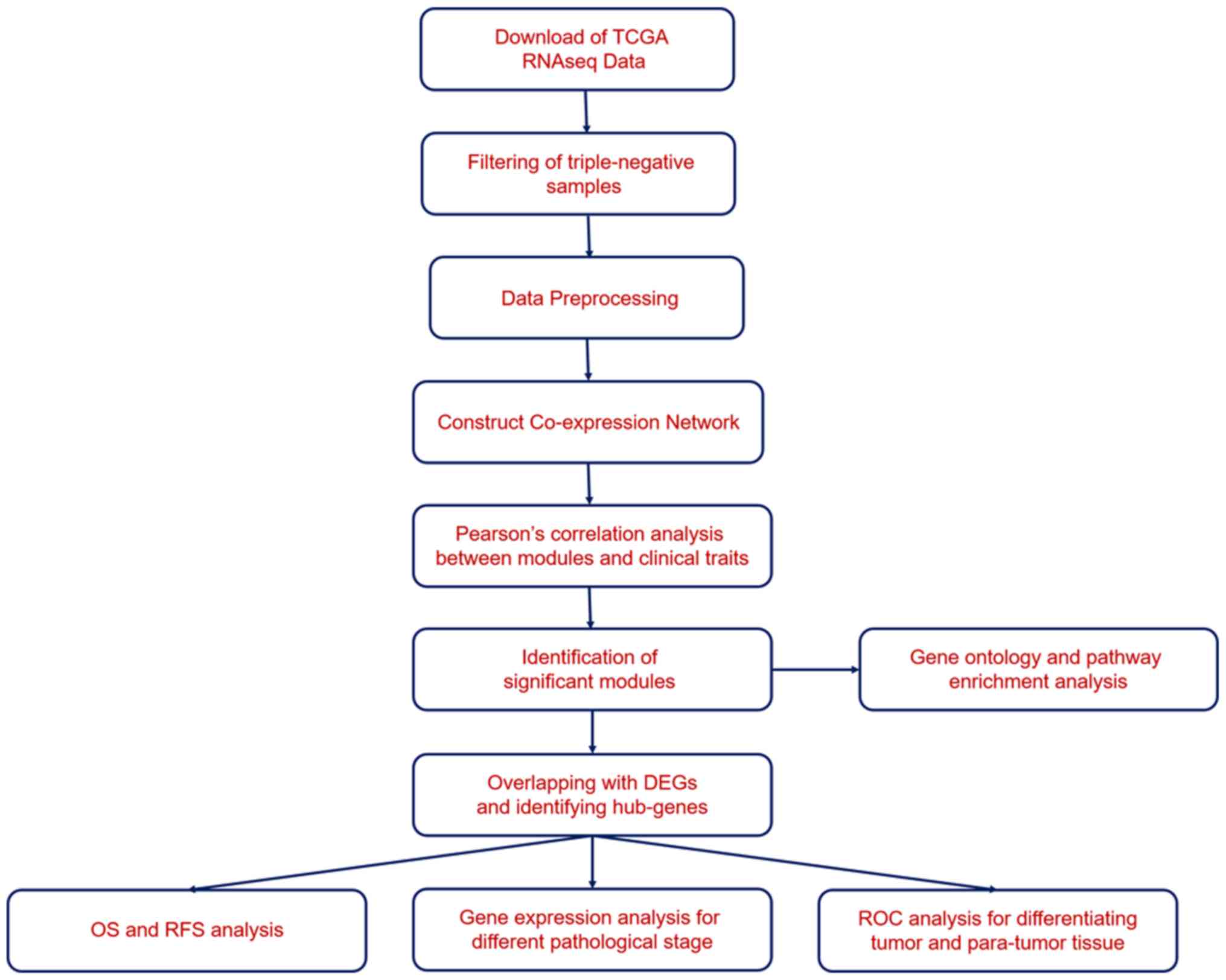
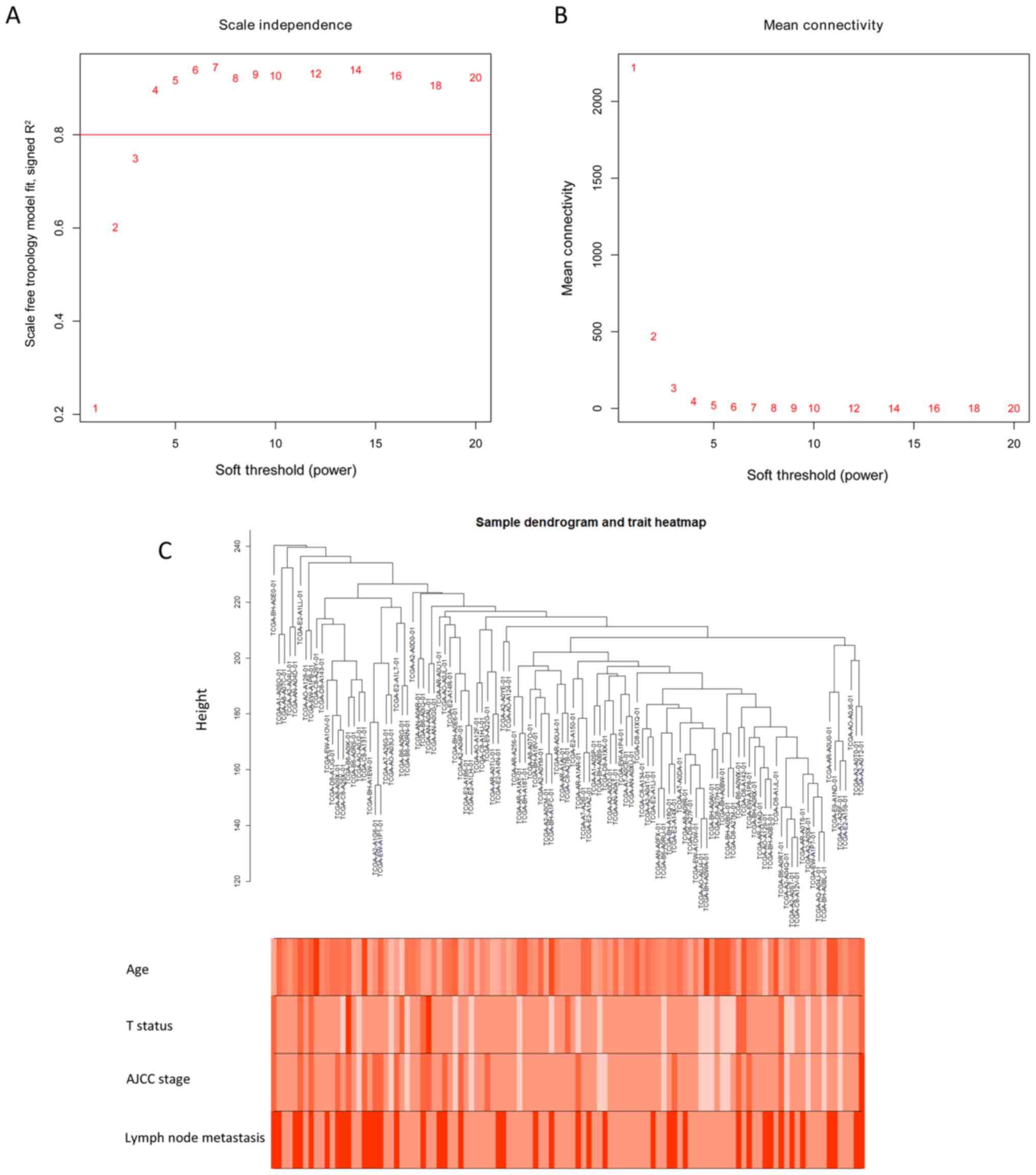
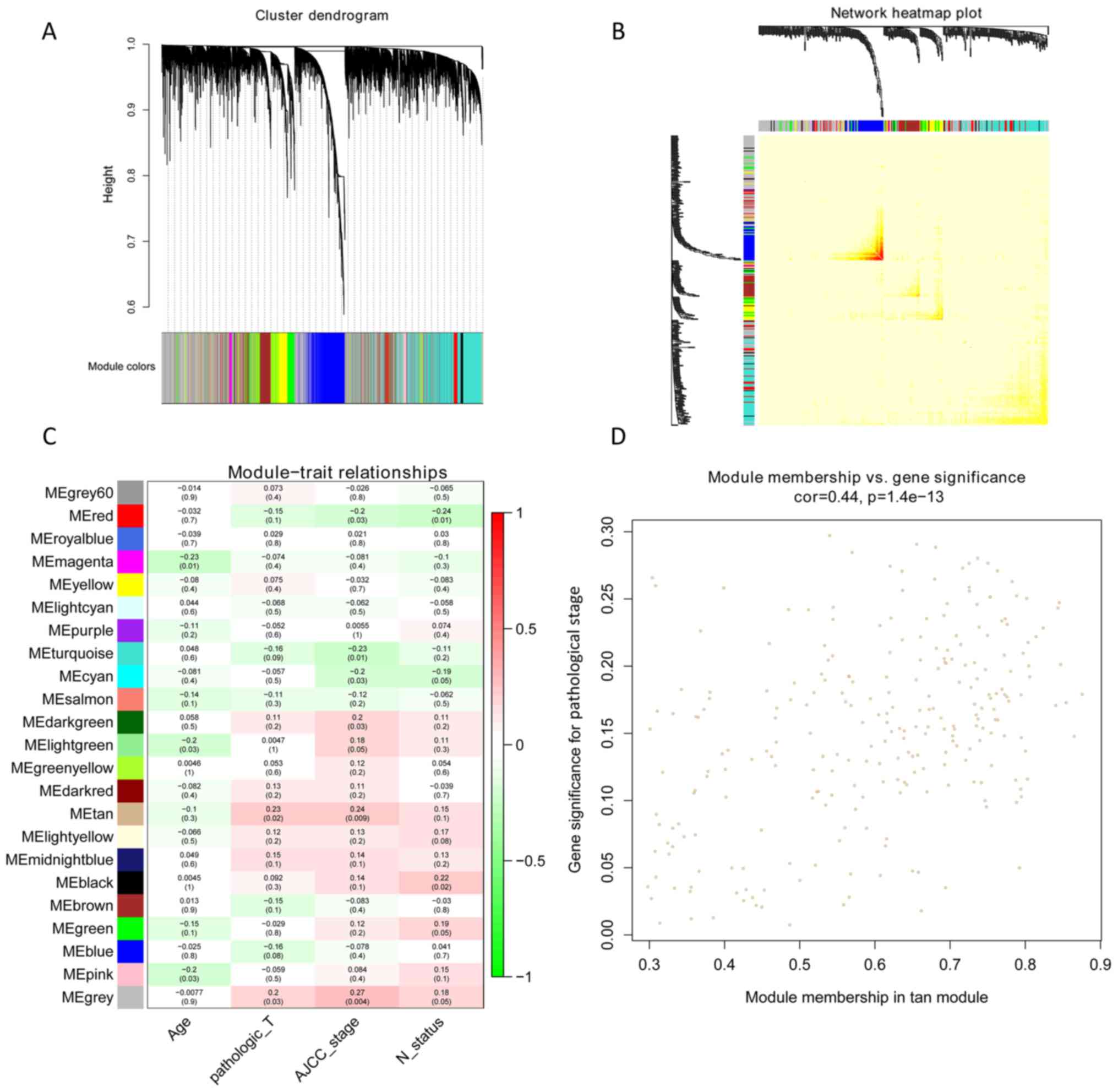
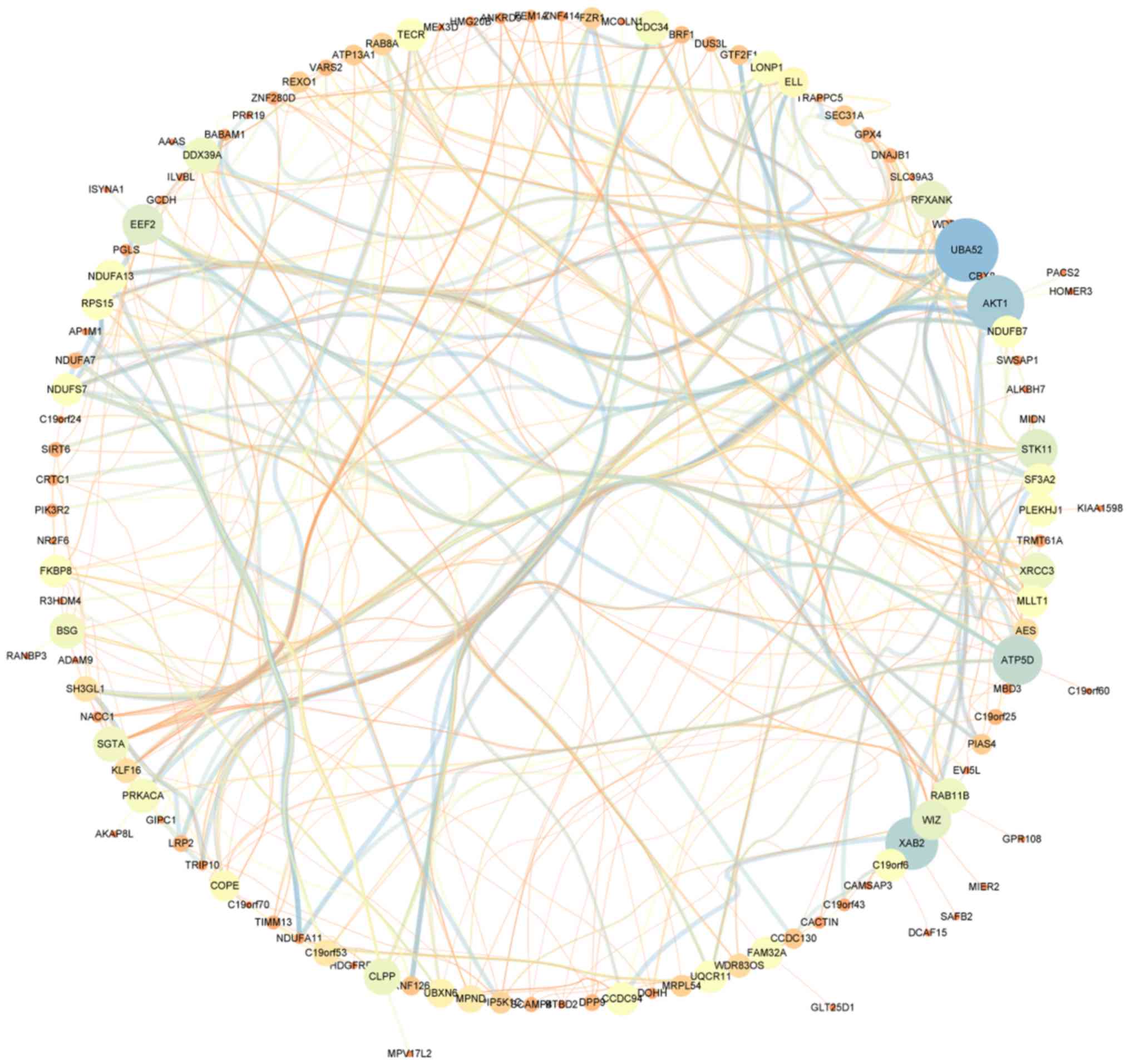
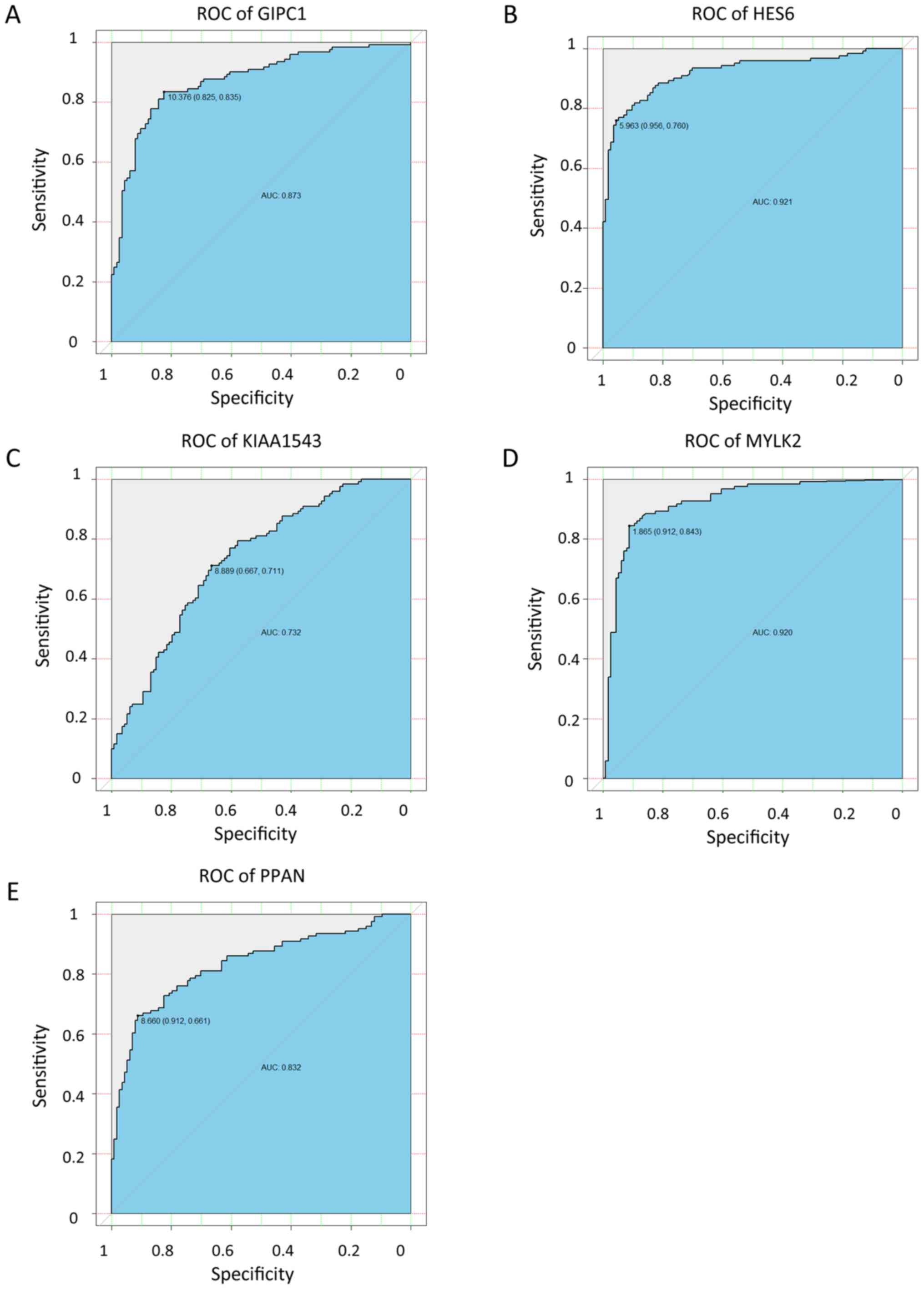
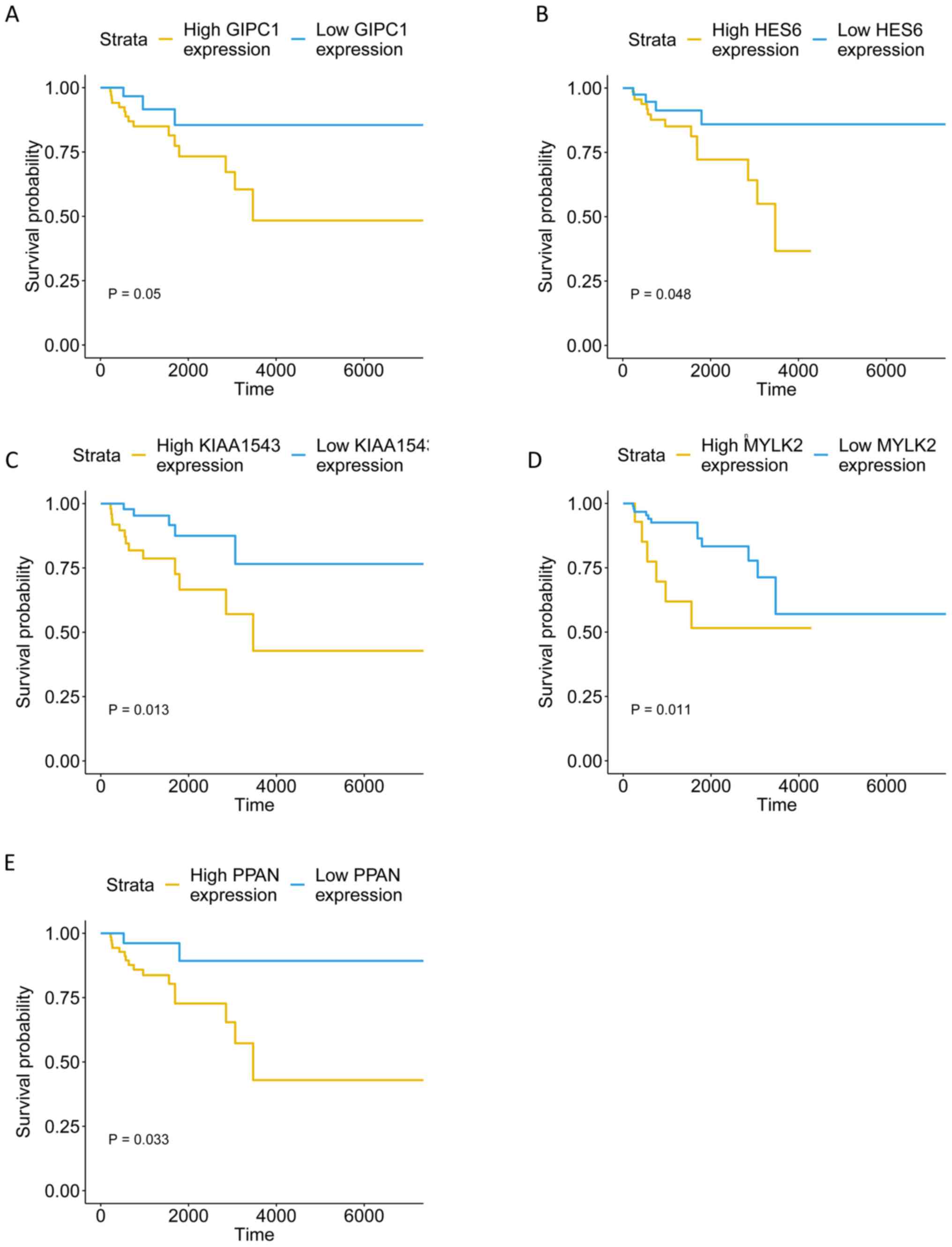
|
1
|
Foulkes WD, Smith IE and Reis-Filho JS:
Triple-negative breast cancer. N Engl J Med. 363:1938–1948. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Dawood S: Triple-negative breast cancer:
Epidemiology and management options. Drugs. 70:2247–2258. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Dent R, Trudeau M, Pritchard KI, Hanna WM,
Kahn HK, Sawka CA, Lickley LA, Rawlinson E, Sun P and Narod SA:
Triple-negative breast cancer: Clinical features and patterns of
recurrence. Clin Cancer Res. 13:4429–4434. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Liedtke C, Mazouni C, Hess KR, André F,
Tordai A, Mejia JA, Symmans WF, Gonzalez-Angulo AM, Hennessy B,
Green M, et al: Response to neoadjuvant therapy and long-term
survival in patients with triple-negative breast cancer. J Clin
Oncol. 26:1275–1281. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Langfelder P and Horvath S: WGCNA: An R
package for weighted correlation network analysis. BMC
Bioinformatics. 9:5592008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wang Y, Zhang Q, Gao Z, Xin S, Zhao Y,
Zhang K, Shi R and Bao X: A novel 4-gene signature for overall
survival prediction in lung adenocarcinoma patients with lymph node
metastasis. Cancer Cell Int. 19:1002019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Huang K, Maruyama T and Fan G: The naive
state of human pluripotent stem cells: A synthesis of stem cell and
preimplantation embryo transcriptome analyses. Cell Stem Cell.
