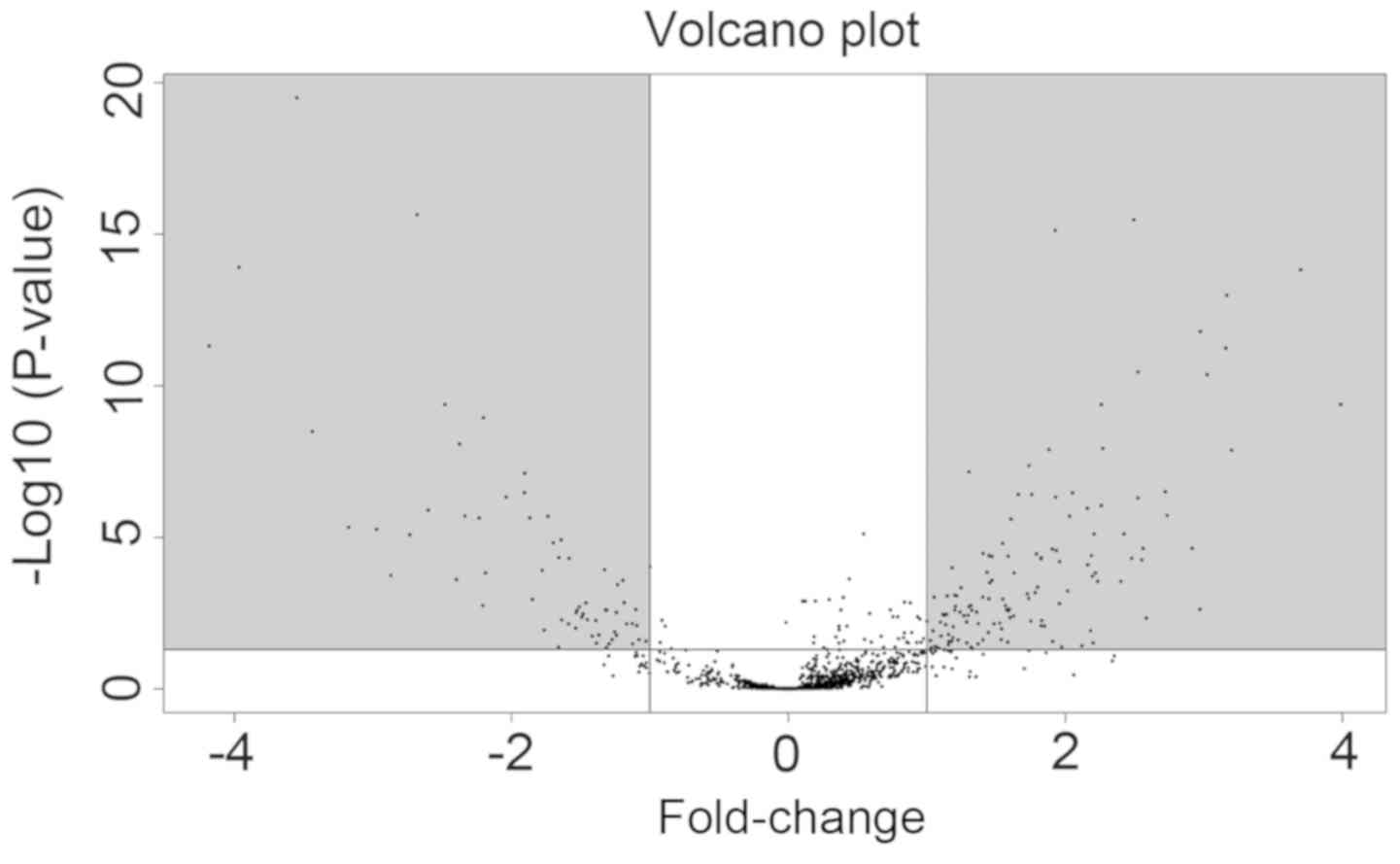
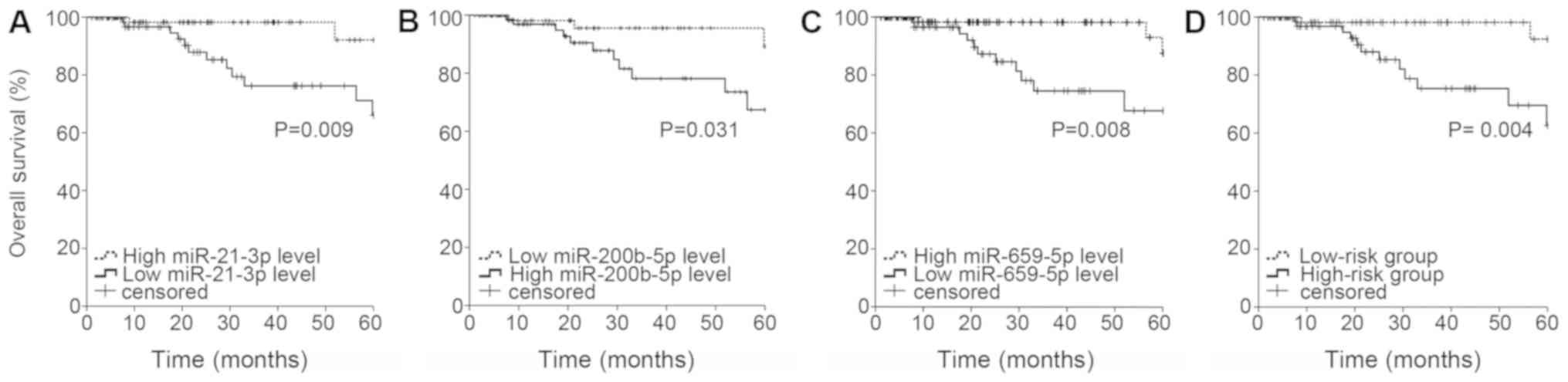
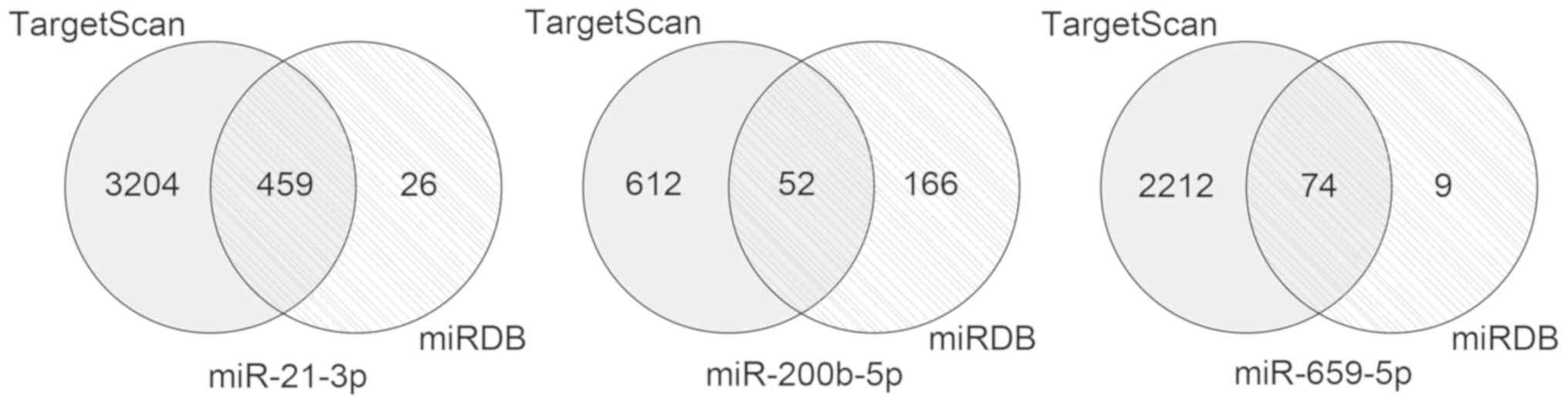
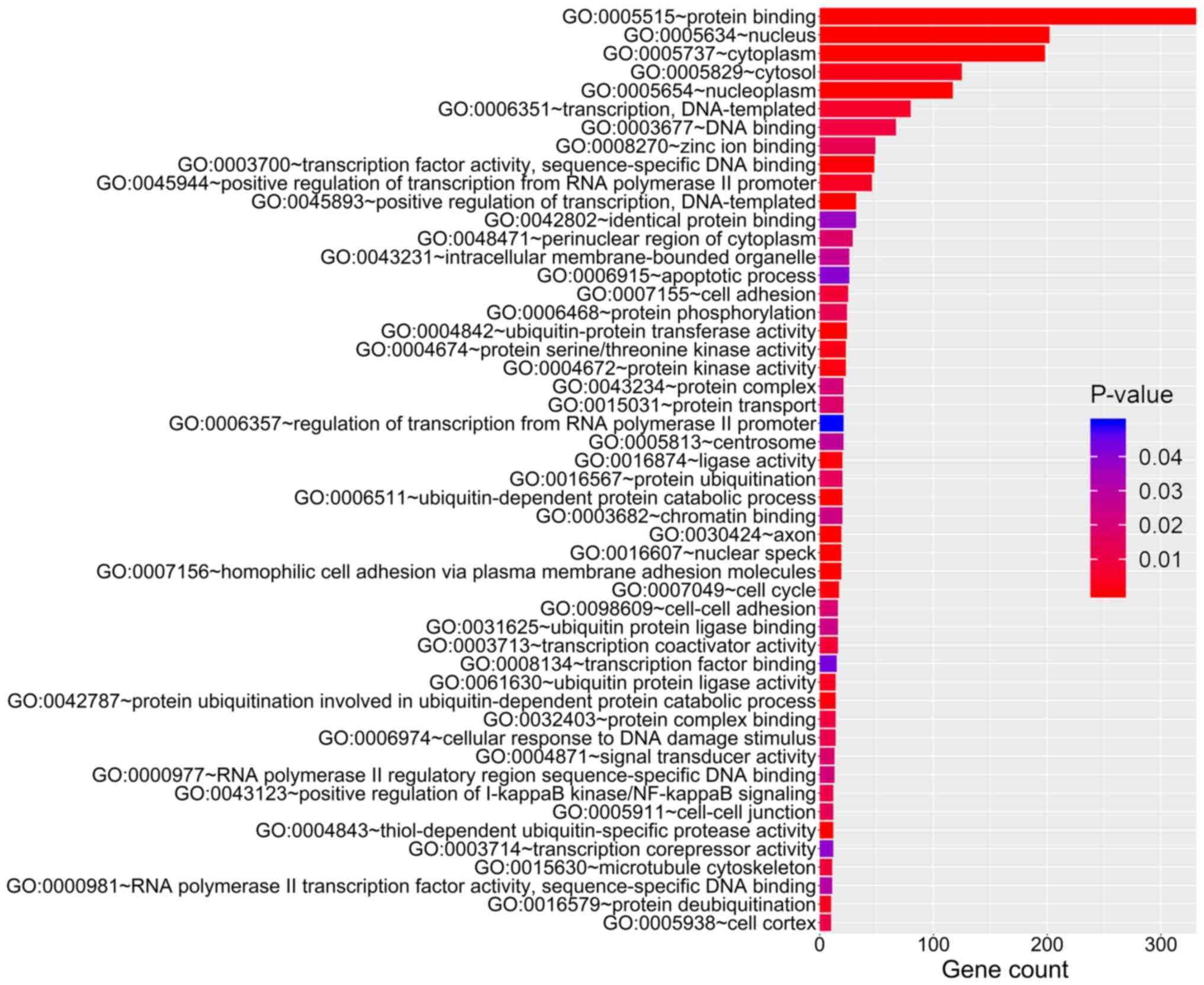
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2018. CA Cancer J Clin. 68:7–30. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Perou CM, Sørlie T, Eisen MB, van de Rijn
M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA,
et al: Molecular portraits of human breast tumours. Nature.
406:747–752. 2000. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Foulkes WD, Smith IE and Reis-Filho JS:
Triple-negative breast cancer. N Engl J Med. 363:1938–1948. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Dent R, Trudeau M, Pritchard KI, Hanna WM,
Kahn HK, Sawka CA, Lickley LA, Rawlinson E, Sun P and Narod SA:
Triple-negative breast cancer: Clinical features and patterns of
recurrence. Clin Cancer Res. 13:4429–4434. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wen YY, Liu WT, Sun HR, Ge X, Shi ZM, Wang
M, Li W, Zhang JY, Liu LZ and Jiang BH: IGF-1-mediated PKM2/β-
catenin/miR-152 regulatory circuit in breast cancer. Sci Rep.
