|
1
|
Dienstmann R, Vermeulen L, Guinney J,
Kopetz S, Tejpar S and Tabernero J: Consensus molecular subtypes
and the evolution of precision medicine in colorectal cancer. Nat
Rev Cancer. 17:79–92. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Torre LA, Siegel RL, Ward EM and Jemal A:
Global cancer incidence and mortality rates and trends-an update.
Cancer Epidemiol Biomarkers Prev. 25:16–27. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
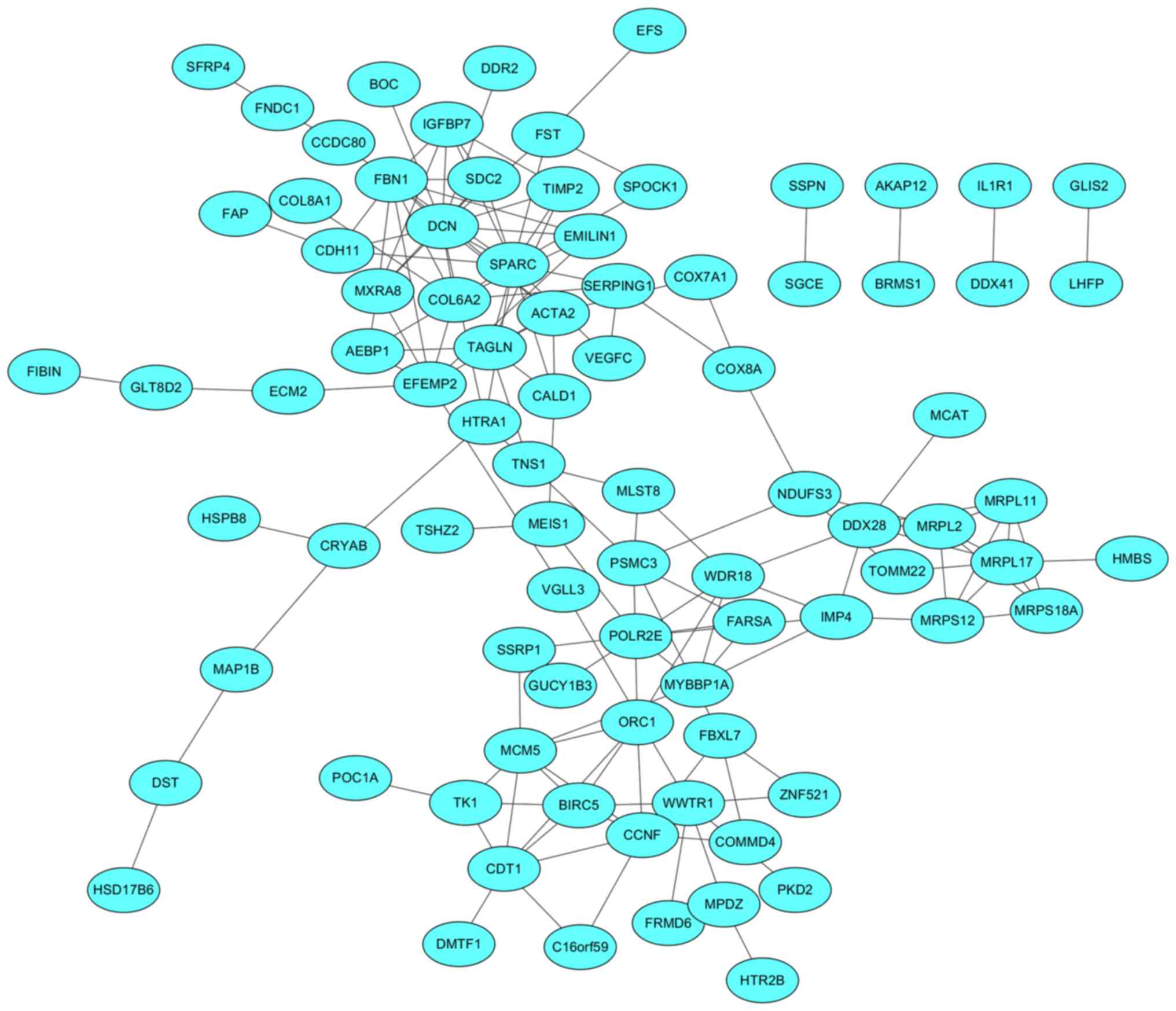
|
Veenstra CM and Krauss JC: Emerging
systemic therapies for colorectal cancer. Clin Colon Rectal Surg.
