Introduction
Chromosomal rearrangements of the lysine
methyltransferase 2A (KMT2A) gene (former MLL) at
11q23 have been reported in ~10% of patients with acute leukemias
(1). Analysis of the KMT2A
recombinome of acute leukemias has identified 135 totally different
KMT2A rearrangements, and 94 related translocation partner genes
have now been identified at the molecular level (2–5). The
MLLT3 super elongation complex subunit (MLLT3) gene (former
AF9) at 9p21.3 is one of the most common translocation
partner genes in acute myeloid leukemia (AML). In the 2016 World
Health Organization (WHO) classification, AML was divided into four
main categories as follows: i) AML with recurrent genetic
aberrations; ii) AML with myelodysplasia-related features; iii)
therapy-related myeloid neoplasms; and iv) AML
not-otherwise-specified (1,6). AML with t(9;11)(p21.3;q23.3)
translocation can be de novo AML according to the WHO
heading of ‘AML with recurrent genetic abnormalities’ and
therapy-related AML (t-AML; mostly caused by DNA topoisomerase II
inhibitors) that were separately categorized into ‘therapy-related
myeloid neoplasms’ (1,7). KMT2A/MLLT3 (former
MLL/AF9) fusion gene resulting from
t(9;11)(p21.3;q23.3) serves a crucial role in malignant clone
proliferation of bone marrow stem cell and leukemogenesis according
to in vitro and in vivo studies (8–10).
Furthermore, secondary chromosome abnormalities to t(9;11) are very
common, especially trisomy 8 or a partial duplication of 8q due to
unbalanced rearrangements (11).
However, trisomy 9 as a secondary chromosome change in patients
with t(9;11) is quite rare. The present study reported a unique
case of AML with an extra chromosome 9 secondary to
t(9;11)(p21.3;q23.3). Using routine G-banded cytogenetics,
fluorescence in situ hybridization (FISH) and array
comparative genomic hybridization (aCGH) analysis, the results
demonstrated that the extra chromosome 9 was the copy of either a
normal chromosome 9 or a derivative chromosome 9.
Case report
In May 2011, a 37-year-old female complaining of
lower back pain was found to have elevated white blood cell (WBC)
with peripheral blasts. The complete blood cell count of the
patient comprised 4.29×1012/l of red blood cell, 124 g/l
of hemoglobin, 152×109/l of platelet and
73.7×109/l of WBC, including 28.0×109/l (38%)
of monocytes, 18.4×109/l (25%) of segmented neutrophils
5.2×109/l (7%) of lymphocytes and 30% of peripheral
blasts. The immunophenotype of peripheral blood blasts was positive
for the cell surface markers Human Leukocyte Antigen-DR isotype,
