Introduction
The completion of the Human Genome Project
illustrated that >90% of the human genome transcribes non-coding
RNAs (ncRNAs). It has been demonstrated that ncRNAs [including
microRNAs (miRNAs) and long non-coding RNAs (lncRNAs)] play a key
role in cancer progression. lncRNAs (non-coding RNA molecules
>200 nucleotides in length) are dysregulated and may function as
potential biomarkers of various malignancies, such as breast
(1), gastric (2), lung (3)
and prostate cancer (4). Moreover,
lncRNAs may act as oncogenes in certain types of cancer. For
example, small nucleolar RNA host gene 15 promotes colon cancer
progression by interacting with, and stabilizing snail family
transcriptional repressor 2 (5) and
differentiation antagonizing non-protein coding RNA (6), increasing the proliferative and
invasive capacities of gastric cancer cells. However, lncRNAs can
also act as tumor suppressors; for instance, lncRNA overexpressed
in colon carcinoma-1 suppressed colorectal cancer-cell
proliferation by destabilizing HuR protein (7), and maternally expressed 3 suppressed
liver cancer-cell proliferation through the inhibition of β-catenin
(8). Therefore, investigation into
the functional roles of lncRNAs in cancer may provide new insights
into the identification of novel diagnostic and therapeutic
targets.
Thyroid cancer (TC) is one of the most common
endocrine malignancies. In previous years, the functional roles of
several lncRNAs in TC have been revealed. Pvt1 oncogene (PVT1) was
the first lncRNA to be reported as having a functional role in TC
(9). Zhou et al (9) found that PVT1 contributed to TC
tumorigenesis through the recruitment of enhancer of zeste 2
polycomb repressive complex 2 subunit and the regulation of thyroid
stimulating hormone receptor expression. Furthermore, lncRNA CDKN2B
antisense RNA 1 was shown to promote TC metastasis through
modulation of the transforming growth factor-β/Smad signaling
pathway (10). lncRNAs were also
found to be involved in the prognosis of TC; lncRNA papillary
thyroid carcinoma susceptibility candidate 3 was identified as a
tumor suppressor gene in TC (11).
Moreover, low expression levels of growth arrest specific 5 were
found to be associated with poor prognosis in patients with TC
(12). High expression levels of
LINC01420 have been associated with poorer overall survival (OS) in
patients with nasopharyngeal carcinoma, and LINC01420-knockdown
inhibited nasopharyngeal carcinoma cell migration. However, the
functions and underlying molecular mechanisms of LINC01420 in TC
progression remain largely unknown.
The present study investigated whether LINC01420 was
of value as a biomarker for TC. The expression of LINC01420 was
evaluated by analyzing a dataset containing TC patient information
retrieved from The Cancer Genome Atlas (TCGA). Additionally,
bioinformatics analysis was performed to reveal the functional
roles of LINC01420 in TC progression. Finally, the effect of
LINC01420 on cell proliferation and cell cycle progression was
investigated. Improved understanding of the role of LINC01420 in TC
progression may indicate its use as either a biomarker, or a
potential therapeutic target.
Materials and methods
TCGA dataset retrieval and
analysis
The TCGA Thyroid carcinoma (THCA) dataset containing
specimens from both TC patients and disease-free subjects was
retrieved from TGCA (https://www.cbioportal.org/study/summary?id=thym_tcga),
and the LINC01420 expression levels were determined (13). In total, 502 PTC tissue samples and
58 normal thyroid tissue samples were included in the TCGA-THCA
dataset. Clinical information regarding LINC01420 was downloaded
using cBioPortal (http://www.cbioportal.org/). The tumor-node-metastasis
classification system (detailed in the American Joint committee on
Cancer Manual) was used to determine disease stage. The inclusion
criteria have been previously reported in TCGA.
Co-expression network, gene ontology
(GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway
analysis
Correlation between the expression of LINC01420 in
cancerous and disease-free tissues was calculated using the
Pearson's correlation coefficient in cBioPortal (http://www.cbioportal.org/). The co-expressed
LINC01420-mRNA pair with an absolute Pearson's correlation
coefficient ≥0.3 was selected for analysis. GO and KEGG pathway
analysis were used to predict the biological functions of LINC01420
using the MAS3.0 system (http://bioinfo.capitalbio.com/mas3/). P<0.05 was
considered to indicate a statistically significant difference.
Furthermore, key positively- and negatively-related gene-mediated
protein-protein interaction networks were identified using the
Search Tool for the Retrieval of Interacting Genes/Proteins
database (http://string.embl.de/) (14). Cytoscape software (version 3.6.1;
http://cytoscape.org/) is a free visualization
software (15). A confidence score
>0.4 was considered as the criterion of judgment.
Cell culture and transfection
All cell lines were obtained from the American Type
Culture Collection. CAL62 and SW579 cells were cultured in L-15
medium supplemented with 10% FBS (Gibco; Thermo Fisher Scientific,
Inc.) in a 37°C incubator with 5% CO2. The short
interfering (si)RNA for LINC01420 (5′-CAUCUCAGGUCUCUUGGCUUUGCCA-3′)
and an siRNA negative control were purchased from Guangzhou RiboBio
Co., Ltd. Cells were transfected with siRNAs (10 nM) using
Lipofectamine™ 3000 reagent (Thermo Fisher Scientific, Inc.).
