma Z, Li
J, Peng P, De W and Wang K: Long non-coding RNA IRAIN suppresses
apoptosis and promotes proliferation by binding to LSD1 and EZH2 in
pancreatic cancer. Tumour Biol. 37:14929–14937. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang H, Tan G, Dong L, Cheng L, Li K, Wang
Z and Luo H: Circulating MiR-125b as a marker predicting
chemoresistance in breast cancer. PLoS One. 7:e342102012.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Shen Y, Xu H, Pan X, Wu W, Wang H, Yan L,
Zhang M, Liu X, Xia S and Shao Q: miR-34a and miR-125b are
upregulated in peripheral blood mononuclear cells from patients
with type 2 diabetes mellitus. Exp Ther Med. 14:5589–5596.
2017.PubMed/NCBI
|
|
20
|
Piatopoulou D, Avgeris M, Marmarinos A,
Xagorari M, Baka M, Doganis D, Kossiva L, Scorilas A and Gourgiotis
D: miR-125b predicts childhood acute lymphoblastic leukaemia poor
response to BFM chemotherapy treatment. Br J Cancer. 117:801–812.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li F, Huang C, Li Q and Wu X: Construction
and comprehensive analysis for dysregulated long non-coding RNA
(lncRNA)-associated competing endogenous RNA (ceRNA) network in
gastric cancer. Med Sci Monit. 24:37–49. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhao Y, Wang H, Wu C, Yan M, Wu H, Wang J,
Yang X and Shao Q: Construction and investigation of
lncRNA-associated ceRNA regulatory network in papillary thyroid
cancer. Oncol Rep. 39:1197–1206. 2018.PubMed/NCBI
|
|
23
|
Zhang S, Dong X, Ji T, Chen G and Shan L:
Long non-coding RNA UCA1 promotes cell progression by acting as a
competing endogenous RNA of ATF2 in prostate cancer. Am J Transl
Res. 9:366–375. 2017.PubMed/NCBI
|
|
24
|
Xia T, Liao Q, Jiang X, Shao Y, Xiao B, Xi
Y and Guo J: Long noncoding RNA associated-competing endogenous
RNAs in gastric cancer. Sci Rep. 4:60882014. View Article : Google Scholar : PubMed/NCBI
|