CD45, CD64, CD13, CD14 and CD15. The peripheral blood smear
presented numerous blasts and immature monocyte-like cells.
The patient had been diagnosed with bilateral,
poorly differentiated and infiltrating ductal breast carcinoma in
2008. Subsequently, she underwent neoadjuvant chemotherapy with
adriamycin (topoisomerase II inhibitor), cytoxan and cytosar-U. The
patient also received radiation and a lumpectomy that detected 0/13
nodes positive for malignancy. Next, she received adjuvant Taxol on
a weekly basis for 12 weeks along with breast radiation and
anticancer endocrine therapy with tamoxifen, which was continued
until the present study. The clinical diagnosis of AML was made in
2011. Following diagnosis, the patient received standard cytarabine
and daunorubicin induction therapy.
Cytogenetic analysis was performed using G-banding
by trypsin using Giemsa (GTG) as a stain technique on 72-h cultures
of leukemic blood collected during the patient's initial clinical
visit for low back pain. The results from the initial G-banding
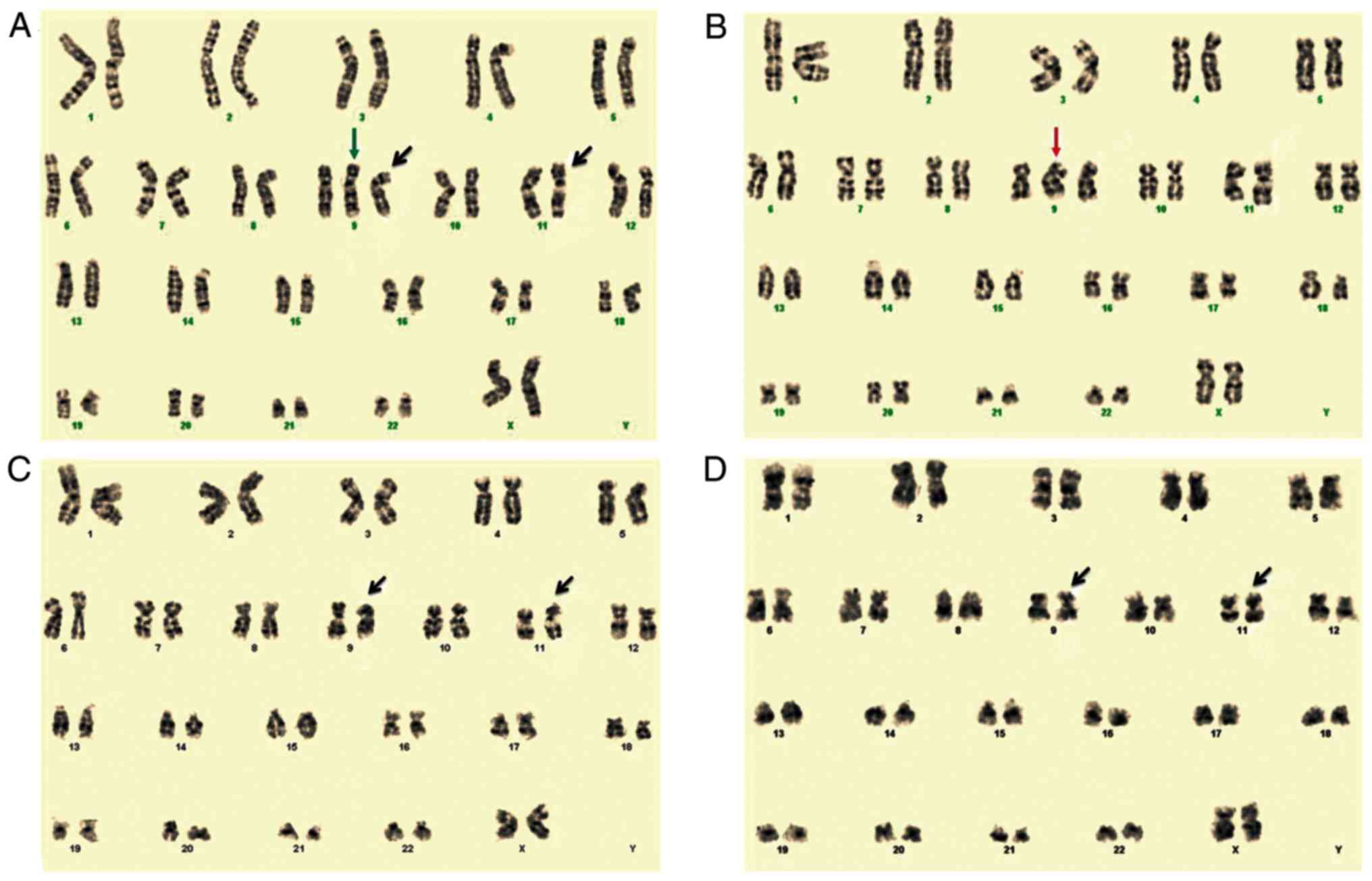
analysis revealed that the patient's karyotype was
47,XX,+9,t(9;11)(p21.3;q23.3)[17]/46,XX[3] (Fig. 1A). Karyotypes were described
according to the International System for Human Cytogenetic
Nomenclature (ISCN 2016) (12).
To confirm t(9;11)(p21.3;q23.3) translocation and
the presence of the extra chromosome 9, FISH analyses were
performed with the combination of the KMT2A Break Apart probe
(Vysis/Abbott, Inc.) and the centromere enumeration probe (CEP) 9
probe (Vysis/Abbott, Inc.). The KMT2A Break Apart probe was mapped
to 11q23, 5′KMT2A was labeled as SpectrumGreen and
3′KMT2A was labeled as SpectrumOrange. The CEP 9 probe
labeled with SpectrumGreen was used to hybridize to the centromeric
region of chromosome 9. A total of 200 cultured leukemic blood
cells including metaphases and interphases were screened. The FISH
results are presented in Table SI.
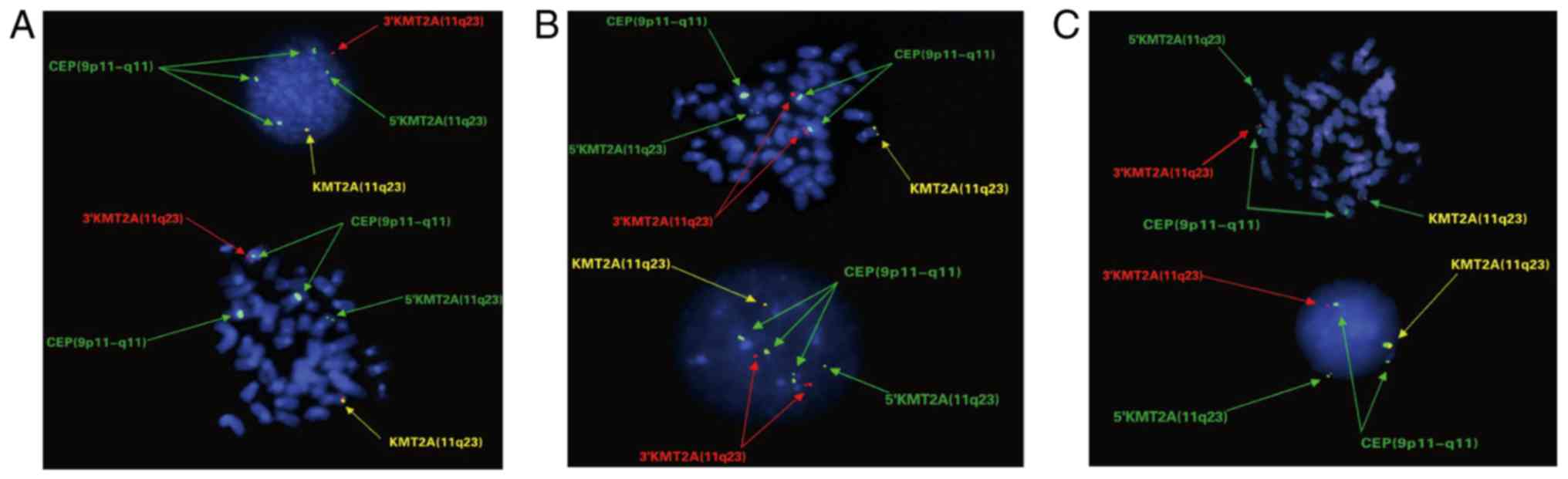
The results demonstrated that 8 normal cells carried two yellow
fusion signals of KMT2A Break Apart probe on 11q23 region and two
green CEP 9 signals on centromeric region of chromosome 9. A total
of 36 cells, including 10 cells in metaphase, presented one yellow
intact KMT2A signal on the normal chromosome 11, one red signal on
the derivative chromosome 9 and four non-fused green signals,
including three CEP 9 signals on centromeric region of chromosome 9
and one 5′KMT2A signal on derivative chromosome 11, which indicated
that these 36 cells may carry an extra normal chromosome 9 in
addition to t(9;11)(p21.3;q23.3) (Fig.
