Introduction
Thyroid cancer is the most prevalent endocrine
malignancy and the incidence rate of thyroid cancer has increased
from 2.4 to 9.4% annually in the last three decades in the USA
(1). Papillary thyroid carcinoma
(PTC), which originates from thyroid epithelial cells, is the most
frequent histopathological subtype of thyroid cancer and has the
highest mortality rate of all types of thyroid cancer in the USA
over the past few decades (2,3).
Effective therapeutic strategies for PTC, including thyroidectomy,
radioactive iodine and thyroid stimulating hormone suppression
therapy have contributed to a five-year survival rate >95% prior
to tumor cell dissemination between 2009 and 2015 in the USA
(4). The majority of patients with
PTC exhibit a good prognosis following comprehensive therapy;
however, distant metastasis and recurrence can occur in certain
subtypes of PTC (5). Therefore,
there is a requirement to increase understanding regarding the
molecular mechanisms that underlie the carcinogenesis and
development of PTC. Improved understanding may promote the use of
gene therapy for PTC and improve the prognosis of patients with
PTC.
MicroRNAs (miRNAs or miRs) are a class of small and
non-coding RNAs that consist of 19–22 nucleotides and regulate
post-transcriptional genes via a number of mechanisms, including
translational repression and mRNA degradation (6). miRNAs are involved in various
biological processes, including tumorigenesis and metastasis, which
indicates a crucial role of miRNAs in the pathogenesis of diverse
human malignancies. Commonly upregulated miRNAs, including
miR-146b, miR-222, miR-221 and miR-151, have been implicated in the
development and metastasis of PTC (7,8). High
levels of circulating miR-222 and miR-146b have been identified to
be associated with PTC recurrence and a poor clinical survival.
Recently, numerous studies have investigated the role of miR-203 in
the carcinogenesis and growth of a number of cancer types,
including colorectal cancer (9),
non-small cell lung cancer (10),
melanoma (11), T-cell lymphoma
(12), endometrial cancer (13) and gastric cancer (14). However, to the best of our knowledge,
the biological functions and molecular mechanisms of miR-203 in PTC
remain unclear. The present study aimed to clarify the biological
role of miR-203 in PTC and investigate possible targets.
It is understood that the occurrence and development
of tumors can be regulated by both genetics and epigenetics.
Certain miRNAs in tumor cells are regulated by epigenetic
modifications, including DNA methylation and histone acetylation,
and protein-coding genes (15,16). It
has been reported that the level of histone acetylation is
associated with tumor grade and the risk of tumor recurrence in
human prostate cancer (17,18). In addition, overexpression of c-Myc
can regulate histone H4 acetylation, which has been revealed to
affect the G2/M cell cycle progression of Raji cells (19). Furthermore, a number of studies have
supported a role of miRNAs as targets and effectors of aberrant
histone acetylation. miR-133a can be regulated by histone
acetylation and promote myocardial fibrosis (20). In addition, an ectopic expression of
miR-200c is associated with the level of histone deacetylase
inhibitors that act as tumor suppressors to inhibit the
proliferation, invasion, and migration of breast cancer cells
(21). Therefore, the present study
aimed to investigate whether the inhibition of histone acetylation
can control tumor growth by regulating the expression of miRNA,
which may provide a potential biological target for the treatment
of PTC.
Materials and methods
Clinical tissue specimens and cell
lines
A total of 30 PTC patients aged from 31 to 70 were
enrolled in this research. The cohort included 19 male and 11
female patients and they had not received radiation therapy or
chemotherapy prior to surgery. All tissue samples were collected at
Ningbo No. 2 Hospital (Ningbo, China) between September 2016 and
May 2017 and were frozen in liquid nitrogen immediately following
duodenopancreatectomy. It was only possible to collect normal
tissue samples from 20 of the 30 participants. All diagnoses were
based on pathological and/or cytological evidence. The present
study was approved by the Ethics Committee of Ningbo No. 2 Hospital
(Ningbo, China), and written informed consent was obtained from all
patients.
Cell lines and culture
The Nthy-ori3-1 cell line, which is a normal human
thyroid follicular cell line, and the three human PTC cell lines
HTH83, NIM-1 and TPC-1 were obtained from the American Type Culture
Collection (Manassas, VA, USA) and cultured according to the
manufacturer's protocol. Briefly, all cells were grown in
Dulbecco's modified Eagle's medium (DMEM; Gibco; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) with 10% fetal bovine serum
(Biological Industries, Kibbutz Beit Haemek, Israel) and 100 U/ml
penicillin and 100 mg/ml streptomycin at 37°C in humidified air
with 5% CO2.
miRNA microarray assay
The RNA of ten PTC tissue sample samples and ten
matched adjacent normal tissue sample samples was analyzed at
Oebiotech Co., Ltd. (Shanghai, China) using the Agilent Human miRNA
Microarray (8×60 K; version 21.0; Agilent Technologies, Inc., Santa
Clara, CA, USA) with capture probes for a total of 2,549 human
miRNAs based on the Sanger miRBase database (version 21.0;
www.mirbase.org). The microarray assay was
performed according to the manufacturer's protocol (Agilent
Technologies GmbH, Waldbronn, Germany). Total RNA was isolated
using TRIzol® reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) and the miRNA molecules in the total RNA sample
were labeled using a miRNA Complete Labeling and Hyb kit (Agilent
Technologies, Inc.). Each microarray slide was hybridized using 100
ng Cy3-labeled RNA (Agilent Technologies, Inc.) in a hybridization
oven. Following hybridization, the slides were washed using a Gene
Expression Wash Buffer kit (Agilent Technologies), scanned using
the Agilent Microarray Scanner (Agilent Technologies) and analyzed
with the Feature Extraction software program (version 10.7; Agilent
Technologies) with default settings. Raw data were normalized using
the quantile algorithm in the Gene Spring software program (v.12.6;
Agilent Technologies). A relative fold-change >3 in the
differential expression of miRNAs and P<0.01 were considered to
indicate a statistically significant result.