31:179–191. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Gonzalez N, Prieto I, Del Puerto-Nevado L,
Portal-Nuñez S, Ardura JA, Corton M, Fernández-Fernández B,
Aguilera O, Gomez-Guerrero C, Mas S, et al: 2017 update on the
relationship between diabetes and colorectal cancer: Epidemiology,
potential molecular mechanisms and therapeutic implications.
Oncotarget. 8:18456–18485. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Jayasekara H, English DR, Haydon A, Hodge
AM, Lynch BM, Rosty C, Williamson EJ, Clendenning M, Southey MC,
Jenkins MA, et al: Associations of alcohol intake, smoking,
physical activity and obesity with survival following colorectal
cancer diagnosis by stage, anatomic site and tumor molecular
subtype. Int J Cancer. 142:238–250. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lichtenstein P, Holm NV, Verkasalo PK,
Iliadou A, Kaprio J, Koskenvuo M, Pukkala E, Skytthe A and Hemminki
K: Environmental and heritable factors in the causation of
cancer-analyses of cohorts of twins from Sweden, Denmark, and
Finland. N Engl J Med. 343:78–85. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ballester V, Rashtak S and Boardman L:
Clinical and molecular features of young-onset colorectal cancer.
World J Gastroenterol. 22:1736–1744. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yeo H, Betel D, Abelson JS, Zheng XE,
Yantiss R and Shah MA: Early-onset colorectal cancer is distinct
from traditional colorectal cancer. Clin Colorectal Cancer.
16:293–299.e6. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Cavestro GM, Mannucci A, Zuppardo RA, Di
Leo M, Stoffel E and Tonon G: Early onset sporadic colorectal
cancer: Worrisome trends and oncogenic features. Dig Liver Dis.
50:521–532. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Burnett-Hartman AN, Powers JD, Chubak J,
Corley DA, Ghai NR, McMullen CK, Pawloski PA, Sterrett AT and
Feigelson HS: Treatment patterns and survival differ between
early-onset and late-onset colorectal cancer patients: The patient
outcomes to advance learning network. Cancer Causes Control.
30:747–755. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Williams SP, Barthorpe AS, Lightfoot H,
Garnett MJ and McDermott U: High-throughput RNAi screen for
essential genes and drug synergistic combinations in colorectal
cancer. Sci Data. 4:1701392017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gao M, Zhong A, Patel N, Alur C and Vyas
D: High throughput RNA sequencing utility for diagnosis and
prognosis in colon diseases. World J Gastroenterol. 23:2819–2825.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Xiong W, Ai YQ and Li YF, Ye Q, Chen ZT,
Qin JY, Liu QY, Wang H, Ju YH, Li WH and Li YF: Microarray analysis
of circular RNA expression profile associated with
5-fluorouracil-based chemoradiation resistance in colorectal cancer
cells. Biomed Res Int. 2017:84216142017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sun G, Li Y, Peng Y, Lu D, Zhang F, Cui X,
Zhang Q and Li Z: Identification of differentially expressed genes
and biological characteristics of colorectal cancer by integrated
bioinformatics analysis. J Cell Physiol. 2019.(Epub ahead of
print). View Article : Google Scholar
|
|
16
|
He M, Lin Y and Xu Y: Identification of
prognostic biomarkers in colorectal cancer using a long non-coding
RNA-mediated competitive endogenous RNA network. Oncol Lett.
17:2687–2694. 2019.PubMed/NCBI
|
|
17
|
Zhang H, Dong S and Feng J: Epigenetic
profiling and mRNA expression reveal candidate genes as biomarkers
for colorectal cancer. J Cell Biochem. 120:10767–10776. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yu C, Hong H, Zhang S, Zong Y, Ma J, Lu A,
Sun J and Zheng M: Identification of key genes and pathways
involved in microsatellite instability in colorectal cancer. Mol
Med Rep. 19:2065–2076. 2019.PubMed/NCBI
|
|
19
|
Marisa L, de Reynies A, Duval A, Selves J,
Gaub MP, Vescovo L, Etienne-Grimaldi MC, Schiappa R, Guenot D,
Ayadi M, et al: Gene expression classification of colon cancer into
molecular subtypes: Characterization, validation, and prognostic
value. PLoS Med. 10:e10014532013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Miller JA, Cai C, Langfelder P, Geschwind
DH, Kurian SM, Salomon DR and Horvath S: Strategies for aggregating
gene expression data: the collapseRows R function. BMC
Bioinformatics. 12:3222011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Giorgi FM, Bolger AM, Lohse M and Usadel
B: Algorithm-driven artifacts in median polish summarization of
microarray data. BMC Bioinformatics. 11:5532010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Langfelder P and Horvath S: WGCNA: An R
package for weighted correlation network analysis. BMC
Bioinformatics. 9:5592008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chung N, Zhang XD, Kreamer A, Locco L,
Kuan PF, Bartz S, Linsley PS, Ferrer M and Strulovici B: Median
absolute deviation to improve hit selection for genome-scale RNAi
screens. J Biomol Screen. 13:149–158. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Horvath S, Zhang B, Carlson M, Lu KV, Zhu
S, Felciano RM, Laurance MF, Zhao W, Qi S, Chen Z, et al: Analysis
of oncogenic signaling networks in glioblastoma identifies ASPM as
a molecular target. Proc Natl Acad Sci USA. 103:17402–17407. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Szklarczyk D, Franceschini A, Wyder S,
Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos
A, Tsafou KP, et al: STRING v10: Protein-protein interaction
networks, integrated over the tree of life. Nucleic Acids Res.