2A). However, 140 cells, including 20 cells in metaphase,
presented two red 3′KMT2A signals on two derivative chromosome 9,
one yellow signal on the normal chromosome 11 and four non-fused
green signals, including three CEP 9 signals on the centromeric
region of chromosome 9 and one 5′KMT2A signal on the derivative
chromosome 11, suggesting that these 140 cells may carry an extra
derivative chromosome 9 in addition to t(9;11)(p21.3;q23.3)
translocation (Fig. 2B).
Furthermore, 16 cells presented one yellow signal on the normal
chromosome 11, one red signal on the derivative chromosome 9 and
three green signals, including two on centromeric region of
chromosome 9 and one on the derivative chromosome 11, which
indicated that these cells would only have a translocation without
an extra chromosome 9 (Fig. 2C).
To confirm the origin of the extra chromosome 9 and
to better identify the region of the rearrangement, additional
assays were performed to define minor and cryptic genomic
imbalances near the translocation break points. Subsequently, aCGH
was performed with genomic DNA isolated from uncultured leukemic
blood specimen collected from the patient during the initial visit.
The results from aCGH revealed the following unique genomic
imbalance patterns: i) Gain of 119.795 Mb (3 copies) at 9p21.3-qter
(chr9: 20,351,514-140,146,188); ii) gain of 19.712 Mb (2–3 copies)
at 9pter-p21.3 (chr9: 638,796-20,351,121) (Fig. 3A); and iii) gain of 16.578 Mb (2–3
copies) at 11q23.3-qter (chr11: 117,867,162-134,444,816) (Fig. 3B), according to the human genome
build NCBI36/hg18 (http://genome-asia.ucsc.edu/cgi-bin/hgTracks?db=hg18).
These results were consistent with the FISH results. The results
from aCGH and FISH collectively suggested that the extra chromosome
9 in certain metaphase cells may be the derivative chromosome 9. In
addition, the derivative extra chromosome 9 was detected in some
metaphase cells by GTG analysis at the initial diagnosis following
one additional screening (Fig. 1B).
The dosage of 9qter appeared to be lower than that of other 9q
regions (Fig. 3A), which may be due
to the general waving feature of the whole baseline of chromosome
9.
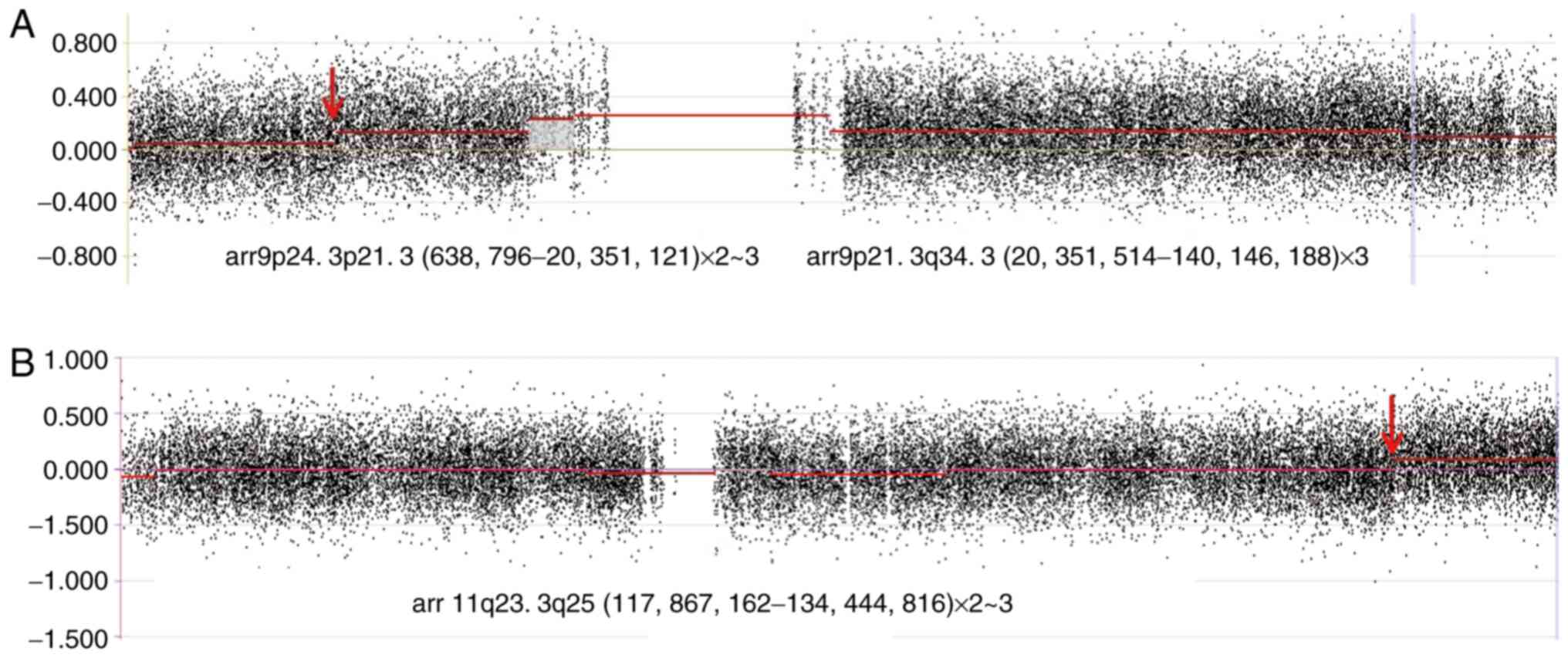 | Figure 3.Results from array comparative genomic
hybridization. The red arrows indicate the breakpoints of
chromosomes 9 and 11 at 9p21.3 and 11q23.3, respectively. (A) Gain
119.795Mb (3 copies) at 9p21.3-q34.3 (hg18.
Chr9:20,351,514-140,146,188) and gain 19.712 Mb (2–3 copies) at
p24.3-p21.3 (hg18. Chr9:638,796-20,351,121). The gain ratio of the
latter is smaller than that of the former. (B) Gain 16.578 Mb (2–3
copies) at 11q23.3-q25 (hg18. Chr11:117,867,162-134,444,816). |
The results from follow-up cytogenetic studies