Reverse transcription-quantitative
(RT-q)PCR analysis
Total RNA was extracted from transfected CAL62 and
SW579 cells using TRIzol® (Sigma-Aldrich; Merck KGaA)
reagent, according to the manufacturer's protocol. Total RNA was
then reverse transcribed into cDNA using the PrimeScript RT Master
Mix (Takara Bio, Inc.), according to the manufacturer's protocol.
qPCR was performed using the AceQ qPCR SYBR® Green
Master Mix (Vazyme) on a Roche LightCycler 480 according to the
manufacturer's protocol. The C value of β-actin was used as an
internal control to calibrate the Cq values of the genes of
interest, in order to determine the differential expression levels.
Relative RNA expression was calculated using the 2−ΔΔCq
method (16), and each sample was
run in triplicate.
Cell proliferation and cell cycle
distribution
The Cell counting kit (CCK)-8 assay (Dojindo
Molecular Technologies, Inc.) was used to detect cell proliferation
following transfection according to the manufacturer's protocol.
After 48 h transfection, cells were seeded in 96 well plate at the
density of 4×103 cells/100 µl per well, and
proliferation was detected at 0, 1, 2, 3 and 4 days. CCK-8 reagent
(10 µl medium/well) was added prior to detection and after
incubation for 1.5 h at 37°C, and the absorbance was measured at
450 nm using a microplate reader. Absorbance at 630 nm was used as
the control.
For the cell cycle assays, 3×105
cells/well were harvested from a 6 well plate and fixed in ice-cold
70% (v/v) ethanol for 24 h at 4°C. The cell pellet was collected
following centrifugation at 500 × g for 10 min at 4°C and
resuspended in PBS. Cells were then stained with a mixture of RNase
(10 µg/ml) and propidium iodide (50 µg/ml; Beyotime Institute of
Biotechnology) in sodium citrate containing 0.5% Triton X-100 for
20 min, in the dark and at room temperature. The cells were
subsequently analyzed using a flow cytometer (Gallios, Beckman