Cell transfection
RNA oligoribonucleotides or small interfering RNA
(siRNA) were transfected into TCP-1 cells using
Lipofectamine® 3000 (Invitrogen; Thermo Fisher
Scientific, Inc.), according to the manufacturer's protocol. The
sequences of the miRs transfected were as follows: miR-203 mimic,
5′-GUGAAAUGUUUAGGACCACUAG-3′; miR-NC, 5′-UUCUCCGAACGUGUCACGUTT-3′;
miR-203 inhibitor, 5′-CUAGUGGUCCUAAACAUUUCAC-3′; and scrambled
miRNA, 5′-CAGUACUUUUGUGUAGUACAA-3′. Subsequent experiments were
performed 48 h after transfection, including assessment of cell
proliferation, apoptosis and migration, and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR) and
western blot analysis.
Cell proliferation assay
PTC cells were transfected with miR-NC or miR-203
mimic for 24 h and then seeded in 96-well plates at a density of
5,000 cells/well. Subsequently, cell proliferation was assessed
using a Cell Counting Kit-8 (CCK-8; Dojindo Molecular Technologies,
Inc., Kumamoto, Japan) at 48 h. Absorbance was determined at 450 nm
using a spectrometer.
Migration assay
Cell migration assay was performed with a 24-well
transwell culture chamber with 8 µm polyester membranes (Merck
KGaA, Darmstadt, Germany). PTC-1 cells were transfected with miR-NC
(miR-NC group) and miR-203 mimics (miR-203 group), or cultured
using complete medium containing Lipofectamine® 3000
(Mock group). For migration assays, cells with different treatments
were plated in serum-free medium in the upper chamber at a density
of 3×104 cells/well, and supplemented medium containing
10% FBS (Invitrogen; Thermo Fisher Scientific, Inc.) to the lower
compartment. After 24 h of incubation, non-migrating cells that had
not migrated through the pores were carefully removed. The filters
were fixed in methanol, stained with crystal violet (Sigma-Aldrich;
Merck KGaA) at room temperature for 20 min and then counted with an
inverted fluorescence microscope at 10× magnification. Average cell
numbers were calculated from five randomly selected fields from
each group.
RT-qPCR
Total RNA was isolated from tissue samples and TPC-1
cells using TRIzol (Invitrogen; Thermo Fisher Scientific, Inc.),
according to the manufacturer's protocol. Total cellular RNA was
used for first strand complementary DNA (cDNA) synthesis with the
Reverse Transcription system (Roche Diagnostics GmbH, Mannheim,
Germany). The cDNA samples were diluted to 100 µl with RNase-free
water and 1 µl cDNA mixture was used for qPCR in a total volume of
10 µl with SYBR Green Reagent (Thermo Fisher Scientific, Inc.) and
sequence-specific primers. The thermocycling conditions for PCR
were 95°C for 15 min followed by 40 cycles of 94°C for 15 sec, 55°C
for 30 sec and 70°C for 30 sec. qPCR was performed with the
following primers: miR-203 forward, 5′-CGGTAGTCTGATACTGTAA-3′ and
reverse, 5′-GTGCTCCGAAGGGGGT-3′; survivin forward,
5′-AGGACCACCGCATCTCTACAT-3′ and reverse,
5′-AAGTCTGGCTCGTTCTCAGTG-3′; miR-101 forward,
5′-GAGGGGTACAGTACTGTGATA-3 and reverse, 5′-TGCGTGTCGTGGAGTC-3′;
miR-195 forward, 5′-GTCGTATCCAGTGCAGGGTCCGAGGT-3′ and reverse,
5′-ATTCGCACTGGATACGACTATAACCG-3′; miR-455 forward,
5′-GTCGTATCCAGTGCAGGGTCCGAGGT-3′ and reverse
5′-ATTCGCACTGGATACGACTCTAACAC-3′; miR-130 forward
5′-AAAGGATCCGCATCAAGCCCGAAGTAT-3′ and reverse
5′-AAAGAATTCGAGGCAGTGTCTATCACAAG-3′; miR-126 forward
5′-TATAAGATCTGAGGATAGGTGGGTTCCCGAGAA-3′ and reverse
5′-ATATGAATTCTCTCAGGGCTATGCCGCCTAAGTAC-3′; U6 forward,
5′-CTCGCTTCGGCAGCACA-3′ and reverse, 5′-AACGCTTCACGAATTTGCGT-3′.
The expression levels of miR-203 and survivin were analyzed using
the 2−ΔΔCq method and normalized to the level of U6
small nuclear RNA expression (22).
Western blot assay
Radioimmunoprecipitation assay lysis buffer (Thermo
Fisher Scientific, Inc.) with a protease inhibitor mixture
(Sigma-Aldrich; Merck KGaA) was used for total protein extraction
from the lysate of PTC patient tissues and TPC-1 cells, and a
bicinchoninic acid assay (Beijing Solarbio Science & Technology
Co., Ltd., Beijing, China) was used to measure the concentration.
Subsequently, 35 µg of protein was separated by a 10% SDS-PAGE and
transferred to polyvinylidene difluoride membranes (Roche
Diagnostics GmbH). Following blocking with 5% skimmed-milk for 1 h
at room temperature, the membranes were incubated overnight at 4°C
with a primary antibody against survivin (1:1,000; cat. no. 2808)
or β-actin (1:1,000; 13E5 rabbit mAb cat. no. 4970; both from Cell
Signaling Technology, Inc., Danvers, MA, USA). Subsequently, the
membranes were washed 3 times (5 min each) with 15 ml TBS-Tween-20
(Beijing Solarbio Science & Technology Co., Ltd., Beijing,
China). The membranes were then incubated with a horseradish
peroxidase-conjugated anti-rabbit IgG antibody (1:2,000; cat. no.
7074, Cell Signaling Technology, Inc., Danvers, MA, USA) in 10 ml
blocking buffer with gentle agitation for 1 h at room temperature.
Membranes were washed three times as described abovee and incubated
with enhanced chemiluminescent substrate (Amersham; GE Healthcare
Life Sciences, Little Chalfont, UK) for 1 min. Excess solution as
removed and exposed to X-ray film (Carestream Health, Inc.,
Rochester, NY, USA).