reported the karyotypes 46,XX,t(9;11)(p22;q23)[3]/46,XX[17]
(Fig. 1C) and
46,XX,t(9;11)(p22;q23)[2]/46,XX[18] (Fig. 1D) for the bone marrow samples
collected on the 17th and the 45th day of AML induction therapy,
respectively. The results from FISH analyses presented one yellow
signal, one red signal and three non-fused green signals, which
confirmed the cytogenetics results (Fig. S1) that the cells with only t(9;11)
persisted, whereas the cells with the extra chromosome 9
disappeared following induction therapy. The patient did not get
complete remission (CR) and succumbed to the disease in March
2012.
Discussion
The present study reported the case of a patient
with AML positive for t(9;11)(p21.3;q23.3) translocation. Trisomy
chromosome 9, either a normal or a derivative chromosome 9, was
detected in the initial sample. This extra chromosome 9 was
hypothesized to be secondary to the primary chromosomal change
t(9;11), since some of the extra chromosome 9 were abnormal
chromosome 9 derived from t(9;11) (p21.3;q23.3). The results from
the follow-up cytogenetic studies indicated that only the balanced
translocation t(9;11), rather than the extra chromosome 9,
persisted in the samples following induction therapy, which
suggested that, in this particular case, chemotherapy may exert
selection pressure against secondary chromosomal changes, but not
against the primary cytogenetic abnormality.
t(9;11)(p21.3;q23.3) translocation is one of the
most common KMT2A-rearrangements in AML which can cause
KMT2A/MLLT3 fusion (8,13,14).
t(9;11) positive AML can occur primarily as a de novo
neoplasm or as a result of previous therapy, for example t-AML,
typically caused by topoisomerase II inhibitors (15–17). It
has been suggested that the topoisomerase II cleavage site and the
DNase I hypersensitive site can colocalize in the break cluster
regions of MLLT3 and KMT2A (16,18,19).
Furthermore, an in vivo experiment reported the cleavage
site of VP-16 (a topoisomerase II-like inhibitor) localized in the
break cluster regions of KMT2A in a patient with AML
(20). The majority of t-AML cases
appeared in patients who had advanced-stage breast cancer and who
had been treated with topoisomerase II inhibitors such as
adriamycin, VP-16 and mitoxantrone. In addition, the latency period
following primary therapy with this type of inhibitors can vary
from 24 to 48 months (15–17). The patient from the present study
suffered from breast carcinoma and received chemotherapy, including
the topoisomerase II inhibitor adriamycin, and radiation straight
after the diagnosis. After three years, the patient was diagnosed
with AML. According to the 2016 WHO classification of myeloid
neoplasms (1), this patient probably
suffered from t-AML.