Coulter, Inc.). The ModFit software version 4.0 (Verity Software
House, Inc.) was used for data analysis.
Statistical analysis
Statistical analysis was performed using the SPSS
software package, version 15.0 (SPSS, Inc.). Significant
differences between two groups were compared using two-tailed
Student's t-test. Statistical comparisons between two paired groups
was performed using a paired t-test. For comparison of >2
groups, one-way ANOVA was used, followed by the Newman-Keuls post
hoc test. Kaplan-Meier and Cox regression analyses were used to
evaluate the association between LINC01420 and disease-free
survival (DFS), as well as the prognosis of patients with TC.
P<0.05 was considered to indicate a statistically significant
difference.
Results
LINC01420 is overexpressed in TC
tissue samples
The TCGA-THCA dataset was analyzed to evaluate the
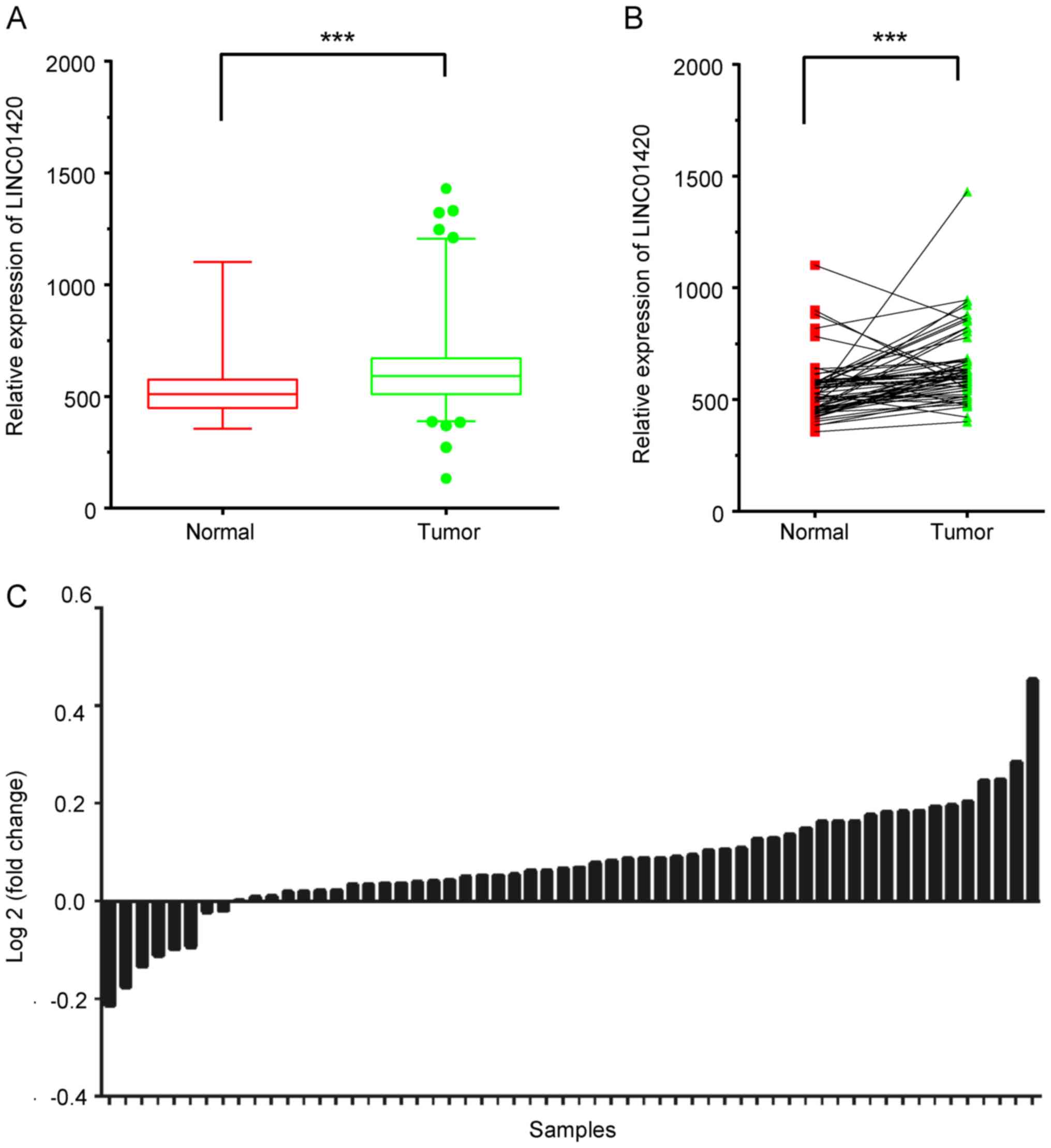
expression levels of LINC01420 in TC. The data suggested that the
expression of LINC01420 was significantly higher in TC tissues,
compared with that in normal tissue samples (Fig. 1A). Furthermore, LINC01420 expression
was analyzed using 50 paired TC samples from TCGA dataset. Of these
paired samples, 80% exhibited a higher expression level of
LINC01420 in TC samples, compared with the adjacent normal tissues
(Fig. 1B and C). The association of
LINC01420 expression levels with clinicopathological data was also
analyzed (Table SI). The results of
these analyses suggest that LINC01420 may play a regulatory role in
TC.
Co-expression analysis of LINC01420 in
TC
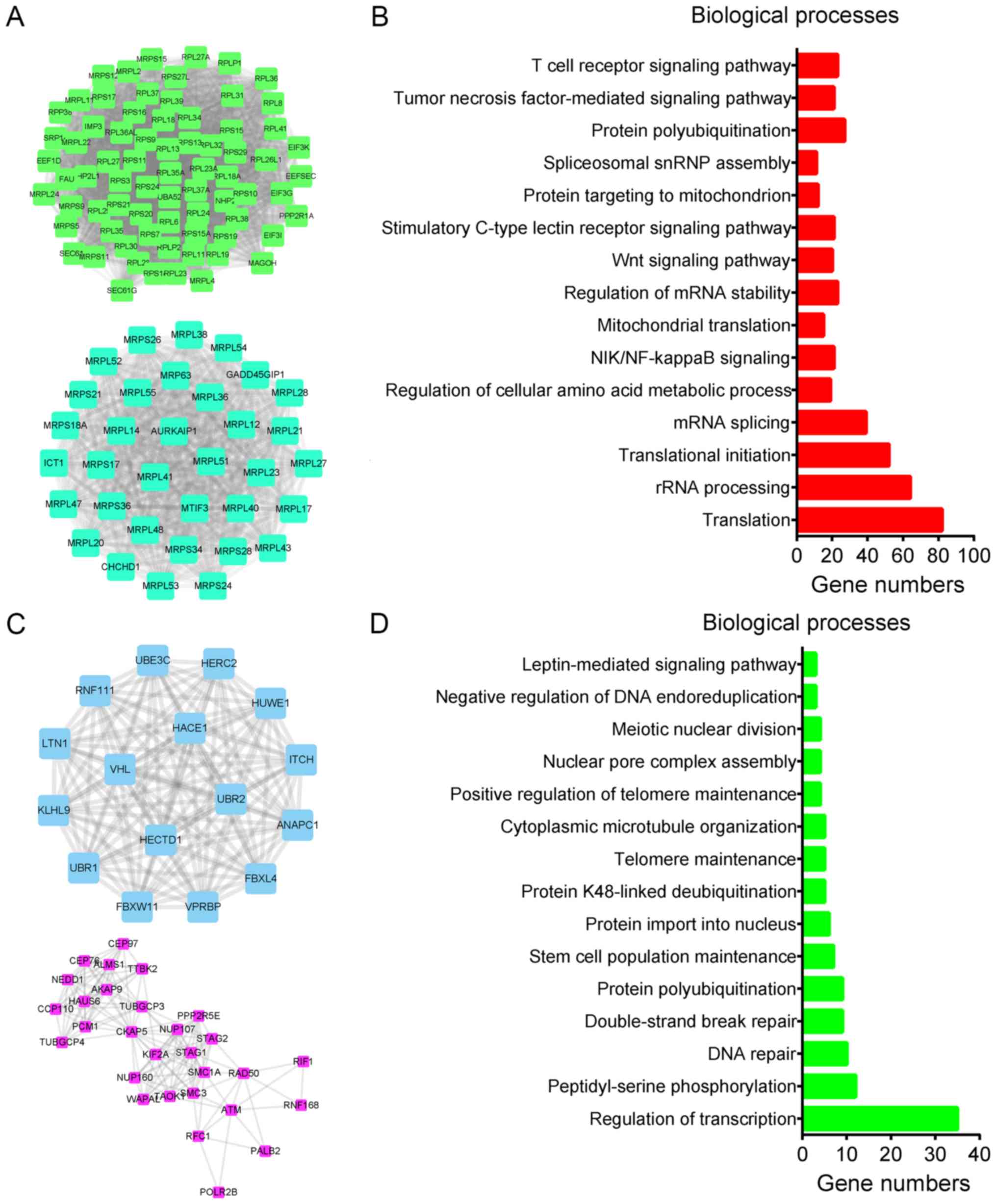
Co-expression analysis is widely used to explore the
potential roles of lncRNAs in human disease. Considering that
LINC01420 is a novel lncRNA implicated in TC, a LINC01420-mediated
co-expression network was constructed to identify its potential
mechanism of action. A Pearson's correlation coefficient value of
≥0.50 was selected as the cut-off for the identification of