7:158972017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Shimelis H, LaDuca H, Hu C, Hart SN, Na J,
Thomas A, Akinhanmi M, Moore RM, Brauch H, Cox A, et al:
Triple-negative breast cancer risk genes identified by multigene
hereditary cancer panel testing. J Natl Cancer Inst. 110:855–862.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lee RC, Feinbaum RL and Ambros V: The C.
elegans heterochronic gene lin-4 encodes small RNAs with antisense
complementarity to lin-14. Cell. 75:843–854. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Fu Q, Yang F, Xiang T, Huai G, Yang X, Wei
L, Yang H and Deng S: A novel microRNA signature predicts survival
in liver hepatocellular carcinoma after hepatectomy. Sci Rep.
8:79332018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Karsli-Ceppioglu S, Dagdemir A, Judes G,
Ngollo M, Penault-Llorca F, Pajon A, Bignon YJ and Bernard-Gallon
D: Epigenetic mechanisms of breast cancer: An update of the current
knowledge. Epigenomics. 6:651–664. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hafez MM, Hassan ZK, Zekri AR, Gaber AA,
Al Rejaie SS, Sayed-Ahmed MM and Al Shabanah O: MicroRNAs and
metastasis-related gene expression in Egyptian breast cancer
patients. Asian Pac J Cancer Prev. 13:591–598. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chang YY, Lai LC, Tsai MH and Chuang EY:
Deep sequencing reveals a MicroRNA expression signature in
triple-negative breast cancer. Methods Mol Biol. 1699:99–111. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lan H, Lu H, Wang X and Jin H: MicroRNAs
as potential biomarkers in cancer: Opportunities and challenges.
Biomed Res Int. 2015:1250942015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zuberi M, Mir R, Das J, Ahmad I, Javid J,
Yadav P, Masroor M, Ahmad S, Ray PC and Saxena A: Expression of
serum miR-200a, miR-200b, and miR-200c as candidate biomarkers in
epithelial ovarian cancer and their association with
clinicopathological features. Clin Transl Oncol. 17:779–787. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Alunni-Fabbroni M, Majunke L, Trapp EK,
Tzschaschel M, Mahner S, Fasching PA, Fehm T, Schneeweiss A, Beck
T, Lorenz R, et al: Whole blood microRNAs as potential biomarkers
in post-operative early breast cancer patients. BMC Cancer.
18:1412018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Garcia-Vazquez R, Ruiz-García E, Meneses
García A, Astudillo-de la Vega H, Lara-Medina F, Alvarado-Miranda
A, Maldonado-Martínez H, González-Barrios JA, Campos-Parra AD,
Rodríguez Cuevas S, et al: A microRNA signature associated with
pathological complete response to novel neoadjuvant therapy regimen
in triple-negative breast cancer. Tumour Biol.
39:10104283177028992017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Han J, Yu J, Dai Y, Li J, Guo M, Song J
and Zhou X: Overexpression of miR-361-5p in triple-negative breast
cancer (TNBC) inhibits migration and invasion by targeting RQCD1
and inhibiting the EGFR/ PI3K/Akt pathway. Bosn J Basic Med Sci.
19:52–59. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ren YQ, Fu F and Han J: MiR-27a modulates
radiosensitivity of triple-negative breast cancer (TNBC) cells by
targeting CDC27. Med Sci Monit. 21:1297–1303. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wahdan-Alaswad RS, Cochrane DR, Spoelstra
NS, Howe EN, Edgerton SM, Anderson SM, Thor AD and Richer JK:
Metformin-induced killing of triple-negative breast cancer cells is
mediated by reduction in fatty acid synthase via miRNA-193b. Horm
Cancer. 5:374–389. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Cascione L, Gasparini P, Lovat F, Carasi
S, Pulvirenti A, Ferro A, Alder H, He G, Vecchione A, Croce CM, et
al: Integrated microRNA and mRNA signatures associated with
survival in triple negative breast cancer. PLoS One. 8:e559102013.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kleivi Sahlberg K, Bottai G, Naume B,
Burwinkel B, Calin GA, Børresen-Dale AL and Santarpia L: A serum
microRNA signature predicts tumor relapse and survival in
triple-negative breast cancer patients. Clin Cancer Res.