Dual-luciferase reporter assay
The wild-type (WT) and mutant (Mut) 3′-untranslated
region (3′-UTR) of survivin was designed and prepared by Shanghai
GenePharma Co., Ltd. (Shanghai, China) and were cloned into the
pMIR-Reprot plasmid (Oligoengine, Seattle, WA, USA). A total of
1×105 293 cells were plated and cultured in 24-well
plates to reach 70% confluency. Cells were cotransfected with 40
pmol miR-203 mimic or miR-NC alongside 300 ng WT survivin 3′-UTR
reporter plasmid or 300 ng Mut survivin 3′-UTR reporter plasmid
using Lipofectamine® 3000 (Invitrogen; Thermo Fisher
Scientific, Inc.), according to the manufacturer's protocol. Each
group of cells was transfected with pMIR-Report β gal control
plasmid for normalization. The sequences of the miRs transfected
were as follows: miR-203 mimic: 5′-GUGAAAUGUUUAGGACCACUAG-3′;
miR-NC: 5′-UUCUCCGAACGUGUCACGUTT-3′; miR-203 inhibitor:
5′-CUAGUGGUCCUAAACAUUUCAC-3′; and scrambled miRNA:
5′-CAGUACUUUUGUGUAGUACAA-3′. MiR-NC was used as the control for the
miR-203 mimic and scrambled miRNA was used as the control for the
miR-203 inhibitor.
Luciferase assays were performed 48 h post
transfection using a dual luciferase reporter gene assay kit
(BioVision, Milpitas, CA, USA). The relative reporter activity was
obtained by normalizing the firefly luciferase activity against the
control luciferase activity using a dual-luciferase reporter assay
system (Promega Corporation, Madison, WI, USA).
Apoptosis and flow cytometry
A total of 3×105 cells/well were seeded
into 6-well plates and incubated for 24 h. After transfection with
miR-203 mimic or miR-NC for 48 h, cells were harvested,
trypsinized, and washed. The cells were stained with FITC Annexin V
Apoptosis Detection Kit (BioLegend, Inc., San Diego, CA, USA) and
propidium iodide (Thermo Fisher Scientific, Inc.) for 30 min at 4°C
in the dark. The samples were evaluated by flow cytometry (BD
FACSCalibur™; BD Biosciences, San Jose, CA, USA) and the data was
analyzed using FlowJo version 7.6 (FlowJo LLC, Ashland, OR,
USA).
Trichostatin A (TSA) treatment
assay
A total of 3×105 TPC-1 cells were plated
and cultured in 12-well plates to reach 70% confluence. Cells were
treated with TSA (Sigma-Aldrich; Merck KGaA) at concentrations of
0, 50, 100 and 200 nM for 12 h, after which cells were collected
and the total mRNA were extracted.
Statistical analysis
GraphPad Prism 6 (GraphPad Software, Inc., La Jolla,
CA, USA) was used for all data analysis. Data are presented as the
mean ± standard deviation from three independent experiments. Two
variables were compared using a two-tailed Student's t-test and
three variables were analyzed by one-way analysis of variance
followed by Tukey's post-hoc test. P<0.05 was considered to
indicate a statistically significant difference. The potential
downstream targets of miRNA were predicted by three online target
prediction software programs, including TargetScan version7.2
(http://www.targetscan.org), miRanda
(http://www.mirbase.org/) and Pic Tar software
(http://pictar.mdc-berlin.de/).
Results
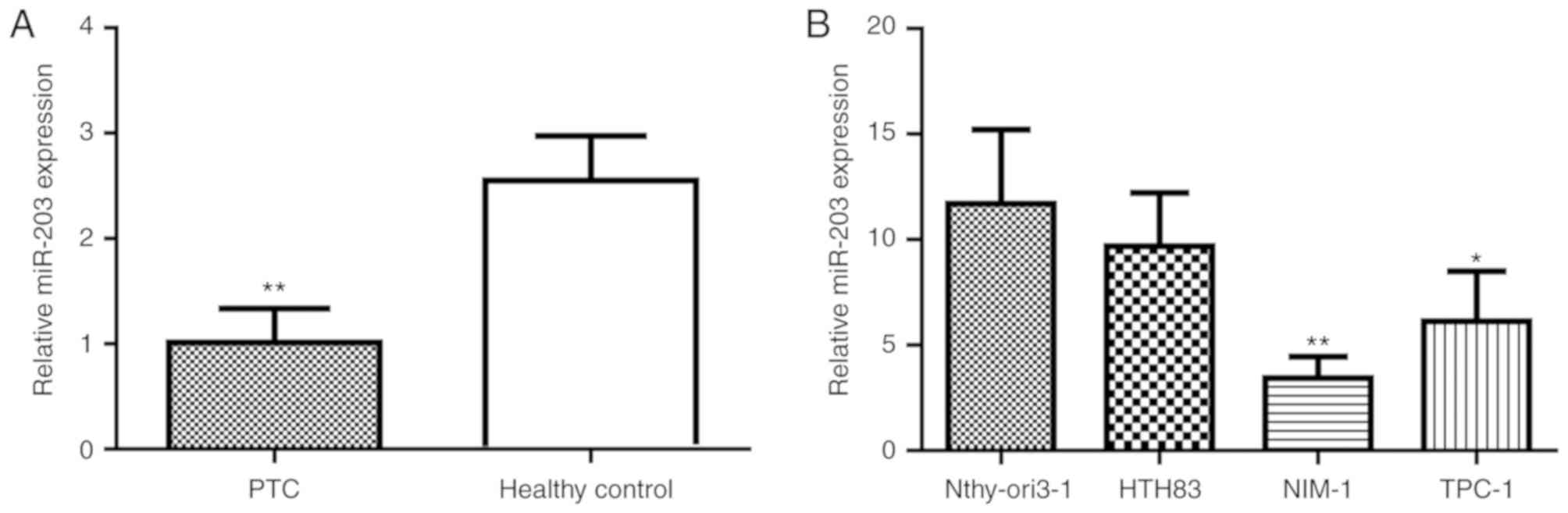
Expression level of miR-203 is lower
in PTC tissues and cell lines
To investigate the role of miRNAs in the
pathogenesis of PTC, Affymetrix microarrays were used to screen for
differently expressed miRNAs in PTC tissues (n=10) and adjacent
normal tissues (n=10). Of the measured miRNAs, the expression
levels of six miRNAs were identified to be significantly lower in
PTC tissues compared with adjacent normal tissues. To validate the
accuracy of the microarray results, the expression levels of the
six different miRNAs including miR-203, miR-101, miR-195, miR-455,
miR-130 and miR-126 were evaluated by RT-qPCR with the PTC tissue
samples (n=30) and adjacent normal tissues (n=20; data not shown).