reliable LINC01420-mRNA pairs. A total of 948 mRNAs were positively
co-expressed and 568 mRNAs were negatively co-expressed in this
network. The two largest hub networks of positively and negatively
co-expressed mRNAs are shown in Fig. 2A
and C, respectively.
Furthermore, key positively- and negatively-related
gene-mediated protein-protein interaction networks were identified
using the Search Tool for the Retrieval of Interacting
Genes/Proteins database. GO analysis revealed that genes positively
related to LINC01420 were significantly associated with
‘translation’, ‘rRNA processing’, ‘translational initiation’, ‘mRNA
splicing’, ‘regulation of cellular amino acid metabolic processes’,
‘NIK/NF-κB signaling’, ‘mitochondrial translation’, ‘regulation of
mRNA stability’, ‘Wnt signaling pathway’, ‘stimulatory C-type
lectin receptor signaling pathway’, ‘protein targeting to
mitochondrion’, ‘spliceosomal snRNP assembly’, ‘protein
polyubiquitination’, ‘TNF-α-mediated signaling pathway’ and ‘T-cell
receptor signaling pathway’ (Fig.
2B). Genes negatively associated with LINC01420 were
significantly involved in ‘regulation of transcription’,
‘peptidyl-serine phosphorylation’, ‘DNA repair’, ‘double-strand
break repair’, ‘protein polyubiquitination’, ‘stem cell population
maintenance’, ‘protein import into nucleus’, ‘protein K48-linked
deubiquitination’, ‘telomere maintenance’ and ‘cytoplasmic
microtubule organization’ (Fig.
2D).
Knockdown of LINC01420 inhibits cell
proliferation in TC
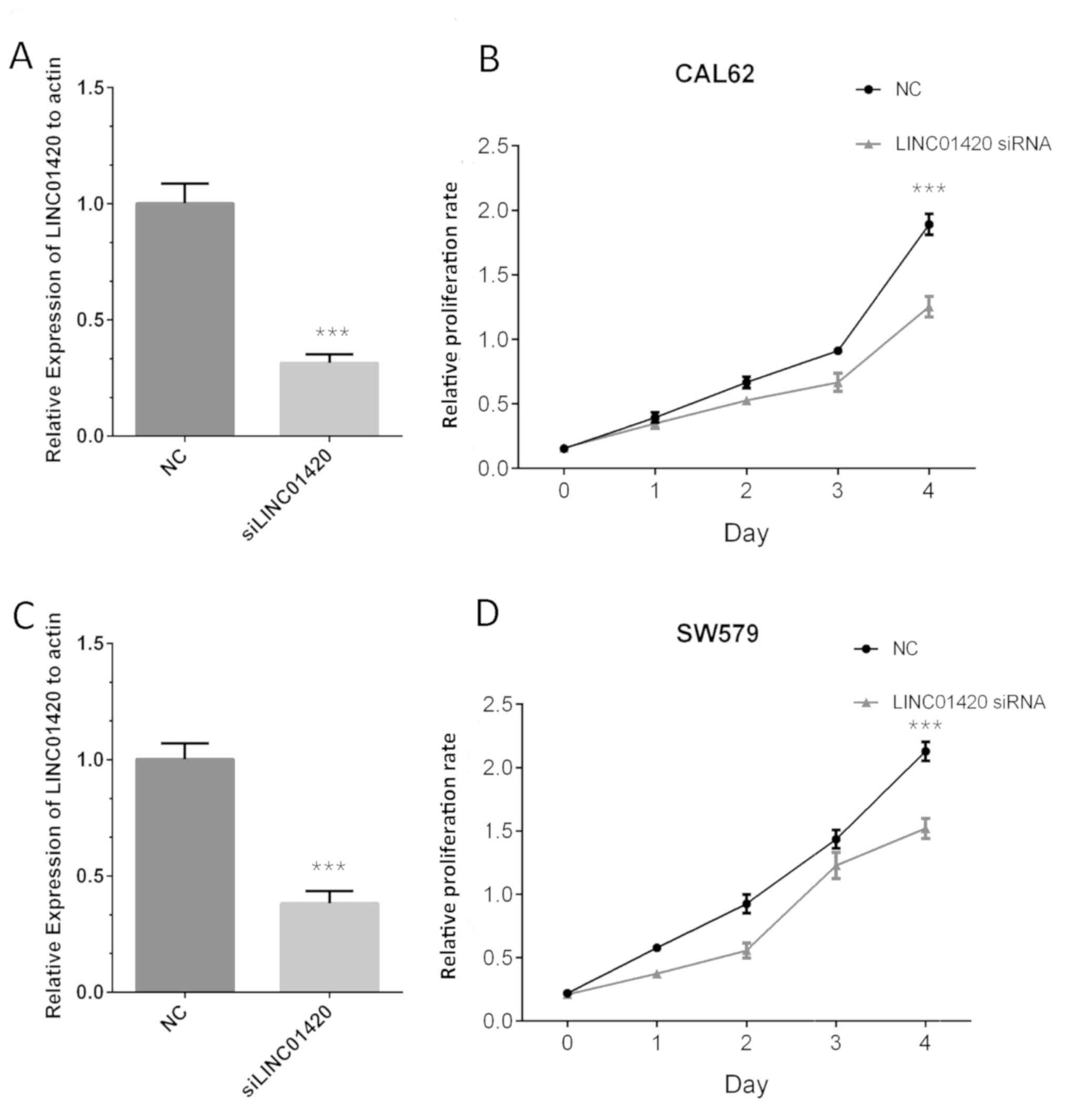
The CCK-8 was used to evaluate the functional roles
of LINC01420 in TC. Following transfection, the expression of
LINC01420 was found to be significantly downregulated in the
LINC01420-knockdown group compared with that in the control group,
in both CAL62 (Fig. 3A) and SW579
cells (Fig. 3C). The results of the
CCK-8 assay indicated that compared with the control group,