43:D447–D452. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Liu C, Chen N, Huang K, Jiang M, Liang H,
Sun Z, Tian J and Wang D: Identifying hub genes and potential
mechanisms associated with senescence in human annulus cells by
gene expression profiling and bioinformatics analysis. Mol Med Rep.
17:3465–3472. 2018.PubMed/NCBI
|
|
28
|
Kawakami H, Zaanan A and Sinicrope FA:
Microsatellite instability testing and its role in the management
of colorectal cancer. Curr Treat Options Oncol. 16:302015.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Connell LC, Mota JM, Braghiroli MI and
Hoff PM: The rising incidence of younger patients with colorectal
cancer: Questions about screening, biology, and treatment. Curr
Treat Options Oncol. 18:232017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ferlay J, Steliarova-Foucher E,
Lortet-Tieulent J, Rosso S, Coebergh JW, Comber H, Forman D and
Bray F: Cancer incidence and mortality patterns in Europe:
Estimates for 40 countries in 2012. Eur J Cancer. 49:1374–1403.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Siegel RL, Jemal A and Ward EM: Increase
in incidence of colorectal cancer among young men and women in the
United States. Cancer Epidemiol Biomarkers Prev. 18:1695–1698.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Stigliano V, Sanchez-Mete L, Martayan A
and Anti M: Early-onset colorectal cancer: A sporadic or inherited
disease? World J Gastroenterol. 20:12420–12430. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Vaz J, Ansari D, Sasor A and Andersson R:
SPARC: A potential prognostic and therapeutic target in pancreatic
cancer. Pancreas. 44:1024–1035. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Feng J and Tang L: SPARC in tumor
pathophysiology and as a potential therapeutic target. Curr Pharm
Des. 20:6182–6190. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Jing Y, Jin Y, Wang Y, Chen S, Zhang X,
Song Y, Wang Z, Pu Y, Ni Y and Hu Q: SPARC promotes the
proliferation and metastasis of oral squamous cell carcinoma by
PI3K/AKT/PDGFB/PDGFRβ axis. 2019.(Epub ahead of print). View Article : Google Scholar
|
|
36
|
Chen Y, Zhang Y, Tan Y and Liu Z: Clinical
significance of SPARC in esophageal squamous cell carcinoma.
Biochem Biophys Res Commun. 492:184–191. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Yu XZ, Guo ZY, Di Y, Yang F, Ouyang Q, Fu
DL and Jin C: The relationship between SPARC expression in primary
tumor and metastatic lymph node of resected pancreatic cancer
patients and patients' survival. Hepatobiliary Pancreat Dis Int.
16:104–109. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Chew A, Salama P, Robbshaw A, Klopcic B,
Zeps N, Platell C and Lawrance IC: SPARC, FOXP3, CD8 and CD45
correlation with disease recurrence and long-term disease-free
survival in colorectal cancer. PLoS One. 6:e220472011. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Neill T, Schaefer L and Iozzo RV: Decorin
as a multivalent therapeutic agent against cancer. Adv Drug Deliv
Rev. 97:174–185. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zhang W, Ge Y, Cheng Q, Zhang Q, Fang L
and Zheng J: Decorin is a pivotal effector in the extracellular
matrix and tumour microenvironment. Oncotarget. 9:5480–5491.