Numerous secondary chromosome abnormalities have
been reported to be associated with t(9;11)(p21.3;q23.3), including
trisomy 8 and modifications to chromosome 11 in the form of
self-insertion or deletion (11,21). To
the best of our knowledge, trisomy 9 as a cytogenetic abnormality
secondary to t(9;11) in AML has rarely been reported and studied
(22). The patient from the present
study was positive for t(9;11)(p21.3;q23.3) translocation with an
extra chromosome 9. In addition, the origin of this extra
chromosome 9 appeared to be either a normal or an abnormal
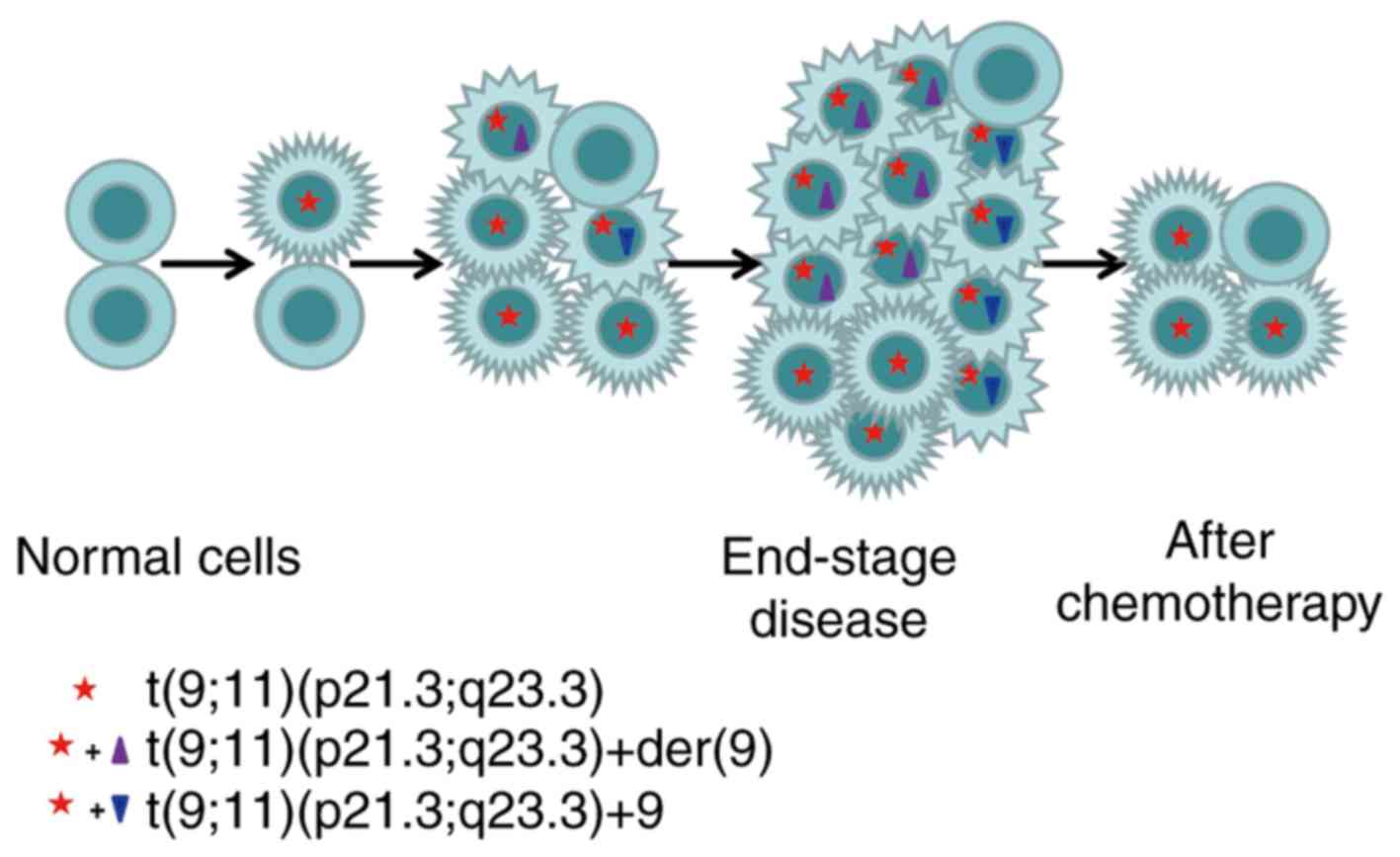
chromosome 9. According to the FISH results, this patient presented
the four following cell clones: i) Normal cells; ii) cells with
t(9;11)(p21.3;q23.3) translocation; iii) cells with
t(9;11)(p21.3;q23.3) and a normal chromosome 9; and iv) cells with
t(9;11)(p21.3;q23.3) and a derivative chromosome 9. The results
from aCGH confirmed that the extra chromosome 9 could either be the
normal chromosome 9 or the derivative chromosome 9. The karyotype
for the initial sample based on the proportion determined by the
FISH result should therefore be
47,XX,t(9;11)(p21.3;q23.3)[2]/47,XX,+9,t(9;11)(p21.3;q23.3)[3]/47,XX,t(9;11)(p21.3;q23.3),+der(9)t(9;11)[14]/46,XX[1].
Previous studies demonstrated that patients with
t(9;11)(p21.3-q23.3) have a favorable outcome compared with
patients with other abnormalities involving 11q23 (23,24),
whereas some other studies suggested that t(9;11)(p21.3;q23.3)
translocation could indicate an intermediate risk (25,26). The
present study did not confirm the reported prognostically favorable
outcome of patients with AML and t(9;11)(p21.3;q23.3). In addition,
previous studies demonstrated that there are few intrinsic
differences between de novo AML and t-AML with
t(9;11)(p21.3;q23.3) translocation, and that t-AML presents minor
worse prognosis compared with patients with de novo
t(9;11)(p21.3;q23.3) positive AML, which could be due to prior
therapy setting or additional karyotypic changes (17,27).