LINC01420 silencing significantly suppressed CAL62 (Fig. 3B) and SW579 (Fig. 3D) cell proliferation at 72 h.
LINC01420-knockdown inhibits cell
cycle progression in TC
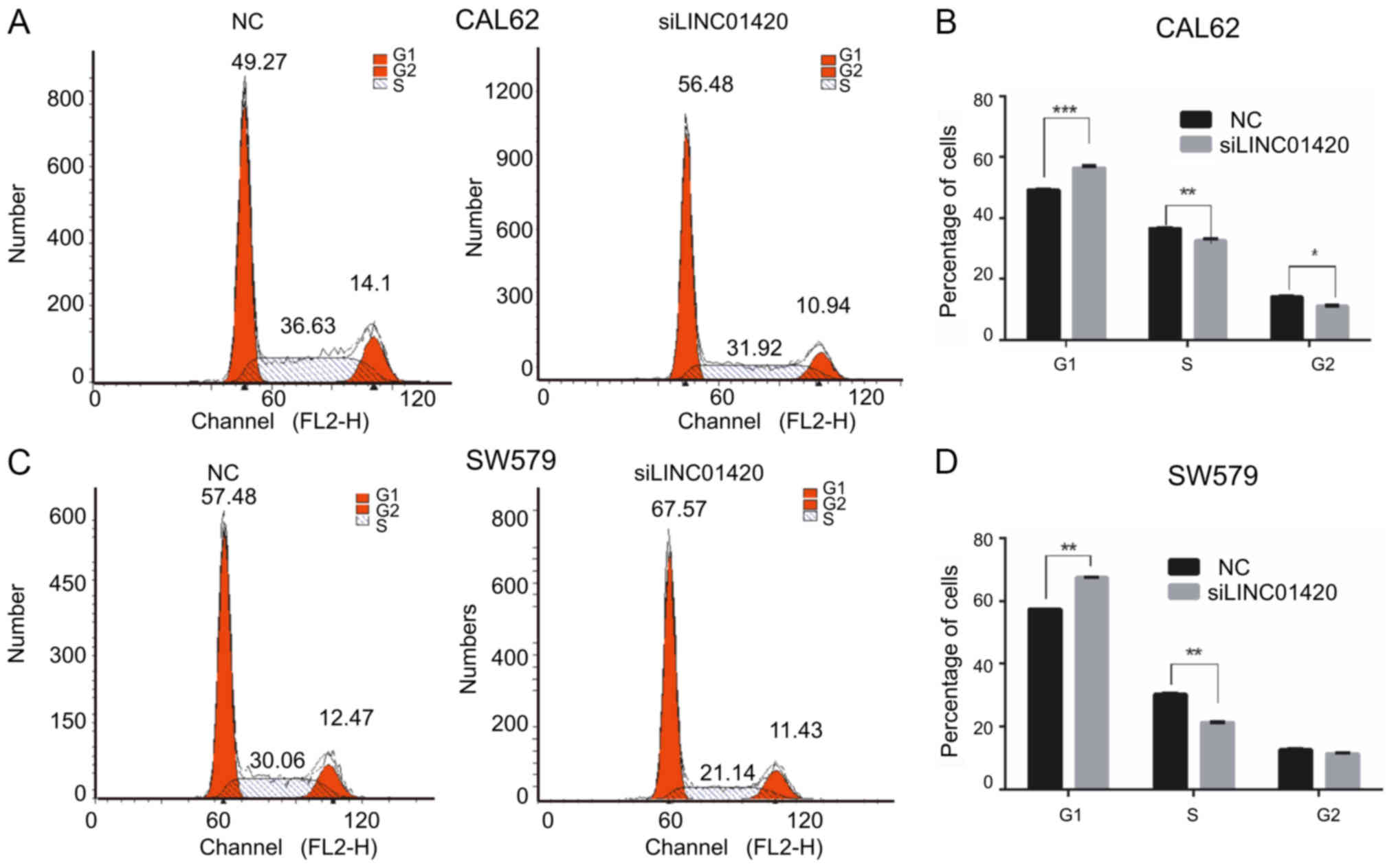
The effect of LINC01420 on the cell cycle was also
investigated. Flow cytometry demonstrated that suppressing
LINC01420 expression modulated the cell cycle by inducing
G0/G1 arrest (compared with the control
groups) in CAL62 (Fig. 4A-B) and
SW579 cells (Fig. 4C-D).
Discussion
lncRNAs have been identified as important regulators
of cancer progression, binding to DNA, RNA and proteins to regulate
epigenetic modification and protein and gene post-transcriptional
regulation (17). lncRNAs act as
both oncogenes and tumor suppressor genes; for example,
epigenetically-induced lncRNA1 was reported to be oncogenic,
promoting cell cycle progression by interacting with the MYC
proto-oncogene (18). Conversely,
testis associated oncogenic lncRNA promotes cancer progression and
mRNA stabilization by interacting with insulin-like growth factor 2
mRNA binding protein 1 (19).
Moreover, certain lncRNAs, such as X-inactive specific transcript,
nuclear paraspeckle assembly transcript 1 and HOX transcript
antisense RNA, were found to act as miRNA molecular sponges, which
are implicated in human cancer progression (20–24). The
current study investigated the role of LINC01420 in TC progression.
LINC01420 expression was previously reported to be upregulated in
nasopharyngeal carcinoma, and LINC01420-knockdown inhibited
nasopharyngeal carcinoma-cell migration (25). In the present study, a
loss-of-function assay was performed and revealed that
LINC01420-knockdown inhibited TC cell proliferation by arresting
cell cycle progression. Taken together, these findings provide
evidence to support the oncogenic nature of LINC01420.
TC is a common endocrine malignancy; however no
sensitive biomarkers are currently available for TC diagnosis.