15:410–415. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wang Y, Xin S, Zhang K, Shi R and Bao X:
Low GAS5 levels as a predictor of poor survival in patients with
lower-grade gliomas. J Oncol. 2019:17850422019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Smyth GK: Limma: linear models for
microarray data. Bioinformatics and computational biology solutions
using R and BioconductorStatistics for Biology and Health.
Gentleman R..Carey V.J..Huber W..Irizarry R.A..Dudoit S.: Springer;
New York, NY: pp. 397–420. 2005, View Article : Google Scholar
|
|
10
|
Kassambara A, Kosinski M and Biecek P:
Survminer: Drawing Survival Curves using ‘ggplot2’. R package
version 03. 2017 1;
|
|
11
|
Haffty BG, Yang Q, Reiss M, Kearney T,
Higgins SA, Weidhaas J, Harris L, Hait W and Toppmeyer D:
Locoregional relapse and distant metastasis in conservatively
managed triple negative early-stage breast cancer. J Clin Oncol.
24:5652–5657. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Atchley DP, Albarracin CT, Lopez A, Valero
V, Amos CI, Gonzalez-Angulo AM, Hortobagyi GN and Arun BK: Clinical
and pathologic characteristics of patients with BRCA-positive and
BRCA-negative breast cancer. J Clin Oncol. 26:4282–4288. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Cleator S, Heller W and Coombes RC:
Triple-negative breast cancer: Therapeutic options. Lancet Oncol.
8:235–244. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Powell SN and Kachnic LA: Roles of BRCA1
and BRCA2 in homologous recombination, DNA replication fidelity and
the cellular response to ionizing radiation. Oncogene.
22:5784–5791. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Evers B, Helleday T and Jonkers J:
Targeting homologous recombination repair defects in cancer. Trends
Pharmacol Sci. 31:372–380. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang M, Wu W, Wu W, Rosidi B, Zhang L,
Wang H and Iliakis G: PARP-1 and Ku compete for repair of DNA
double strand breaks by distinct NHEJ pathways. Nucleic Acids Res.
34:6170–6182. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang Y, Deng H, Xin S, Zhang K, Shi R and
Bao X: Prognostic and predictive value of three DNA methylation
signatures in lung adenocarcinoma. Front Genet. 10:3492019.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Baylin SB and Jones PA: Epigenetic
determinants of cancer. Cold Spring Harb Perspect Biol.
8:a0195052016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Tang J, Kong D, Cui Q, Wang K, Zhang D,
Gong Y and Wu G: Prognostic genes of breast cancer identified by
gene co-expression network analysis. Front Oncol. 8:3742018.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lan L, Xu B, Chen Q, Jiang J and Shen Y:
Weighted correlation network analysis of triple-negative breast
cancer progression: Identifying specific modules and hub genes
based on the GEO and TCGA database. Oncol Lett. 18:1207–1217.