Out of the six miRNAs, a significantly lower expression level of
miR-203 was identified in the PTC tissue samples compared with the
adjacent normal tissue sample (P<0.001 Fig. 1A). Furthermore, as presented in
Fig. 1B, compared with the normal
thyroid cell line Nthy-ori3-1, the miR-203 expression level was
significantly lower in NIM-1 (P<0.01) and TPC-1 cells
(P<0.05); however, a significantly lower expression level was
not identified in HTH83 cells. These results indicate that a lower
expression level of miR-203 may be associated with PTC development
and progression.
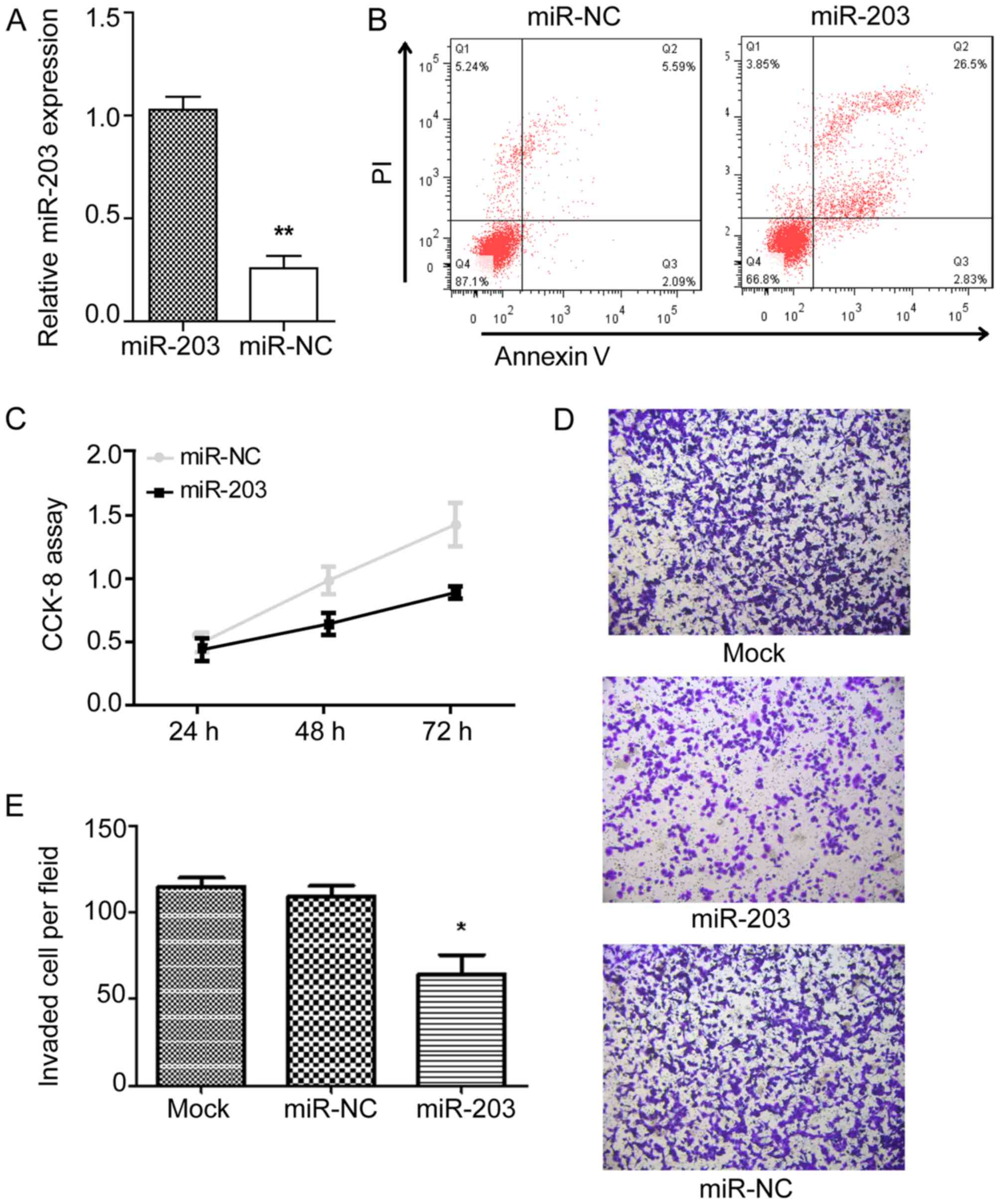
miR-203 overexpression regulates cell
proliferation, apoptosis and motility in vitro
Based on the aforementioned results, the present
study investigated whether miR-203 regulates PTC proliferation.
miR-negative control (miR-NC) and miR-203 mimic were transfected
into NIM-1 cells and the effects on proliferation were observed.
The relative expression level of miR-203 was significantly higher
in the miR-203 mimic group compared with the miR-NC group following
transfection (Fig. 2A). An apoptosis
assay revealed that transfection with miR-203 mimic decreased the
cell viability compared with transfection with miR-NC (Fig. 2B). In addition, a CCK-8 assay
demonstrated that following transfection with miR-203 mimic, the
proliferation rate of NIM-1 cells was significantly lower compared
with the miR-NC group (Fig. 2C).
Furthermore, Transwell assays were performed to assess whether
miR-203 could inhibit the migration of PTC cells. As presented in
Fig. 2C and D, the number and state
of TPC-1 cells was similar between the Mock and miR-NC groups.
However, as presented in Fig. 2E,
overexpression of miR-203 significantly reduced the number of
migratory TPC-1 cells numbers compared with the Mock (P<0.05)
and miR-NC groups, which indicates that miR-203 overexpression may
inhibit the migration of TPC-1 cells. In summary, these results
indicate that overexpression of miR-203 suppresses the
proliferation and motility of PTC cells and induces apoptosis in
vitro.