21:1207–1214. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Braicu C, Raduly L, Morar-Bolba G,
Cojocneanu R, Jurj A, Pop LA, Pileczki V, Ciocan C, Moldovan A,
Irimie A, et al: Aberrant miRNAs expressed in HER-2 negative breast
cancers patient. J Exp Clin Cancer Res. 37:2572018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Tomczak K, Czerwińska P and Wiznerowicz M:
The Cancer Genome Atlas (TCGA): An immeasurable source of
knowledge. Contemp Oncol (Pozn). 19:A68–A77. 2015.PubMed/NCBI
|
|
23
|
No authors listed: The future of cancer
genomics. Nat Med. 21:992015. View
Article : Google Scholar : PubMed/NCBI
|
|
24
|
Dorman SN, Viner C and Rogan PK: Splicing
mutation analysis reveals previously unrecognized pathways in lymph
node-invasive breast cancer. Sci Rep. 4:70632014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lossos IS, Czerwinski DK, Alizadeh AA,
Wechser MA, Tibshirani R, Botstein D and Levy R: Prediction of
survival in diffuse large-B-cell lymphoma based on the expression
of six genes. N Engl J Med. 350:1828–1837. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lewis BP, Shih IH, Jones-Rhoades MW,
Bartel DP and Burge CB: Prediction of mammalian microRNA targets.
Cell. 115:787–798. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wong N and Wang X: miRDB: An online
resource for microRNA target prediction and functional annotations.
Nucleic Acids Res. 43((Database Issue)): D146–D152. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang X: Improving microRNA target
prediction by modeling with unambiguously identified
microRNA-target pairs from CLIP-ligation studies. Bioinformatics.
32:1316–1322. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Huang da W, Sherman BT and Lempicki RA:
Bioinformatics enrichment tools: Paths toward the comprehensive
functional analysis of large gene lists. Nucleic Acids Res.
37:1–13. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Liang B, Li Y and Wang T: A three miRNAs
signature predicts survival in cervical cancer using bioinformatics
analysis. Sci Rep. 7:56242017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Phipson B, Lee S, Majewski IJ, Alexander
WS and Smyth GK: Robust hyperparameter estimation protects against
hypervariable genes and improves power to detect differential
expression. Ann Appl Stat. 10:946–963. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ouyang M, Li Y, Ye S, Ma J, Lu L, Lv W,
Chang G, Li X, Li Q, Wang S and Wang W: MicroRNA profiling implies
new markers of chemoresistance of triple-negative breast cancer.
PLoS One. 9:e962282014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Mavrogiannis AV, Kokkinopoulou I, Kontos
CK and Sideris DC: Effect of vinca alkaloids on the expression
levels of microRNAs targeting apoptosis-related genes in breast
cancer cell lines. Curr Pharm Biotechnol. 19:1076–1086. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Calvano Filho CM, Calvano-Mendes DC,
Carvalho KC, Maciel GA, Ricci MD, Torres AP, Filassi JR and Baracat
EC: Triple-negative and luminal A breast tumors: Differential
expression of miR-18a-5p, miR-17-5p, and miR-20a-5p. Tumour Biol.