2019.PubMed/NCBI
|
|
21
|
Valdembri D, Caswell PT, Anderson KI,
Schwarz JP, König I, Astanina E, Caccavari F, Norman JC, Humphries
MJ, Bussolino F and Serini G: Neuropilin-1/GIPC1 signaling
regulates alpha5beta1 integrin traffic and function in endothelial
cells. PLoS Biol. 7:e252009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Rudchenko S, Scanlan M, Kalantarov G,
Yavelsky V, Levy C, Estabrook A, Old L, Chan GL, Lobel L and Trakht
I: A human monoclonal autoantibody to breast cancer identifies the
PDZ domain containing protein GIPC1 as a novel breast
cancer-associated antigen. BMC Cancer. 8:2482008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Westbrook JA, Cairns DA, Peng J, Speirs V,
Hanby AM, Holen I, Wood SL, Ottewell PD, Marshall H, Banks RE, et
al: CAPG and GIPC1: Breast cancer biomarkers for bone metastasis
development and treatment. J Natl Cancer Inst. 108:2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chittenden TW, Pak J, Rubio R, Cheng H,
Holton K, Prendergast N, Glinskii V, Cai Y, Culhane A, Bentink S,
et al: Therapeutic implications of GIPC1 silencing in cancer. PLoS
One. 5:e155812010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Bae S, Bessho Y, Hojo M and Kageyama R:
The bHLH gene Hes6, an inhibitor of Hes1, promotes neuronal
differentiation. Development. 127:2933–2943. 2000.PubMed/NCBI
|
|
26
|
Hartman J, Lam EW, Gustafsson JA and Ström
A: Hes-6, an inhibitor of Hes-1, is regulated by 17beta-estradiol
and promotes breast cancer cell proliferation. Breast Cancer Res.
11:R792009. View
Article : Google Scholar : PubMed/NCBI
|
|
27
|
Akhmanova A and Hoogenraad CC: Microtubule
minus-end-targeting proteins. Curr Biol. 25:R162–R171. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Fu Y, Zheng S, An N, Athanasopoulos T,
Popplewell L, Liang A, Li K, Hu C and Zhu Y: β-catenin as a
potential key target for tumor suppression. Int J Cancer.
129:1541–1551. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Castellana B, Escuin D, Pérez-Olabarria M,
Vázquez T, Muñoz J, Peiró G, Barnadas A and Lerma E: Genetic
up-regulation and overexpression of PLEKHA7 differentiates invasive
lobular carcinomas from invasive ductal carcinomas. Hum Pathol.
43:1902–1909. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Tanaka N, Meng W, Nagae S and Takeichi M:
Nezha/CAMSAP3 and CAMSAP2 cooperate in epithelial-specific
organization of noncentrosomal microtubules. Proc Natl Acad Sci
USA. 109:20029–20034. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Soung YH, Lee JW, Kim SY, Nam SW, Park WS,
Lee JY, Yoo NJ and Lee SH: Mutational analysis of the kinase domain
of MYLK2 gene in common human cancers. Pathol Res Pract.
202:137–140. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Rong Y, Jin D, Hou C, Hu J, Wu W, Ni X,
Wang D and Lou W: Proteomics analysis of serum protein profiling in
pancreatic cancer patients by DIGE: Up-regulation of
mannose-binding lectin 2 and myosin light chain kinase 2. BMC
Gastroenterol. 10:682010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Damasco C, Lembo A, Somma MP, Gatti M, Di
Cunto F and Provero P: A signature inferred from drosophila mitotic
genes predicts survival of breast cancer patients. PLoS One.
6:e147372011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Tabernero J, Cervantes A, Rivera F,
Martinelli E, Rojo F, von Heydebreck A, Macarulla T,
Rodriguez-Braun E, Eugenia Vega-Villegas M, Senger S, et al:
Pharmacogenomic and pharmacoproteomic studies of cetuximab in
metastatic colorectal cancer: Biomarker analysis of a phase I
dose-escalation study. J Clin Oncol. 28:1181–1189. 2010. View Article : Google Scholar : PubMed/NCBI
|