Other studies reported that over-representation of 3′KMT2A
could serve a crucial role in leukemia progression (28). Subsequently, most leukemia cells from
the present case gained an extra copy of the terminal portion of
chromosome 11, from band q23 to its distal end, including the 3′
end of KMT2A. In addition, one previous study proposed three
stages of abnormal clone evolution: i) Appearance of balanced
rearrangement; ii) trisomy; and iii) loss of chromosomal material
(29). The appearance of an
unbalanced genome could provide an advantage in proliferative
activity and may be associated with the poor outcome of
chemotherapy (29). Based on these
studies, the chromosome 9 trisomy in the present study may be
derived from chromosome segregation errors with the presence of the
translocation. The gain of the Janus kinase 2 gene and other genes
on chromosome 9 may contribute to a proliferation advantage to the
cells with trisomy 9 (30). In the
present study, because cells with an extra chromosome 9 disappeared
following chemotherapy, cells with the extra chromosome 9 or
partial trisomy 9 were likely to be sensitive to the chemotherapy
(Fig. 4). To the best of our
knowledge, the present study was the first to report a case of
trisomy 9 as a secondary chromosome abnormality to
t(9;11)(p21.3;q23.3) with the observation of clonal evolution
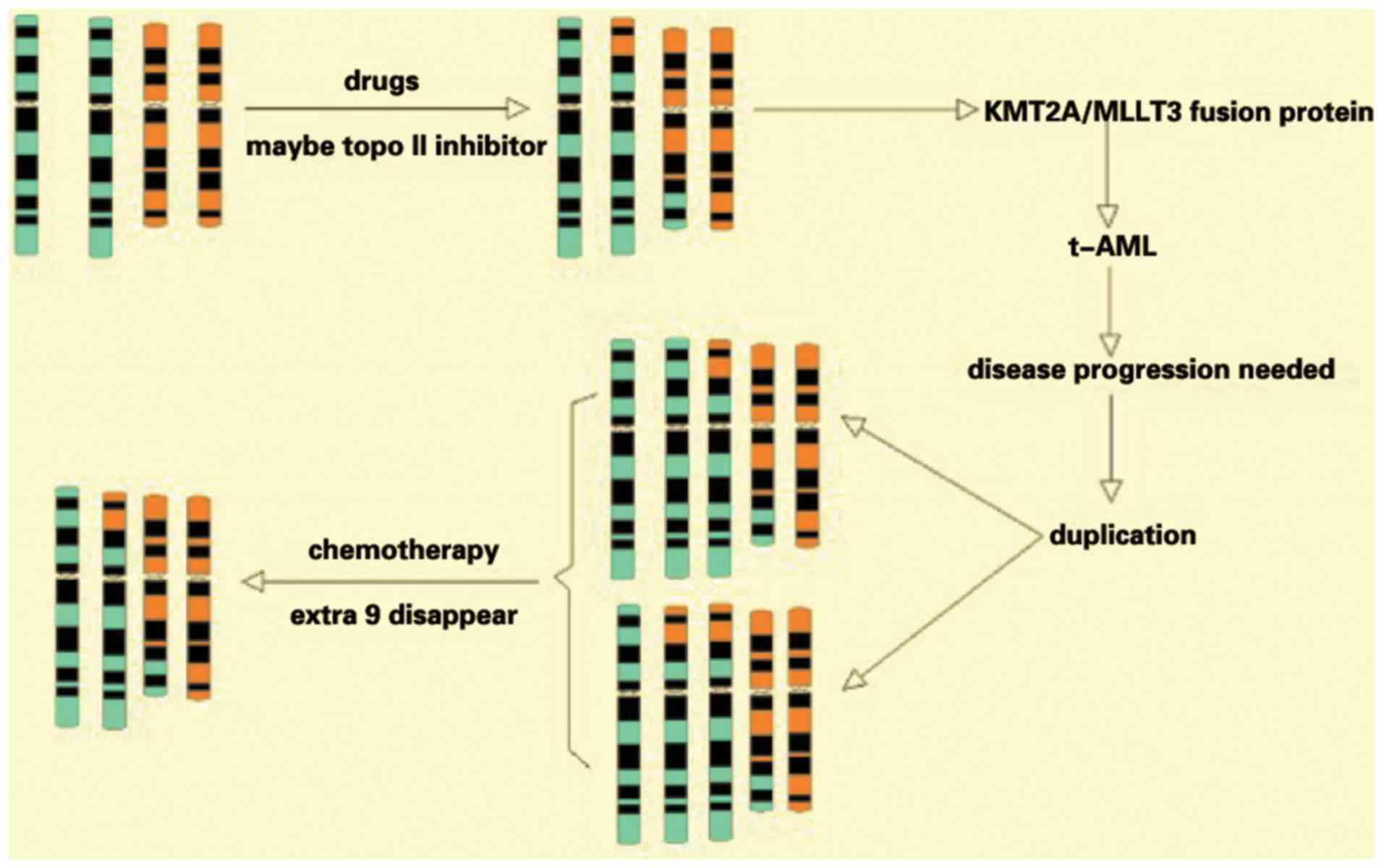
during disease progression and AML treatments. The results from the
present study suggested a likely progression course of chromosomal
constitution (Fig. 5).