35:7733–7741. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Farazi TA, Horlings HM, Ten Hoeve JJ,
Mihailovic A, Halfwerk H, Morozov P, Brown M, Hafner M, Reyal F,
van Kouwenhove M, et al: MicroRNA sequence and expression analysis
in breast tumors by deep sequencing. Cancer Res. 71:4443–4453.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Aure MR, Leivonen SK, Fleischer T, Zhu Q,
Overgaard J, Alsner J, Tramm T, Louhimo R, Alnæs GI, Perälä M, et
al: Individual and combined effects of DNA methylation and copy
number alterations on miRNA expression in breast tumors. Genome
Biol. 14:R1262013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Doberstein K, Bretz NP, Schirmer U, Fiegl
H, Blaheta R, Breunig C, Müller-Holzner E, Reimer D, Zeimet AG and
Altevogt P: miR-21-3p is a positive regulator of L1CAM in several
human carcinomas. Cancer Lett. 354:455–466. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Lo TF, Tsai WC and Chen ST:
MicroRNA-21-3p, a berberine-induced miRNA, directly down-regulates
human methionine adenosyltransferases 2A and 2B and inhibits
hepatoma cell growth. PLoS One. 8:e756282013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Jiao W, Leng X, Zhou Q, Wu Y, Sun L, Tan
Y, Ni H, Dong X, Shen T, Liu Y and Li J: Different miR-21-3p
isoforms and their different features in colorectal cancer. Int J
Cancer. 141:2103–2111. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Yeh TS, Wang F, Chen TC, Yeh CN, Yu MC,
Jan YY and Chen MF: Expression profile of microRNA-200 family in
hepatocellular carcinoma with bile duct tumor thrombus. Ann Surg.
259:346–354. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Sun Y, Yang D, Xi L, Chen Y, Fu L, Sun K,
Yin J, Li X, Liu S, Qin Y, et al: Primed atypical ductal
hyperplasia-associated fibroblasts promote cell growth and polarity
changes of transformed epithelium-like breast cancer MCF-7 cells
via miR-200b/c-IKKβ signaling. Cell Death Dis. 9:1222018.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Pang Y, Liu J, Li X, Xiao G, Wang H, Yang
G, Li Y, Tang SC, Qin S, Du N, et al: MYC and DNMT3A-mediated DNA
methylation represses microRNA-200b in triple negative breast
cancer. J Cell Mol Med. 22:6262–6274. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Choi SK, Kim HS, Jin T, Hwang EH, Jung M
and Moon WK: Overexpression of the miR-141/200c cluster promotes
the migratory and invasive ability of triple-negative breast cancer
cells through the activation of the FAK and PI3K/AKT signaling
pathways by secreting VEGF-A. BMC Cancer. 16:5702016. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Luo G, Zhou Y, Yi W and Yi H: Expression
levels of JNK associated with polymorphic lactotransferrin
haplotypes in human nasopharyngeal carcinoma. Oncol Lett.
12:1085–1094. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Piscopo P, Albani D, Castellano AE,
Forloni G and Confaloni A: Frontotemporal lobar degeneration and
MicroRNAs. Front Aging Neurosci. 8:172016. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Zitman-Gal T, Green J, Pasmanik-Chor M,
Golan E, Bernheim J and Benchetrit S: Vitamin D manipulates
miR-181c, miR-20b and miR-15a in human umbilical vein endothelial
cells exposed to a diabetic-like environment. Cardiovasc Diabetol.
13:82014. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Dmitriev P, Barat A, Polesskaya A,
O'Connell MJ, Robert T, Dessen P, Walsh TA, Lazar V, Turki A,
Carnac G, et al: Simultaneous miRNA and mRNA transcriptome
profiling of human myoblasts reveals a novel set of myogenic
differentiation-associated miRNAs and their target genes. BMC
Genomics. 14:2652013. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Asher G, Tsvetkov P, Kahana C and Shaul Y:
A mechanism of ubiquitin-independent proteasomal degradation of the
tumor suppressors p53 and p73. Genes Dev. 19:316–321. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Zhao Q, Song W, He DY and Li Y:
Identification of key gene modules and pathways of human breast
cancer by co-expression analysis. Breast Cancer. 25:213–223. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Zhang Y, Wei L, Yu J, Li G, Zhang X, Wang
A, He Y, Li H and Yin D: Targeting of the β6 gene to suppress
degradation of ECM via inactivation of the MAPK pathway in breast
adenocarcinoma cells. Oncol Rep. 32:1787–1795. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Shah SP, Roth A, Goya R, Oloumi A, Ha G,
Zhao Y, Turashvili G, Ding J, Tse K, Haffari G, et al: The clonal
and mutational evolution spectrum of primary triple-negative breast
cancers. Nature. 486:395–399. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Bartholomeusz C, Gonzalez-Angulo AM, Liu
P, Hayashi N, Lluch A, Ferrer-Lozano J and Hortobágyi GN: High ERK
protein expression levels correlate with shorter survival in
triple-negative breast cancer patients. Oncologist. 17:766–774.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Masuda H, Zhang D, Bartholomeusz C,
Doihara H, Hortobagyi GN and Ueno NT: Role of epidermal growth
factor receptor in breast cancer. Breast Cancer Res Treat.