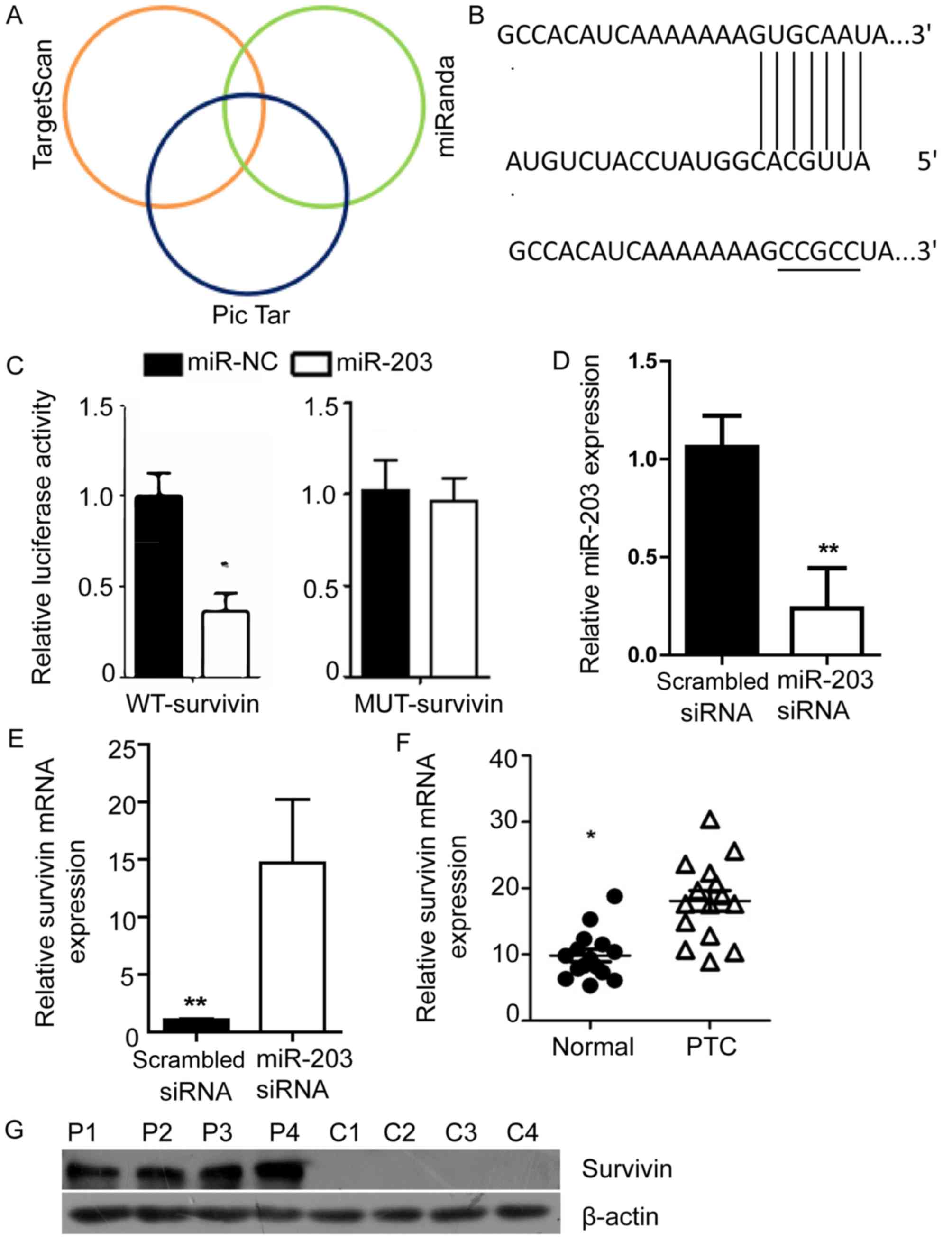
Survivin is a downstream target gene
of miR-203
To identify potential targets of miR-203, the
network-based target prediction software programs miRanda, Pic Tar,
and TargetScan were applied. All three programs identified survivin
as a downstream target of miR-203 (Fig.
3A), which suggests that miR-203 may be an upstream regulator
of survivin. Therefore, the present study mutated the 3′-UTR of
survivin to investigate whether survivin is a target of miR-203
(Fig. 3B). Subsequently, luciferase
assays were performed with the WT and MUT survivin. As presented in
Fig. 3C, transfection with miR-203
mimic significantly inhibited the luciferase activity of the
reporter gene containing the WT survivin 3′-UTR; however, miR-203
mimic did not inhibit the reporter gene with the mutated
3′-UTR.
The effect of miR-203 on survivin expression was
then investigated by transfection with miR-203 siRNA. The relative
expression level of miR-203 was significantly lower in the miR-203
siRNA group compared with the scrambled siRNA group following
transfection (Fig. 3D). It was
identified that inhibition of miR-203 was associated with a
significantly lower expression level of survivin in TPC-1 cells
compared with the scrambled siRNA (P<0.01; Fig. 3E). The expression levels of survivin
in patient tissue samples were then evaluated. RNA was extracted
from 30 PTC tissue samples and 20 matched adjacent normal tissue
samples for RT-qPCR. The results revealed that the expression level
of survivin was significantly higher in PTC tissue samples compared
with in adjacent normal tissue samples (P<0.05; Fig. 3F). In addition, western blot analysis
demonstrated that the survivin expression levels were markedly
higher in the PTC tissue samples compared with the adjacent normal
tissue samples (Fig. 3G).