2018.PubMed/NCBI
|
|
41
|
Ho TH, Serie DJ, Parasramka M, Cheville
JC, Bot BM, Tan W, Wang L, Joseph RW, Hilton T, Leibovich BC, et
al: Differential gene expression profiling of matched primary renal
cell carcinoma and metastases reveals upregulation of extracellular
matrix genes. Ann Oncol. 28:604–610. 2017.PubMed/NCBI
|
|
42
|
Bi X, Pohl NM, Qian Z, Yang GR, Gou Y,
Guzman G, Kajdacsy-Balla A, Iozzo RV and Yang W: Decorin-mediated
inhibition of colorectal cancer growth and migration is associated
with E-cadherin in vitro and in mice. Carcinogenesis. 33:326–330.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Milewicz DM, Guo DC, Tran-Fadulu V, Lafont
AL, Papke CL, Inamoto S, Kwartler CS and Pannu H: Genetic basis of
thoracic aortic aneurysms and dissections: Focus on smooth muscle
cell contractile dysfunction. Annu Rev Genomics Hum Genet.
9:283–302. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Cierna Z, Mego M, Jurisica I, Machalekova
K, Chovanec M, Miskovska V, Svetlovska D, Kalavska K, Rejlekova K,
Kajo K, et al: Fibrillin-1 (FBN-1) a new marker of germ cell
neoplasia in situ. BMC Cancer. 16:5972016. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Yang D, Zhao D and Chen X: MiR-133b
inhibits proliferation and invasion of gastric cancer cells by
up-regulating FBN1 expression. Cancer Biomark. 19:425–436. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Guo Q, Song Y, Zhang H, Wu X, Xia P and
Dang C: Detection of hypermethylated fibrillin-1 in the stool
samples of colorectal cancer patients. Med Oncol. 30:6952013.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Wei J, Wang L, Zhu J, Sun A, Yu G, Chen M,
Huang P, Liu H, Shao G, Yang W and Lin Q: The Hippo signaling
effector WWTR1 is a metastatic biomarker of gastric cardia
adenocarcinoma. Cancer Cell Int. 19:742019. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Ruan H, Bao L, Song Z, Wang K, Cao Q, Tong
J, Cheng G, Xu T, Chen X, Liu D, et al: High expression of TAZ
serves as a novel prognostic biomarker and drives cancer
progression in renal cancer. Exp Cell Res. 376:181–191. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Wang Y, Han Y, Guo Z, Yang Y and Ren T:
Nuclear TAZ activity distinctly associates with subtypes of
non-small cell lung cancer. Biochem Biophys Res Commun.
509:828–832. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Yuen HF, McCrudden CM, Huang YH, Tham JM,
Zhang X, Zeng Q, Zhang SD and Hong W: TAZ expression as a
prognostic indicator in colorectal cancer. PLoS One. 8:e542112013.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Dos Santos Hidalgo G, Meola J, Rosa E
Silva JC, Paro de Paz CC and Ferriani RA: TAGLN expression is
deregulated in endometriosis and may be involved in cell invasion,
migration, and differentiation. Fertil Steril. 96:700–703. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Liu Y, Wu X, Wang G, Hu S, Zhang Y and
Zhao S: CALD1, CNN1, and TAGLN identified as potential prognostic
molecular markers of bladder cancer by bioinformatics analysis.
Medicine (Baltimore). 98:e138472019. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Li Q, Shi R, Wang Y and Niu X: TAGLN
suppresses proliferation and invasion, and induces apoptosis of
colorectal carcinoma cells. Tumor Biol. 34:505–513. 2013.
View Article : Google Scholar
|
|
54
|
Tu YT and Barrientos A: The Human
mitochondrial DEAD-box protein DDX28 resides in RNA granules and
functions in mitoribosome assembly. Cell Rep. 10:854–864. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Loo LW, Cheng I, Tiirikainen M, Lum-Jones
A, Seifried A, Dunklee LM, Church JM, Gryfe R, Weisenberger DJ,
Haile RW, et al: cis-Expression QTL analysis of established
colorectal cancer risk variants in colon tumors and adjacent normal
tissue. PLoS One. 7:e304772012. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Place RF, Li LC, Pookot D, Noonan EJ and
Dahiya R: MicroRNA-373 induces expression of genes with
complementary promoter sequences. Proc Natl Acad Sci USA.
105:1608–1613. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Vallejo G, Mestre-Citrinovitz AC,
Winterhager E and Saragueta PE: CSDC2, a cold shock domain
RNA-binding protein in decidualization. J Cell Physiol.
234:740–748. 2018. View Article : Google Scholar : PubMed/NCBI
|