136:331–345. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Tilch E, Seidens T, Cocciardi S, Reid LE,
Byrne D, Simpson PT, Vargas AC, Cummings MC, Fox SB, Lakhani SR, et
al: Mutations in EGFR, BRAF and RAS are rare in triple-negative and
basal-like breast cancers from Caucasian women. Breast Cancer Res
Treat. 143:385–392. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Park HS, Jang MH, Kim EJ, Kim HJ, Lee HJ,
Kim YJ, Kim JH, Kang E, Kim SW, Kim IA and Park SY: High EGFR gene
copy number predicts poor outcome in triple-negative breast cancer.
Mod Pathol. 27:1212–1222. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Burstein MD, Tsimelzon A, Poage GM,
Covington KR, Contreras A, Fuqua SA, Savage MI, Osborne CK,
Hilsenbeck SG, Chang JC, et al: Comprehensive genomic analysis
identifies novel subtypes and targets of triple-negative breast
cancer. Clin Cancer Res. 21:1688–1698. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
López-Ozuna VM, Hachim IY, Hachim MY,
Lebrun JJ and Ali S: Prolactin Pro-differentiation pathway in
triple negative breast cancer: Impact on prognosis and potential
therapy. Sci Rep. 6:309342016. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Martignetti L, Tesson B, Almeida A,
Zinovyev A, Tucker GC, Dubois T and Barillot E: Detection of miRNA
regulatory effect on triple negative breast cancer transcriptome.
BMC Genomics. 16 (Suppl):S42015. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Pham TND, Perez White BE, Zhao H,
Mortazavi F and Tonetti DA: Protein kinase C α enhances migration
of breast cancer cells through FOXC2-mediated repression of
p120-catenin. BMC Cancer. 17:8322017. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Di Modica M, Regondi V, Sandri M, Iorio
MV, Zanetti A, Tagliabue E, Casalini P and Triulzi T: Breast
cancer-secreted miR-939 downregulates VE-cadherin and destroys the
barrier function of endothelial monolayers. Cancer Lett.
384:94–100. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Lim HC, Multhaupt HA and Couchman JR: Cell
surface heparan sulfate proteoglycans control adhesion and invasion
of breast carcinoma cells. Mol Cancer. 14:152015. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Li Q and Mattingly RR: Restoration of
E-cadherin cell-cell junctions requires both expression of
E-cadherin and suppression of ERK MAP kinase activation in
Ras-transformed breast epithelial cells. Neoplasia. 10:1444–1458.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Wendt MK, Taylor MA, Schiemann BJ and
Schiemann WP: Down-regulation of epithelial cadherin is required to
initiate metastatic outgrowth of breast cancer. Mol Biol Cell.
22:2423–2435. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Mali AV, Joshi AA, Hegde MV and Kadam SS:
Enterolactone modulates the ERK/NF-κB/Snail signaling pathway in
triple-negative breast cancer cell line MDA-MB-231 to revert the
TGF-β-induced epithelial-mesenchymal transition. Cancer Biol Med.
15:137–156. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Javle MM, Gibbs JF, Iwata KK, Pak Y,
Rutledge P, Yu J, Black JD, Tan D and Khoury T:
Epithelial-mesenchymal transition (EMT) and activated extracellular
signal-regulated kinase (p-Erk) in surgically resected pancreatic
cancer. Ann Surg Oncol. 14:3527–3533. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Wen S, Hou Y, Fu L, Xi L, Yang D, Zhao M,
Qin Y, Sun K, Teng Y and Liu M: Cancer-associated fibroblast
(CAF)-derived IL32 promotes breast cancer cell invasion and
metastasis via integrin β3-p38 MAPK signalling. Cancer Lett.
442:320–332. 2019. View Article : Google Scholar : PubMed/NCBI
|