Previous studies have primarily focused on identifying diagnostic
and therapeutic targets for TC. For example, Read et al
(26) reported that higher PTTG1
regulator of sister chromatid separation, securin and zinc finger
protein 395 expression predicted poor patient outcomes in TC.
Downregulation of serum dickkopf WNT signaling pathway inhibitor 1
was also found to be associated with a poor prognosis in PTC
patients (27). Furthermore, lncRNAs
are often dysregulated in TC; NEAT1_2 (28), TNRC6C-AS1 (29), CNALPTC1 (30) and AFAP1-AS1 (31) were found to be overexpressed in TC.
However, GAS8-AS1 (32), GAS5
(12) and BANCR (33) were all found to be downregulated. To
the best of our knowledge, the present study is the first to
demonstrate the upregulation of LINC01420 in TC compared with
normal tissues, and to determine a correlation between the
aforementioned upregulation and poor prognosis. Furthermore, high
LINC01420 expression was also associated with shorter DFS time.
Thus the results of the present study suggest that LINC01420 may
play a regulatory role in TC development, and consequently, may be
a useful biomarker.
The functional roles and underlying molecular
mechanisms of LINC01420 in TC progression remain largely unclear.
Consequently, GO and KEGG pathway analysis were also performed in
the present study. The findings demonstrated that LINC01420 was
significantly associated with translation, rRNA processing,
translational initiation, mRNA splicing, regulation of
transcription, DNA repair and double-strand break repair.
In conclusion, the present study determined that
LINC01420 is implicated in proliferation and cell cycle progression
in TC. It was also observed that LINC01420 expression was
upregulated in human TC tissues. Bioinformatics analyses
demonstrated that LINC01420 was associated with translation, rRNA
processing, translational initiation, mRNA splicing, regulation of
transcription, DNA repair and double-strand break repair.