In conclusion, this study investigated, to the best
of our knowledge, for the first time the case of t(9;11) with
secondary trisomy 9 derived from either the normal chromosome 9 or
a derivative chromosome 9 in a patient with AML. The extra
chromosome 9 may be a consequence of AML progression and may
contribute to cell sensitivity to subsequent induction therapy. To
better explain the phenomenon of an extra chromosome 9, further
studies are required, especially on 9p21-9q34 genes, which may help
clarify the pathogenic mechanism of the extra chromosomal region in
the progression of AML.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by the grant from
the National Natural Science Foundation of China (Grant. No.
81700205).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
MG performed experiments, contributed to the
analysis of the data and drafted the manuscript. HP performed cell
culture, contributed to the interpretation of the data and prepared
figures and tables. YMK performed karyotype and contributed to the
interpretation of the data. XL performed fluorescence in
situ hybridization and contributed to the interpretation of the
data. XW performed array comparative genomic hybridization and
contributed to the interpretation of the data. JL participated in
the data analysis and helped with the drafting of the manuscript.
MW collected and interpreted the clinical information. FM designed
the study, analyzed data and helped with the interpretation of the
clinical information. SL designed the study, analyzed data and
revised the manuscript. All of the authors read and approved the
manuscript.
Ethics approval and consent to
participate
The study was approved by the Institutional Medical
Ethics Review Board of the First Hospital of Jilin University in
compliance with the Declaration of Helsinki. Written informed
consent was obtained from the patient for publication of the
present study.
Patient consent for publication
Written informed consent was obtained from the
patient for publication of the present study.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Arber DA, Orazi A, Hasserjian R, Thiele J,
Borowitz MJ, Le Beau MM, Bloomfield CD, Cazzola M and vardiman JW:
The 2016 revision to world health organization classification of
myeloid neoplasms and acute leukemia. Blood. 127:2391–2405. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Meyer C, Schneider B, Reichel M,
Angermuneller S, Strehl S, Schnittger S, Schoch C, Jansen MW, van
Dongen JJ, Pieters R, et al: Diagnostic tool for the identification
of MLL rearrangements including unknown partner genes. Proc Natl
Acad Sci USA. 102:449–454. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Meyer C, Kowarz E, Hofmann J, Rennevilla
A, Zuna J, Trka J, Ben Abdelali R, Macintyre E, De Braekeleer E, De
Braekeleer M, et al: New insights to the MLL recombinome of acute
leukemias. Leukemia. 23:1490–1499. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Meyer C, Hofmann J, Burmeister T, Gröger
D, Park TS, Emerenciano M, Pombo de Oliveira M, Renneville A,
Villarese P, Macintyre E, et al: The MLL recombinome of acute
leukemias in 2013. Leukemia. 27:2165–2176. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Meyer C, Burmeister T, Gröger D, Tsaur G,
Fechina L, Renneville A, Sutton R, Venn NC, Emerenciano M,
Pombo-de-Oliveira MS, et al: The MLL recombinome of acute leukemias
in 2017. Leukemia. 32:273–284. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Asou N: Myeloid neoplasms in the world
health organization 2016 classification. Rinsho Ketsueki.
58:2178–2187. 2017.(In Japanese). PubMed/NCBI
|
|
7
|
Biondi A, Cimino G, Pieters R and Pui CH:
Biological and therapeutic aspects of infant leukemia. Blood.
96:24–33. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Corral J, Lavenir I, Impey H, Warren AJ,
Forster A, Larson TA, Bell S, McKenzie AN, King G and Rabbitts TH:
An MLL-AF9 fusion gene made by homologous recombination causes
acute leukemia in chimeric mice: A method to create fusion
oncogenes. Cell. 85:853–861. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Dobson CL, Warren AJ, Pannel R, Forster A,
Lavenir I, Corral J, Smith AJ and Rabbitts TH: The MLL-AF9 gene
fusion in mice controls myeloproliferation and specifies acute
myeloid leukaemogenesis. EMBO J. 18:3564–3574. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Somervaille TC and Cleary ML:
Identification and characterization of leukemia stem cells in
murine MLL-AF9 acute myeloid leukemia. Cancer Cell. 10:257–268.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Anguita E, Barrio CG, González FA, Ferro
MT, del Potro E, Ropero P and Villegas A: Association of
t(9;11)-MLL AF9 and trisomy 8 in an AML-M5 preceded by
pancytopenia. Cancer Genet Cytogenet. 120:144–147. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
McGowan-Jordan J, Simons A and Schmid M:
ISCN 2016: An International System for Human Cytogenomic
NomenclatureReprint of Cytogenetic and Genome Research. 149. 1st.