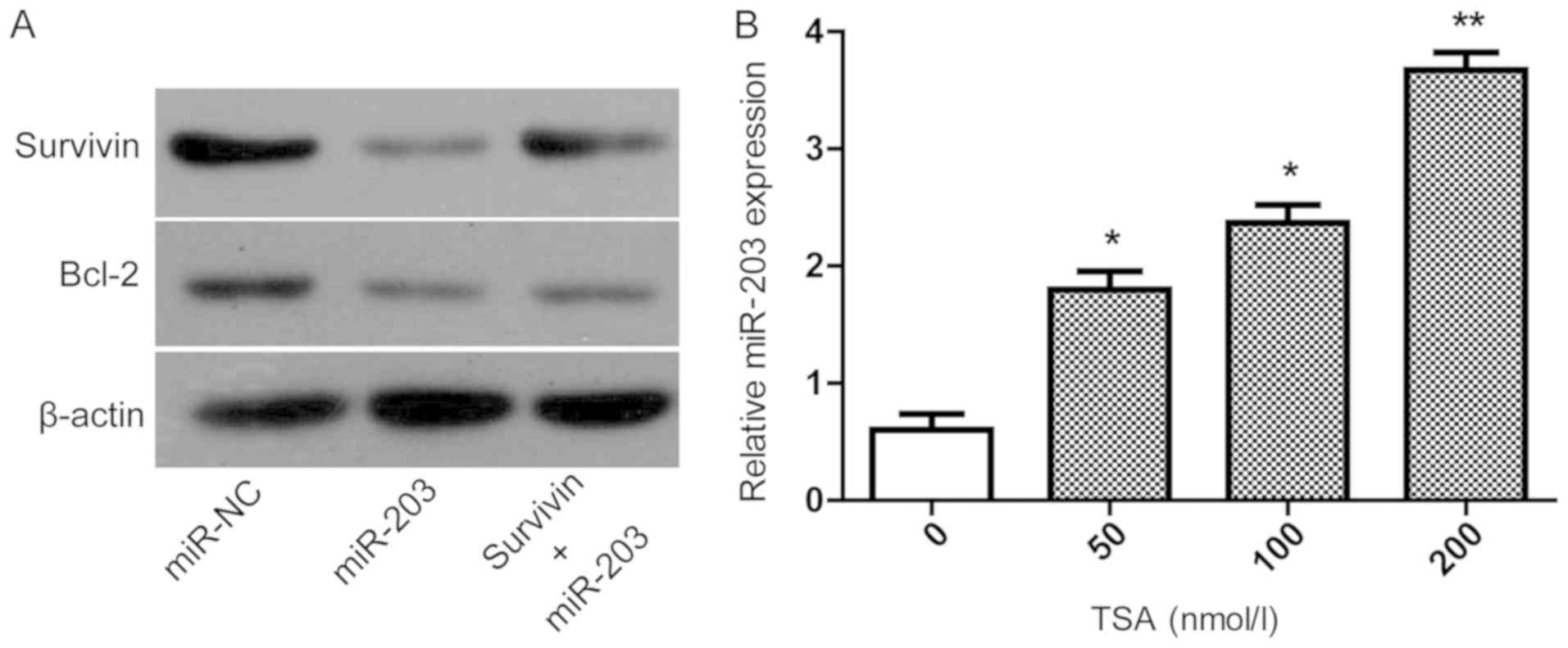
Inhibition of histone acetylation can
upregulate miR-203
The mechanisms by which miR-203 inhibits the
proliferation and motility of and induces apoptosis in TPC-1 cells
was investigated. It has been previously reported that Bcl-2
protein serves an important role in cell proliferation and
apoptosis (23). TPC-1 cells were
transfected with miR-NC or miR-203 mimic, followed by protein
extraction for western blot analysis. As presented in Fig. 4A, the expression levels of survivin
and Bcl-2 demonstrated a similar trend. It has been reported that
histone acetylation can regulate the expression of miRNA. To
demonstrate that the decreased expression of miR-203 in PTC
resulted in the reduction of the target protein survivin, which
affects the expression of Bcl-2, miR-NC and miR-203, mimics were
transfected into TPC-1 cells and the protein expression levels were
determined by western blotting. As shown in Fig. 4A, survivin and Bcl-2 showed a similar
trend suggesting that miR-203 functions by indirectly affecting the
expression of Bcl-2. Histone acetylation can regulate the
expression of miRNA (24).
Therefore, to investigate the low expression level of miR-203 in
patients with PTC, TPC-1 cells were treated with TSA, an inhibitor
of histone deacetylases. It was identified that the expression
level of miR-203 was higher following treatment with TSA in a
dose-dependent manner. Compared with untreated cells the miR-203
expression level was significantly higher in cells treated with 50
nmol/l TSA (P<0.05), 100 nmol/l TSA (P<0.05) and 200 nmol/l
TSA (P<0.01). In summary, these data suggest that miR-203
inhibits the proliferation and motility, and induces apoptosis of
TPC-1 cells, which may occur via a regulation of the expression of
Bcl-2.
Discussion
PTC is the most common thyroid malignancy (25). In the early stages of PTC no specific
symptoms are observed, which makes the diagnosis of early-stage PTC
difficult (26). miRNAs,
post-transcriptional gene regulators, have been reported to serve a
key role in thyroid cancer progression by regulating numerous
cellular events (27,28). A numbers of miRNAs, including miR-146
(29), miR-222 (30) and miR-20b (31), have been identified to promote or
suppress the progression of PTC.
The present results revealed that the expression
level of miR-203 was significantly lower in PTC tumor tissues
compared with adjacent normal tissues. miR-203 has been reported to
be downregulated in metastatic melanoma (11) and non-small-cell lung cancer
(32), which indicates miR-203
serves a role in different types of cancer. Consistent with
previous studies, the present study observed similar results in
different PTC cell lines, which indicates a low expression level of
miR-203 in PTC. Therefore, the present study selected miR-203 for
further investigation.
It is understood that cancer cells are characterized
by a limitless replicative potential, resistance to cell death,
tissue invasion and metastasis (33). In the present study, the CCK-8 assay
demonstrated that an overexpression of miR-203 inhibits the
proliferation of NIM-1 cells and an apoptosis assay revealed that
the apoptosis rate was significantly higher following transfection
of NIM-1 cells with miR-203 mimic. In addition, the effect of
miR-203 on the function of TPC-1 cells was evaluated, which
revealed that an upregulation of miR-203 inhibits cell migration.
In summary, the present results indicate that miR-203 upregulation
can inhibit cell proliferation, induce cell apoptosis and suppress
the motility of PTC cells.
Furthermore, survivin was identified as a potential
downstream regulator of miR-203. To verify this hypothesis, a
luciferase assay was performed to detect survivin gene expression.
Upregulation of miR-203 decreased the level of survivin gene
expression. However, when survivin was mutated, the gene expression
level of survivin was not altered following miR-203 upregulation,
which further suggests that miR-203 serves as an upstream regulator
of survivin. In addition, inhibition of miR-203 was identified to
increase the expression level of survivin in TPC-1 cells, which
further supports our hypothesis. Consequently, the gene and protein
expression levels of survivin were revealed to be significantly
higher in PTC tissues compared with adjacent normal tissues.