Therefore, the results of the present study indicate that LINC01420
may serve as a potential therapeutic target, and that increased
LINC01420 levels may be used as a novel prognostic biomarker for
TC.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets generated and/or analyzed during the
current study are available in the TCGA-THCA repository (https://www.cbioportal.org/study/summary?id=thym_tcga).
Authors' contributions
Experimental conception and design were conducted by
JZL and LJZ. JZL, LQ and LJZ developed the methodology, and
analyzed and interpreted the data. JZL, LQ and LJZ wrote, reviewed
and revised the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
lncRNAs
|
long non-coding RNAs
|
|
TC
|
thyroid cancer
|
|
ncRNA
|
non-coding RNAs
|
|
miRNA
|
microRNAs
|
|
TCGA
|
The Cancer Genome Atlas
|
|
PTC
|
papillary thyroid carcinoma
|
|
GO
|
gene ontology
|
|
KEGG
|
Kyoto Encyclopedia of Genes and
Genomes
|
References
|
1
|
Lv M, Xu P, Wu Y, Huang L, Li W, Lv S, Wu
X, Zeng X, Shen R, Jia X, et al: LncRNAs as new biomarkers to
differentiate triple negative breast cancer from non-triple
negative breast cancer. Oncotarget. 7:13047–13059. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zhou X, Yin C, Dang Y, Ye F and Zhang G:
Identification of the long non-coding RNA H19 in plasma as a novel
biomarker for diagnosis of gastric cancer. Sci Rep. 5:115162015.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wang HM, Lu JH, Chen WY and Gu AQ:
Upregulated lncRNA-UCA1 contributes to progression of lung cancer
and is closely related to clinical diagnosis as a predictive
biomarker in plasma. Int J Clin Exp Med. 8:11824–11830.
2015.PubMed/NCBI
|
|
4
|
Crea F, Watahiki A, Quagliata L, Xue H,
Pikor L, Parolia A, Wang Y, Lin D, Lam WL, Farrar WL, et al:
Identification of a long non-coding RNA as a novel biomarker and
potential therapeutic target for metastatic prostate cancer.
Oncotarget. 5:764–774. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Jiang H, Li T, Qu Y, Wang X, Li B, Song J,
Sun X, Tang Y, Wan J, Yu Y, et al: Long non-coding RNA SNHG15
interacts with and stabilizes transcription factor Slug and
promotes colon cancer progression. Cancer Lett. 425:78–87. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Mao Z, Li H, Du B, Cui K, Xing Y, Zhao X
and Zai S: LncRNA DANCR promotes migration and invasion through
suppression of lncRNA-LET in gastric cancer cells. Biosci Rep.
37(pii): BSR201710702017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Pibouin L, Villaudy J, Ferbus D, Muleris
M, Prosperi MT, Remvikos Y and Goubin G: Cloning of the mRNA of
overexpression in colon carcinoma-1: A sequence overexpressed in a
subset of colon carcinomas. Cancer Genet Cytogenet. 133:55–60.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
He Y, Wu YT, Huang C, Meng XM, Ma TT, Wu
BM, Xu FY, Zhang L, Lv XW and Li J: Inhibitory effects of long
noncoding RNA MEG3 on hepatic stellate cells activation and liver
fibrogenesis. Biochim Biophys Acta. 1842:2204–2215. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhou Q, Chen J, Feng J and Wang J: Long
noncoding RNA PVT1 modulates thyroid cancer cell proliferation by
recruiting EZH2 and regulating thyroid-stimulating hormone receptor
(TSHR). Tumour Biol. 37:3105–3113. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhao JJ, Hao S, Wang LL, Hu CY, Zhang S,
Guo LJ, Zhang G, Gao B, Jiang Y, Tian WG and Luo DL: Long
non-coding RNA ANRIL promotes the invasion and metastasis of
thyroid cancer cells through TGF-β/Smad signaling pathway.
Oncotarget. 7:57903–57918. 2016.PubMed/NCBI
|
|
11
|
Fan M, Li X, Jiang W, Huang Y, Li J and
Wang Z: A long non-coding RNA, PTCSC3, as a tumor suppressor and a
target of miRNAs in thyroid cancer cells. Exp Ther Med.
5:1143–1146. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Guo LJ, Zhang S, Gao B, Jiang Y, Zhang XH,
Tian WG, Hao S, Zhao JJ, Zhang G, Hu CY, et al: Low expression of
long non-coding RNA GAS5 is associated with poor prognosis of
patients with thyroid cancer. Exp Mol Pathol. 102:500–504. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Cancer Genome Atlas Research Network, .
Integrated genomic characterization of papillary thyroid carcinoma.
Cell. 159:676–690. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Szklarczyk D, Franceschini A, Wyder S,
Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos
A, Tsafou KP, et al: STRING v10: Protein-protein interaction
networks, integrated over the tree of life. Nucleic Acids Res.