Karger Publishers; Basel, Switzerland: 2016
|
|
13
|
Krivtsov AV, Twomey D, Feng Z, Stubbs MC,
Wang Y, Faber J, Levine JE, Wang J, Hahn WC, Gilliland DG, et al:
Transformation from committed progenitor to leukemia stem cell
initiated by MLL-AF9. Nature. 442:818–822. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Schneidawind C, Jeong J, Schneidawind D,
Kim IS, Duque-Afonso J, Wong SHK, Iwasaki M, Breese EH, Zehnder JL,
Porteus M and Cleary ML: MLL leukemia induction by t(9;11)
chromosomal translocation in human hematopoietic stem cells using
genome editing. Blood Adv. 2:832–845. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bredeson CN, Barnett MJ, Horsman DE, Dalal
BI, Ragaz J and Phillips GL: Therapy-related acute myologenous
leukemia associated with 11q23 chromosomal abnormalities and
topoisomerase II inhibitors: Report of four additional cases and
brief commentary. Leuk Lymphoma. 11:141–145. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Langer T, Metzler M, Reinhardt D, Viehmann
S, Borkhardt A, Reichel M, Stanulla M, Schrappe M, Creutzig U,
Ritter J, et al: Analysis of t(9;11) chromosomal breakpoint
sequences in childhood acute leukemia: Almost identical MLL
breakpoints in therapy-related AML after treatment without
etoposides. Genes Chromosomes Cancer. 36:393–401. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chandra P, Luthra R, Zuo Z, Yao H, Ravandi
F, Reddy N, Garcia-Manero G, Kantarjian H and Jones D: Acute
myeloid leukemia with t(9;11)(p21-22;q23): Common properties of
dysregulated ras pathway signaling and genomic progression
characterize de novo and therapy-related cases. Am J Clin Pathol.
133:686–693. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Strick R, Strissel PL, Borgers S, Smith SL
and Rowley JD: Dietary bioflavonoids induce cleavage in the MLL
gene and may contribute to infant leukemia. Proc Natl Acad Sci USA.
97:4790–4795. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bariar B, Vestal CG, Deem B, Goodenow D,
Ughetta M, Engledove RW, Sahyouni M and Richardson C: Bioflavonoids
promote stable translocation between MLL-AF9 breakpoint cluster
regions independent of normal chromosomal context: Model system to
screen environmental risks. Environ Mol Mutagen. 60:154–167. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Strissel PL, Strick R, Tomek RJ, Roe BA,
Rowley JD and Zeleznik-Le NJ: DNA structural properties of AF9 are
similar to MLL and could act as recombination hot spots resulting
in MLL/AF9 translocations and leukemogenesis. Hum Mol Genet.
9:1671–1679. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Johansson B, Moorman AV and Secker-Walker
LM: Derivative chromosomes of 11q23-translocations in hematologic
malignancies. European 11q23 Workshop participants. Leukemia.
12:828–833. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Krauter J, Peter W, Pascheberg U, Heinze
B, Bergmann L, Hoelzer D, Lübbert M, Schlimok G, Arnold R, Kirchner
H, et al: Detection of karyotypic aberrations in acute myeloblastic
leukaemia: A prospective comparison between PCR/FISH and standard
cytogenetics in 140 patients with de novo AML. Br J Haematol.
103:72–78. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Mrózek K, Heinonen K, Lawrence D, Carroll
AJ, Koduru PR, Rao KW, Strout MP, Hutchison RE, Moore JO, Mayer RJ,
et al: Adult patient with de novo acute myeloid leukemia and
t(9;11)(p22;q23) have a superior outcome to patient with other
translocation involving band 11q23: A cancer and leukemia group B
study. Blood. 90:4532–4538. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Rubnitz JE, Raimondi SC, Tong X,
Srivastava DK, Razzouk BI, Shurtleff SA, Downing JR, Pui CH,
Ribeiro RC and Behm FG: Favorable impact of the t(9;11) in
childhood acute myeloid leukemia. J Clin Oncol. 20:2302–2309. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Balgobind BV, Raimondi SC, Harbott J,
Zimmermann M, Alonzo TA, Auvrignon A, Beverloo HB, Chang M,
Creutzig U, Dworzak MN, et al: Novel prognostic subgroups in
childhood 11q23/MLL-rearranged acute myeloid leukemia: Results of
an international retrospective study. Blood. 114:2489–2496. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Stӧlzel F, Mohr B, Kramer M, Oelschlӓgel
U, Bochtler T, Berdel WE, Kaufmann M, Baldus CD, Schӓfer-Eckart K,
Stuhlmann R, et al: Karyotype complexity and prognosis in acute
myeloid leukemia. Blood Cancer J. 6:e3862016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Pession A, Martino V, Tonelli R,
Beltramini C, Locatelli F, Biserni G, Franzoni M, Freccero F,
Montemrro L, Pattacini L and Paolucci G: MLL-AF9 oncogene
expression affects cell growth but not terminal differentiation and
is downregulated during monocyte-macrophage maturation in AML-M5
THP-1 cells. Oncogene. 22:8671–8676. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Sambani C, La Starza R, Roumier C,
Crescenzi B, Stavropoulou C, Katsarou O Karafoulidou A, Dhalle JH,
Lai JL, Preudhomme C, et al: Partial duplication of the MLL
oncogene in patients with aggressive acute myeloid leukemia.
Haematologica. 89:403–407. 2004.PubMed/NCBI
|
|
29
|
Andreeva SV, Drozdova VD and Kavardakova
NV: Phenomenon of the evolution of clonal chromosomal abnormalities
in childhood acute myeloid leukemia. Tsitol Genet. 44:41–52.
2010.(In Russian). PubMed/NCBI
|
|
30
|
Li M, Wen L, Cen J, Feng Y and Chen S:
JAK2V617F allele burden in patients with myeloproliferative
neoplasms carrying trisomy 9 and its relationship with clinical
phenotypes. Int J Hematol. 103:599–601. 2016. View Article : Google Scholar : PubMed/NCBI
|