Previous studies have suggested that survivin is highly expressed
in PTC and may be associated with disease occurrence, lymph node
metastasis and clinical staging of PTC (34–37).
It has been reported that Bcl-2 can regulate
proliferation and apoptosis (38,39).
This raises the question of whether survivin can functionally
inhibit the expression of Bcl-2. The present study revealed a
consistent trend between the expression levels of survivin and
Bcl-2, which suggests that survivin and Bcl-2 may interact in the
occurrence and development of PTC. miR-203 can regulate cell
apoptosis and a low expression level of miR-203 in PTC may be
associated with abnormal proliferation in the tumor. Finally, the
present study investigated the influence of the expression of
miR-203 from the perspective of epigenetics. It was revealed that
the expression level of miR-203 is higher following treatment with
TSA, which suggests that TSA inhibition of histone acetylation
upregulates miR-203 expression.
In conclusion, the present study identified a low
expression level of miR-203 in PTC tissue and cell lines. miR-203
was demonstrated to inhibit cell proliferation and migration, and
induce apoptosis. Overexpression of miR-203 may serve a role in PTC
tumor cells by down regulating survivin, which may regulate Bcl-2
expression. Furthermore, inhibition of histone acetylation was
demonstrated to upregulate miR-203 expression, which suggests that
deacetylation and epigenetics may serve a role in PTC therapy.
These results indicate that miR-203 serves a role in the
pathogenesis of PTC, and may act as a novel biomarker and a target
for the development of novel therapeutic strategies against
PTC.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Ningbo
Natural Science Foundation (grant no. 2015A610227).
Availability of data and materials
Not applicable.
Authors' contributions
XJW, LD, ZJZ and JRZ performed the experiments. XJW
interpreted the results and drafted the manuscript. JPZ examined
the archives and identified the subjects included in the study, and
revised the manuscript thoroughly prior to submission.
Ethics approval and consent to
participate
All studies associated with patient samples were
approved by the Ethics Committee of Ningbo Medical University, and
informed consent was obtained from all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Pellegriti G, Frasca F, Regalbuto C,
Squatrito S and Vigneri R: Worldwide increasing incidence of
thyroid cancer: Update on epidemiology and risk factors. J Cancer
Epidemiol. 2013:9652122013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Patil VS, Vijayakumar A and Natikar N:
Unusual presentation of cystic papillary thyroid carcinoma. Case
Rep Endocrinol. 2012:7327152012.PubMed/NCBI
|
|
3
|
Anil S, Radu B and Ricardo L: Metastatic
papillary thyroid carcinoma with absence of tumor focus in thyroid
gland. Am J Case Rep. 14:73–75. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Haugen BR, Alexander EK, Bible KC, Doherty
GM, Mandel SJ, Nikiforov YE, Pacini F, Randolph GW, Sawka AM,
Schlumberger M, et al: 2015 American thyroid association management
guidelines for adult patients with thyroid nodules and
differentiated thyroid cancer: The American thyroid association
guidelines task force on thyroid nodules and differentiated thyroid
cancer. Thyroid. 26:1–133. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Leboulleux S, Rubino C, Baudin E, Caillou
B, Hartl DM, Bidart JM, Travagli JP and Schlumberger M: Prognostic
factors for persistent or recurrent disease of papillary thyroid
carcinoma with neck lymph node metastases and/or tumor extension
beyond the thyroid capsule at initial diagnosis. J Clin Endocrinol
Metab. 90:5723–5729. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Valinezhad Orang A, Safaralizadeh R and
Kazemzadeh-Bavili M: Mechanisms of miRNA-mediated gene regulation
from common downregulation to mRNA-specific upregulation. Int J
Genomics. 2014:9706072014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yu S, Liu Y, Wang J, Guo Z, Zhang Q, Yu F,
Zhang Y, Huang K, Li Y, Song E, et al: Circulating microRNA
profiles as potential biomarkers for diagnosis of papillary thyroid
carcinoma. J Clin Endocrinol Metab. 97:2084–2092. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Swierniak M, Wojcicka A, Czetwertynska M,
Stachlewska E, Maciag M, Wiechno W, Gornicka B, Bogdanska M,
Koperski L, de la Chapelle A and Jazdzewski K: In-depth
characterization of the microRNA transcriptome in normal thyroid
and papillary thyroid carcinoma. J Clin Endocrinol Metab.
98:E1401–E1409. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Takano Y, Masuda T, Iinuma H, Yamaguchi R,
Sato K, Tobo T, Hirata H, Kuroda Y, Nambara S, Hayashi N, et al:
Circulating exosomal microRNA-203 is associated with metastasis
possibly via inducing tumor-associated macrophages in colorectal
cancer. Oncotarget. 8:78598–78613. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hu Y, Wang L, Gu J, Qu K and Wang Y:
Identification of microRNA differentially expressed in three
subtypes of non-small cell lung cancer and in silico functional
analysis. Oncotarget. 8:74554–74566. 2017.PubMed/NCBI
|
|
11
|
Lohcharoenkal W, Mahapatra Das Mahapatra
K, Pasquali L, Crudden C, Kular L, Akkaya Ulum YZ, Zhang L, Xu
Landén N, Girnita L, Jagodic M, et al: Genome-wide screen for
MicroRNAs reveals a role for miR-203 in melanoma metastasis. J
Invest Dermatol. 138:882–892. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Dusílková N, Bašová P, Polívka J, Kodet O,
Kulvait V, Pešta M, Trněný M and Stopka T: Plasma miR-155, miR-203,
and miR-205 are biomarkers for monitoring of primary cutaneous
T-cell lymphomas. Int J Mol Sci. 18(pii): E21362017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Benati M, Montagnana M, Danese E, Paviati
E, Giudici S, Franchi M and Lippi G: Evaluation of mir-203
expression levels and DNA promoter methylation status in serum of
patients with endometrial cancer. Clin Lab. 63:1675–1681. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zheng Y, Liu W, Guo L and Yang X: The
expression level of miR-203 in patients with gastric cancer and its
clinical significance. Pathol Res Pract. 213:1515–1518. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li E: Chromatin modification and
epigenetic reprogramming in mammalian development. Nat Rev Genet.