43:D447–D452. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Smoot ME, Ono K, Ruscheinski J, Wang PL
and Ideker T: Cytoscape 2.8: New features for data integration and
network visualization. Bioinformatics. 27:431–432. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Schmitt AM and Chang HY: Long noncoding
RNAs in cancer pathways. Cancer Cell. 29:452–463. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang Z, Yang B, Zhang M, Guo W, Wu Z, Wang
Y, Jia L, Li S; Cancer Genome Atlas Research Network, ; Xie W and
Yang D: lncRNA epigenetic landscape analysis identifies EPIC1 as an
oncogenic lncRNA that interacts with MYC and promotes cell-cycle
progression in cancer. Cancer Cell. 33:706–720.e9. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chen W, Chen M, Xu Y, Chen X, Zhou P, Zhao
X, Pang F and Liang W: Long non-coding RNA THOR promotes human
osteosarcoma cell growth in vitro and in vivo. Biochem Biophys Res
Commun. 499:913–919. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chen DL, Ju HQ, Lu YX, Chen LZ, Zeng ZL,
Zhang DS, Luo HY, Wang F, Qiu MZ, Wang DS, et al: Long non-coding
RNA XIST regulates gastric cancer progression by acting as a
molecular sponge of miR-101 to modulate EZH2 expression. J Exp Clin
Cancer Res. 35:1422016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kong Q, Zhang S, Liang C, Zhang Y, Kong Q,
Chen S, Qin J and Jin Y: LncRNA XIST functions as a molecular
sponge of miR-194-5p to regulate MAPK1 expression in hepatocellular
carcinoma cell. J Cell Biochem. 119:4458–4468. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhang J, Li Y, Dong M and Wu D: Long
non-coding RNA NEAT1 regulates E2F3 expression by competitively
binding to miR-377 in non-small cell lung cancer. Oncol Lett.
14:4983–4988. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Liu XH, Sun M, Nie FQ, Ge YB, Zhang EB,
Yin DD, Kong R, Xia R, Lu KH, Li JH, et al: Lnc RNA HOTAIR
functions as a competing endogenous RNA to regulate HER2 expression
by sponging miR-331-3p in gastric cancer. Mol Cancer. 13:922014.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ren K, Li Y, Lu H, Li Z, Li Z, Wu K, Li Z
and Han X: Long noncoding RNA HOTAIR controls cell cycle by
functioning as a competing endogenous RNA in esophageal squamous
cell carcinoma. Transl Oncol. 9:489–497. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yang L, Tang Y, He Y, Wang Y, Lian Y,
Xiong F, Shi L, Zhang S, Gong Z, Zhou Y, et al: High expression of
LINC01420 indicates an unfavorable prognosis and modulates cell
migration and invasion in nasopharyngeal carcinoma. J Cancer.
8:97–103. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Read ML, Fong JC, Modasia B, Fletcher A,
Imruetaicharoenchoke W, Thompson RJ, Nieto H, Reynolds JJ, Bacon A,
Mallick U, et al: Elevated PTTG and PBF predicts poor patient
outcome and modulates DNA damage response genes in thyroid cancer.
Oncogene. 36:5296–5308. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhao YP, Wang W, Wang XH, Xu Y, Wang Y,
Dong ZF and Zhang JJ: Downregulation of serum DKK-1 predicts poor
prognosis in patients with papillary thyroid cancer. Genet Mol Res.
14:18886–18894. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Sun W, Lan X, Zhang H, Wang Z, Dong W, He
L, Zhang T, Zhang P, Liu J and Qin Y: NEAT1_2 functions as a
competing endogenous RNA to regulate ATAD2 expression by sponging
microRNA-106b-5p in papillary thyroid cancer. Cell Death Dis.
9:3802018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hou S, Lin Q, Guan F and Lin C: LncRNA
TNRC6C-AS1 regulates UNC5B in thyroid cancer to influence cell
proliferation, migration and invasion as a competing endogenous RNA
of miR-129-5p. J Cell Biochem. 119:8304–8316. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chen C, Zhou L, Wang H, Chen J, Li W, Liu
W, Shen M, Liu H and Fu X: Long noncoding RNA CNALPTC1 promotes
cell proliferation and migration of papillary thyroid cancer via
sponging miR-30 family. Am J Cancer Res. 8:192–206. 2018.PubMed/NCBI
|
|
31
|
Dai W, Tian Y, Jiang B and Chen W:
Down-regulation of long non-coding RNA AFAP1-AS1 inhibits tumor
growth, promotes apoptosis and decreases metastasis in thyroid
cancer. Biomed Pharmacother. 99:191–197. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhang D, Liu X, Wei B, Qiao G, Jiang T and
Chen Z: Plasma lncRNA GAS8-AS1 as a potential biomarker of
papillary thyroid carcinoma in Chinese patients. Int J Endocrinol.
2017:26459042017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Liao T, Qu N, Shi RL, Guo K, Ma B, Cao YM,
Xiang J, Lu ZW, Zhu YX, Li DS and Ji QH: BRAF-activated LncRNA
functions as a tumor suppressor in papillary thyroid cancer.
Oncotarget. 8:238–247. 2017.PubMed/NCBI
|