3:662–673. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhou VW, Goren A and Bernstein BE:
Charting histone modifications and the functional organization of
mammalian genomes. Nat Rev Genet. 12:7–18. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Bianco-Miotto T, Chiam K, Buchanan G,
Jindal S, Day TK, Thomas M, Pickering MA, O'Loughlin MA, Ryan NK,
Raymond WA, et al: Global levels of specific histone modifications
and an epigenetic gene signature predict prostate cancer
progression and development. Cancer Epidemiol Biomarkers Prev.
19:2611–2622. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Seligson DB, Horvath S, Shi T, Yu H, Tze
S, Grunstein M and Kurdistani SK: Global histone modification
patterns predict risk of prostate cancer recurrence. Nature.
435:1262–1266. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yang Y, Xue K, Li Z, Zheng W, Dong W, Song
J, Sun S, Ma T and Li W: c-Myc regulates the CDK1/cyclin B1
dependent-G2/M cell cycle progression by histone H4 acetylation in
Raji cells. Int J Mol Med. 41:3366–3378. 2018.PubMed/NCBI
|
|
20
|
Chaturvedi P, Kalani A, Givvimani S, Kamat
PK, Familtseva A and Tyagi SC: Differential regulation of DNA
methylation versus histone acetylation in cardiomyocytes during
HHcy in vitro and in vivo: An epigenetic mechanism. Physiol
Genomics. 46:245–255. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Bian X, Liang Z, Feng A, Salgado E and
Shim H: HDAC inhibitor suppresses proliferation and invasion of
breast cancer cells through regulation of miR-200c targeting CRKL.
Biochem Pharmacol. 147:30–37. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Swierczynski S, Klieser E, LLLig R,
Alinger-Scharinger B, Kiesslich T and Neureiter D: Histone
deacetylation meets miRNA: Epigenetics and post-transcriptional
regulation in cancer and chronic diseases. Expert Opin Biol Ther.
15:651–664. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Xu L, Jia Y, Yang XH, Han F, Zheng Y, Ni
Y, Chen X, Hong J, Liu JQ, Li Q, et al: MicroRNA-130b
transcriptionally regulated by histone H3 deacetylation renders Akt
ubiquitination and apoptosis resistance to 6-OHDA. Biochim Biophys
Acta Mol Basis Dis. 1863:1678–1689. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liebner DA and Shah MH: Thyroid cancer:
Pathogenesis and targeted therapy. Ther Adv Endocrinol Metab.
2:173–195. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Baldane S, Ipekci SH, Sozen M and
Kebapcilar L: Mean platelet volume could be a possible biomarker
for papillary thyroid carcinomas. Asian Pac J Cancer Prev.
16:2671–2674. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Aragon Han P, Weng CH, Khawaja HT,
Nagarajan N, Schneider EB, Umbricht CB, Witwer KW and Zeiger MA:
MicroRNA expression and association with clinicopathologic features
in papillary thyroid cancer: A systematic review. Thyroid.
25:1322–1329. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
de la Chapelle A and Jazdzewski K:
MicroRNAs in thyroid cancer. J Clin Endocrinol Metab. 96:3326–3336.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Chou CK, Yang KD, Chou FF, Huang CC, Lan
YW, Lee YF, Kang HY and Liu RT: Prognostic implications of miR-146b
expression and its functional role in papillary thyroid carcinoma.
J Clin Endocrinol Metab. 98:E196–E205. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Visone R, Russo L, Pallante P, De Martino
I, Ferraro A, Leone V, Borbone E, Petrocca F, Alder H, Croce CM and
Fusco A: MicroRNAs (miR)-221 and miR-222, both overexpressed in
human thyroid papillary carcinomas, regulate p27Kip1 protein levels
and cell cycle. Endocr Relat Cancer. 14:791–798. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Hong S, Yu S, Li J, Yin Y, Liu Y, Zhang Q,
Guan H, Li Y and Xiao H: MiR-20b displays tumor-suppressor
functions in papillary thyroid carcinoma by regulating the MAPK/ERK
signaling pathway. Thyroid. 26:1733–1743. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Chi Y, Jin Q, Liu X, Xu L, He X, Shen Y,
Zhou Q, Zhang J and Jin M: miR-203 inhibits cell proliferation,
invasion, and migration of non-small-cell lung cancer by
downregulating RGS17. Cancer Sci. 108:2366–2372. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Selemetjev S, Savin S, Paunovic I, Tatic S
and Cvejic D: Concomitant high expression of survivin and vascular
endothelial growth factor-C is strongly associated with metastatic
status of lymph nodes in papillary thyroid carcinoma. J Cancer Res
Ther. 14 (Suppl):S114–S119. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Liu J, Sun W, Dong W, Wang Z, Qin Y, Zhang
T and Zhang H: HSP90 inhibitor NVP-AUY922 induces cell apoptosis by
disruption of the survivin in papillary thyroid carcinoma cells.
Biochem Biophys Res Commun. 487:313–319. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Yan Y, Hu F, Wu W, Ma R and Huang H:
Expression characteristics of proteins of IGF-1R, p-Akt, and
survivin in papillary thyroid carcinoma patients with type 2
diabetes mellitus. Medicine (Baltimore). 96:e63932017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wang YX, Li ML, Yu SG, Liu CM, Han Y and
Wang XY: The association between the Survivin A9194G exon
polymorphisms and papillary thyroid carcinoma risk in the Han
Chinese population. Pathol Res Pract. 209:151–154. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Li H, Li X, Bai M, Suo Y, Zhang G and Cao
X: Matrine inhibited proliferation and increased apoptosis in human
breast cancer MCF-7 cells via upregulation of Bax and
downregulation of Bcl-2. Int J Clin Exp Pathol. 8:14793–14799.
2015.PubMed/NCBI
|
|
39
|
Ding JH, Yuan LY, Huang RB and Chen GA:
Aspirin inhibits proliferation and induces apoptosis of multiple
myeloma cells through regulation of Bcl-2 and Bax and suppression
of VEGF. Eur J Haematol. 93:329–339. 2014. View Article : Google Scholar : PubMed/NCBI
|